Abstract
The biosynthesis of the FeMo cofactor (FeMoco) of Azotobacter vinelandii nitrogenase presumably starts with the production of its Fe/S core by NifB (the nifB gene product). This core is subsequently processed on the α2β2 tetrameric NifEN complex (formed by the nifE and nifN gene products). In this article, we identify a NifEN-bound FeMoco precursor form that can be converted to fully assembled FeMoco in a so-called FeMoco-maturation assay containing only purified components. We also establish that only molybdate, homocitrate, MgATP, and Fe protein are essential for FeMoco maturation. The FeMoco-maturation assay described here will further address the remaining questions related to the assembly mechanism of the ever-intriguing FeMoco.
Keywords: Fe protein, FeMoco, MoFe protein
The biochemical machinery for the reduction of dinitrogen to ammonia is provided by the metalloenzyme nitrogenase (for recent reviews, see refs. 1–7). This enzyme is composed of two proteins, the Fe and the MoFe protein. The homodimeric Fe protein couples ATP hydrolysis to interprotein electron transfer, serving as an obligate electron donor to the catalytically active component, the MoFe protein. The α2β2 tetrameric MoFe protein contains two copies of unique metal clusters, designated the P-cluster and the FeMoco, respectively. Whereas the [8Fe—7S] P-cluster (8) likely participates in interprotein electron transfer, the FeMoco serves as the active site of substrate binding and reduction.
There is a vast amount of interest in elucidating the mechanism by which the metalloclusters of the MoFe protein are synthesized in vivo because of their importance in N2 fixation and because they are biologically and chemically unprecedented. In particular, the structure and assembly of FeMoco, which provides the site of substrate binding and reduction, has attracted considerable attention for more than a decade. FeMoco is a heterometallic double cubane consisting of one [4Fe—3S] and one [Mo—3Fe—3S] partial cubane that are bridged by three sulfides and share a μ6-central atom of which the identity is unknown but is considered to be C, O, or N (9). Situated entirely in the α-subunit, FeMoco is attached to the protein by only two ligands: a Cys that is bound to the Fe at one end of the cluster and a His that is bound to the Mo at the opposite end of the cluster. The Mo is also coordinated by homocitrate.
Progress has been made in understanding the biosynthesis of FeMoco in Azotobacter vinelandii, which starts with the production of an Fe/S core of the FeMoco (designated NifB-co) by NifB,† the nifB gene product (11, 12). NifB-co, which probably contains all of the Fe and S that ends up in the FeMoco, is then transferred to the α2β2 tetrameric scaffold NifEN protein (encoded by the nifE and nifN gene products) (13, 14). Subsequently, NifB-co is further processed on NifEN by an unknown mechanism and forms the completed FeMoco. The process of FeMoco formation also requires the Fe protein and MgATP (15–17). The completed FeMoco is then presumably transferred to a nafY-encoded protein, called γ (18, 19), which delivers the FeMoco to a FeMoco-deficient form of MoFe protein. However, whether γ is essential for MoFe protein assembly process has been questioned recently (19).
Sequence similarity of nifE and nifN to nifD and nifK (encoding the α- and β-subunits of the MoFe protein, respectively) has led to the hypothesis that NifE and NifN form a complex (20) that is structurally homologous to the MoFe protein (21). Thus, by analogy to the MoFe protein, it has been speculated that NifEN could also contain two types of metallocluster sites. One such site within the NifEN complex could be analogous to the MoFe protein P-cluster site, whereas the other site may provide a place for FeMoco assembly (13). Although the proposed P-cluster analogue has been identified as a [4Fe—4S] cluster that is likely coordinated at the NifE–NifN interface by NifE–Cys-37, NifE–Cys-62, NifE–Cys-124, and NifN–Cys-44 (13), identification of the speculated FeMoco precursor has proven to be elusive so far, hampering the possibility to answer such questions as follow. (i) How and when is Mo and homocitrate incorporated into the cluster? (ii) What are the functions of Fe protein and MgATP in the process? (iii) What is the role of the nonessential γ protein in the MoFe protein assembly? There was evidence that such a FeMoco precursor was attached to the NifEN complex in the early stages of its purification from a ΔnifHDK background (14). However, the NifEN complexes isolated from either a ΔnifB or a ΔnifHDK background appeared to be identical and “FeMoco precursor-less” (14, 22). In other words, the FeMoco precursor was lost from the NifEN complex during its time-consuming purification (13).
In recent years, we have successfully improved the one-step purification procedure of the His-tagged, fragile MoFe protein variants, and the results of such improvement include increased yield (in some cases, >10-fold) and improved integrity of the metalloproteins in terms of their metal clusters [a recent example being the identification of a P-cluster precursor, which like the proposed FeMoco precursor on NifEN, was easily “lost” during the conventional protein purification (23–25)]. Here, we apply this procedure to the purification of His-tagged NifEN and ΔnifB NifEN from A. vinelandii strains DJ1041 and YM9A, respectively. We report a large-scale isolation of NifEN that contains a FeMoco precursor. Also, we show that through a so-called FeMoco-maturation assay that requires Fe protein and, likely, the hydrolysis of MgATP, this NifEN-bound FeMoco precursor can be converted to mature FeMoco with the addition of molybdate and homocitrate. We believe that this assay can be used as a test system that allows us to further address questions related to the assembly mechanism of FeMoco.
Materials and Methods
Unless noted otherwise, all chemicals and reagents were obtained from Fisher Scientific, Baxter Scientific Products (McGaw Park, IL), or Sigma.
Construction of Variant A. vinelandii Strains. Construction of the A. vinelandii nifB-deletion strain DJ1143 (AvDJ1143) producing a His-tagged MoFe protein (designated ΔnifB MoFe protein) and nifHDKTY-deletion strain DJ1041 (AvDJ1041) producing a His-tagged NifEN (designated NifEN) has been described earlier (13, 26). By following the methods described in ref. 26, pHR18, a pGemT(Easy)-originated plasmid carrying a fragment of nifB gene was constructed. This fragment has an internal portion of nifB (flanked by SphI sites) removed and replaced by a 1.3-kb kanamycin-resistance cartridge. The plasmid pHR18 was then transformed into A. vinelandii DJ1041, and the resulting kanamycin-resistant strain is designated A. vinelandii YM9A (AvYM9A), which produces a His-tagged NifEN (designated ΔnifB NifEN) and has a deletion/kanamycin resistance cartridge insertion in nifB. Construction of A. vinelandii strains producing A157S, A157G, and M156C Fe proteins has been published (27–29).
Cell Growth and Protein Purification. All A. vinelandii strains were grown in 180-liter batches in a 200-liter New Brunswick fermentor in Burke's minimal medium supplemented with 2 mM ammonium acetate. The growth rate was measured by cell density at 436 nm by using a Spectronic 20 Genesys (Spectronic, Westbury, NY). After the consumption of the ammonia, the cells were derepressed for 3 h, followed by harvesting using a flow-through centrifugal harvester (Cepa, Lahr/Schwarzwald, Germany). The cell paste was washed with 50 mM Tris·HCl (pH 8.0). Published methods were used for the purification of all Fe proteins (28), WT MoFe protein (30), His-tagged ΔnifB MoFe protein (26), His-tagged NifEN, and His-tagged ΔnifB NifEN (23). The purification procedure of all His-tagged proteins was improved by (i) adding 10% glycerol to all buffers; (ii) limiting the purification process to <15 h; and (iii) performing cell rupture at <10,000 psi (1 psi = 6.89 kPa).
EPR Spectroscopy. All EPR samples were prepared in a Vacuum Atmospheres (Hawthorne, CA) dry box with an O2 level of <4 ppm. All dithionite-reduced samples were in 25 mM Tris·HCl (pH 8.0), 10% glycerol, and 2 mM Na2S2O4. Indigo disulfonate (IDS)-oxidized samples were prepared as described (28). Samples were either used as they were or concentrated in a Centricon-30 (Amicon) in anaerobic centrifuge tubes outside of the dry box. All EPR spectra were recorded by using an ESP 300 Ez spectrophotometer (Bruker, Billerica, MA), interfaced with an ESR-9002 liquid helium continuous-flow cryostat (Oxford Instruments). Except for power- and temperature-dependent EPR experiments, all spectra were recorded at 13 K by using a microwave power of 50 mW, a gain of 5 × 104, a modulation frequency of 100 kHz, and a modulation amplitude of 5 G. A microwave frequency of 9.43 GHz was used to record 10 scans for each sample. Spin quantitation of EPR signals was carried out under nonsaturating conditions as described (24).
FeMoco Maturation and Insertion Assays. The assays designed to determine the maximum possible FeMoco maturation contained (total volume, 0.8 ml) 25 mM Tris·HCl (pH 8.0), 20 mM Na2S2O4, 0.5 mg of purified FeMoco-deficient ΔnifB MoFe protein from strain AvDJ1143 (26), 1.4 mg of Fe protein, 0.3 mM homocitrate, 0.3 mM sodium molybdate, 0.8 mM ATP, 1.6 mM MgCl2, 10 mM creatine phosphate, and 8 units of creatine phosphokinase. The FeMoco maturation was initiated with the addition of 0.04–4 mg of isolated NifEN to the mixture mentioned above. Such reaction mixtures were incubated at 30°C for 30 min and stopped by the addition of 40 nmol of (NH4)2MoS4 (31, 32), and the enzymatic activity was then determined as described (27, 30, 33). (NH4)2MoS4 is known to block FeMoco insertion into the FeMoco-deficient MoFe protein, probably by occupying the FeMoco site (33). Homocitrate lactone (Sigma) containing an undefined mixture of stereochemical configurations was converted to the free acid as described in ref. 34.
Experiments designed to determine the minimum requirements for FeMoco maturation were carried out as described above, except that the reconstitution was initiated with the addition of 2 mg of isolated NifEN. FeMoco-maturation assays evaluating the function of Fe protein variants had the same composition as described above but contained 0.14 mg of Fe protein. The activities of the reconstitution assays were determined as described in ref. 30. FeMoco-maturation assays evaluating the function of various nucleotides contained, in the same buffer as described above, 0.5 mg of ΔnifB MoFe protein, 1.4 mg of Fe protein (≈33 μM), 0.3 mM homocitrate, 0.3 mM sodium molybdate, 0.6–6.6 mM MgCl2, and 0.3–3.3 mM of the following nucleotides: ATP, ADP, ATPγS [adenosine 5′-O-(3-thiotriphosphate)], or AMPPNP (5′-adenylylimidodiphosphate). The FeMoco maturation was initiated with the addition of 2 mg of isolated NifEN and stopped as described above. Subsequently, the activities of the maturation assays were determined as described in ref. 30, except that the ATP concentration was increased to a 25-fold molar excess relative to the concentrations of various nucleotides tested in the maturation assay.
The EPR sample of reconstituted ΔnifB MoFe protein contained all components of the FeMoco-maturation assay as described above, except that the concentrations of all ingredients were up-scaled by the same factor to yield a final MoFe protein concentration of 15 mg/ml. The EPR spectrum described in Results was corrected by subtracting the spectrum of an identical assay containing ΔnifB NifEN instead of NifEN.
The reaction products H2 and C2H4 were analyzed as described in ref. 27, whereas ammonium was determined by using an HPLC fluorescence method (35).
Metal Analysis. Mo (36) and Fe (37) were determined according to published protocols.
Results
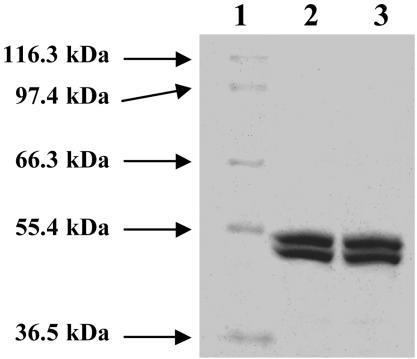
By using the methods described in refs. 23–25, up to 750 mg of the His-tagged NifEN was purified from 250 g of cells of AvDJ1041, whereas up to 300 mg of the His-tagged ΔnifB NifEN was purified from 250 g of cells of AvYM9A. As shown in Fig. 1 (lanes 2 and 3) both NifEN and ΔnifB NifEN are composed of α (≈52 kDa)- and β (≈49 kDa)-subunits. The molecular masses of both proteins are ≈200 kDa based on their elution profiles on gel filtration Sephacryl S-200 HR column (data not shown), indicating that both proteins are α2β2 tetramers. In contrast to what has been reported (14), at the same protein concentration, purified NifEN is much darker in color than ΔnifB NifEN, indicating that NifEN contains an additional cluster (likely the FeMoco precursor), which is missing from ΔnifB NifEN because of the absence of nifB.‡
Fig. 1.
Coomassie blue stained 7.5% SDS/PAGE of purified NifEN and ΔnifB NifEN. Lane 1, 5 μg of protein standard; lane 2, 10 μg of purified NifEN; lane 3, 10 μg of purified ΔnifB NifEN.
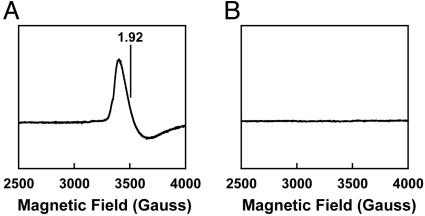
Consistent with the possible presence of an additional cluster on NifEN, the EPR spectroscopic features of NifEN clearly differ from those of ΔnifB NifEN. IDS-oxidized NifEN shows a g = 1.92 EPR signal (Fig. 2A), which is not present in the case of IDS-oxidized ΔnifB NifEN (Fig. 2B). This g = 1.92 signal is discernible only at temperatures of <30 K, with maximum intensity observed at 13 K. The presence of two [4Fe—4S] clusters (one at each NifE–NifN interface) has been reported for an earlier preparation of NifEN (13). These clusters exhibit a midpoint potential of -350 mV (13) and can be oxidized to an EPR silent state by dyes such as IDS (Eo′ = -125 mV). Therefore, it is likely that the ΔnifB NifEN in this study resembles the previously purified NifEN (13), which contains two permanent [4Fe—4S] clusters that become EPR silent upon IDS oxidation (Fig. 2B). Meanwhile, the g = 1.92 EPR signal of IDS-oxidized NifEN (Fig. 2 A) likely originates from an additional cluster of unknown structure, which is bound to NifEN but absent from ΔnifB NifEN. This cluster may have been lost in the earlier preparation of NifEN (13), rendering it in a state identical or similar to the ΔnifB NifEN in this study.
Fig. 2.
EPR spectra of IDS-oxidized NifEN (A) and ΔnifB NifEN (B). The spectra were measured at a protein concentration of 10 mg/ml, as described in Materials and Methods. The g value is indicated.
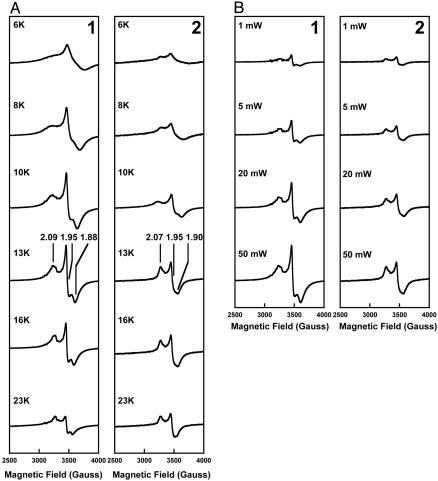
At 10 K, the dithionite-reduced NifEN and ΔnifB NifEN show similar S = 1/2 EPR signals of slightly rhombic line shape in the g = 2 region (Fig. 3A, 1 and 2). However, at temperatures other than 10 K (and, in particular, temperatures of >10 K), the S = 1/2 EPR signals exhibited by NifEN and ΔnifB NifEN appear to be different from each other (Fig. 3A, 1 and 2). The S = 1/2 signal of NifEN adopts a more rhombic line shape at >10 K, with an additional feature between g values of 1.95 and 1.88 becoming more evident (Fig. 3A, 1), whereas the S = 1/2 signal of ΔnifB NifEN maintains nearly the same line shape at >10 K, and the additional feature of NifEN at these temperatures is not apparent in this case (Fig. 3A, 2). The same difference between the line shapes of the S = 1/2 signals of NifEN and ΔnifB NifEN can be observed in spectra collected at 13 K and various microwave powers (ranging 1–50 mW) (Fig. 3B, 1 and 2). In contrast to the S = 1/2 signal of ΔnifB NifEN, which saturates slightly at 50 mW (Fig. 3B, 2), the signal of NifEN is not easily saturated within this power range (Fig. 3B, 1). These results again indicate a different cluster composition of NifEN than that of ΔnifB NifEN. Furthermore, our spin integration under nonsaturating conditions indicates the presence of approximately two spins per ΔnifB NifEN and approximately four spins per NifEN. Therefore, it is likely that in addition to the previously reported two [4Fe—4S] clusters that are present in both ΔnifB NifEN and NifEN, there is an additional metal cluster bound on NifEN. Consistent with this observation, the Fe content of NifEN (16.1 ± 2.4 Fe atoms per molecule) is nearly twice as much as that of ΔnifB NifEN (8.5 ± 1.0 Fe atoms per molecule) (Table 1). If two [4Fe—4S] clusters (eight Fe atoms) are assigned to each molecule of ΔnifB NifEN or NifEN, then the additional Fe atoms per molecule of NifEN could be explained by the possible presence of a FeMoco precursor on NifEN.
Fig. 3.
Temperature (A) and power (B) dependence of the EPR signal exhibited by dithionite-reduced NifEN (1) and ΔnifB NifEN (2). The spectra were measured at a protein concentration of 10 mg/ml between 6 and 23 K as well as between 1 and 50 mW at 13 K, as described in Materials and Methods. The g values are indicated.
Table 1. Metal contents of purified NifEN and ΔnifB NifEN.
| Metal
|
||
|---|---|---|
| Protein | Fe | Mo |
| MoFe protein | 29.5 ± 2.0 | 1.8 ± 0.1 |
| NifEN | 16.1 ± 2.4 | <0.01 |
| ΔnifB NifEN | 8.5 ± 1.0 | <0.01 |
Data are expressed as mol of metal per mol of protein.
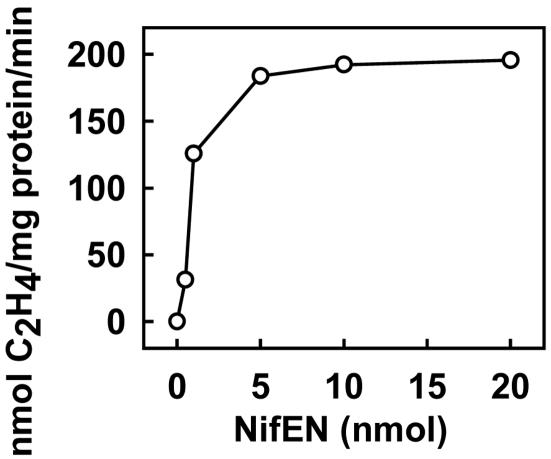
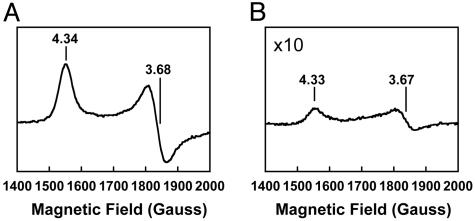
It has been reported that the FeMoco-deficient, His-tagged ΔnifB MoFe protein forms fully active holo-MoFe protein upon the addition of isolated FeMoco (26, 38). Here, we show that the His-tagged ΔnifB MoFe protein [in crude extract of AvDJ1143 (data not shown) or purified form (Fig. 4)] can also be activated in a so-called FeMoco-maturation assay containing purified NifEN and other factors known to be required for FeMoco maturation, such as the Fe protein, MgATP, homocitrate, and Mo. Like the WT MoFe protein (Fig. 5A), ΔnifB MoFe protein activated by the maturation assay shows a well characterized S = 3/2 EPR signal (Fig. 5B) that arises from the FeMoco center of the protein (1). These results strongly point to the presence of a FeMoco precursor on NifEN, which can be converted to mature FeMoco and inserted subsequently into the ΔnifB MoFe protein, resulting in the formation of the active, holo-MoFe protein. Upon the addition of increasing amounts of NifEN, a maximum activity of 217 nmol of C2H4 formation per mg of ΔnifB MoFe protein per min can be observed in the assay containing only purified proteins (Fig. 4). Activated MoFe protein in this assay shows not only C2H4-formation but also H2-formation and N2-fixation activities (Table 2), which is consistent with the presence of a normal, catalytically active FeMoco in the protein. No activities can be observed if NifEN in the assay is replaced by equimolar amounts of ΔnifB NifEN or if any of the components, such as homocitrate, Mo, Fe protein, MgATP, NifEN, or ΔnifB MoFe protein, are omitted (Table 2). Several conclusions can be drawn from these results. (i) Accumulation of the FeMoco precursor on NifEN requires the presence of the nifB gene product. This result is consistent with the model that NifB is essential for FeMoco biosynthesis and represents the starting point of the pathway of FeMoco formation (10, 12, 13). (ii) The identified FeMoco precursor on NifEN does not contain homocitrate and Mo. Therefore, the addition of both components occurs either on the NifEN scaffold during the FeMoco-maturation process or in a later step. The requirement of Mo for FeMoco maturation in NifEN is also consistent with the absence of Mo from purified NifEN (Table 1). (iii) Conversion of the FeMoco precursor to the mature FeMoco requires Fe protein and MgATP because activities can be observed only when both components are present (Table 2).
Fig. 4.
FeMoco-maturation assay based on the reconstitution of purified FeMoco-deficient ΔnifB MoFe protein. The assays also contained Fe protein, homocitrate, molybdate, MgATP, and increasing amounts of NifEN, as described in Materials and Methods.
Fig. 5.
EPR spectra of WT MoFe protein (A) and reconstituted ΔnifB MoFe protein (B) in the FeMoco-maturation assay. The samples (15 mg of protein per ml) were prepared and measured as described in Materials and Methods. The g values are indicated.
Table 2. Determination of factors required for FeMoco maturation.
| Activities*
|
||||
|---|---|---|---|---|
| Assay condition | C2H4 formation under C2H2/Ar | H2 formation under Ar | NH3 formation under N2 | H2 formation under N2 |
| Complete† | 191 ± 26 (100) | 316 ± 11 (100) | 96 ± 6 (100) | 65 ± 13 (100) |
| Complete plus (NH4)2MoS4‡ | 0 (0) | 2 ± 0.04 (<1) | 0 (0) | 2 ± 0.2 (3) |
| Complete minus MgATP | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Complete minus Fe protein | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Complete minus homocitrate | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Complete minus molybdate§ | 10 ± 1 (5) | 15 ± 3 (5) | 0 (0) | 0 (0) |
| Complete minus NifEN | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Complete minus ΔnifB MoFe protein | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Complete minus NifEN, plus ΔnifB NifEN¶ | 0 (0) | 1 ± 0.1 (<1) | 0 (0) | 0 (0) |
| ΔnifB MoFe protein alone | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| NifEN alone | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ΔnifB NifEN alone | 0 (0) | 1 ± 0.1 (<1) | 0 (0) | 0 (0) |
Data are expressed as nmol per min per mg of protein. Percentages are given in parentheses.
The lower detection limits were 0.01, 0.02, 0.001, and 0.02 nmol per min per mg of protein for C2H4 formation under C2H2/Ar, H2 formation under Ar, NH3 formation under N2, and H2 formation under N2, respectively
The complete assay contains purified NifEN, purified ΔnifB MoFe protein, purified Fe protein, molybdate, homocitrate, and MgATP at concentrations described in Materials and Methods
Insertion of FeMoco into ΔnifB MoFe protein was inhibited by the addition of (NH4)2MoS4 at the beginning of the experiment, as described in Materials and Methods
The minor activities were likely caused by molybdenum contamination in the assays
Assay contained the same components as described in †, except that NifEN was replaced by equimolar amounts of ΔnifB NifEN
Table 3 shows that various Fe protein variants, which are able to bind MgATP but are defective in MgATP hydrolysis, are not able to support full FeMoco maturation in our FeMoco-maturation assays. The extent of FeMoco maturation by these Fe protein variants is <10% compared with that by the WT Fe protein, suggesting that MgATP hydrolysis is likely required for FeMoco maturation. Consistent with this hypothesis, neither ADP nor nonhydrolyzable analogues of ATP, such as ATPγS [adenosine 5′-O-(3-thiotriphosphate)] or AMPPNP (5′-adenylylimidodiphosphate), are able to support FeMoco maturation in our FeMoco-maturation assays (Table 3).§,¶
Table 3. Effect of Fe protein variants and various nucleotides on FeMoco maturation.
| Factors | Activities* |
|---|---|
| Fe protein | |
| No Fe protein | 0 (0) |
| WT Fe protein | 196 ± 6 (100) |
| A157S Fe protein†‡ | 18 ± 4 (9) |
| A157G Fe protein†§ | 11 ± 1 (6) |
| M156C Fe protein†¶ | 6 ± 2 (3) |
| Nucleotide∥ | |
| No nucleotide | 0 (0) |
| ATP | 186 ± 10 (100) |
| ADP** | 0 (0) |
| ATPγS** | 0 (0) |
| AMPPNP** | 0 (0) |
Data are expressed as nmol of C2H4 evolution per min per mg of protein. Percentages are given in parentheses. ATPγS, adenosine 5′-O-(3-thiotriphosphate; AMPPNP, 5′-adenylylimidodiphosphate.
The lower detection limit was 0.01 nmol of C2H4 evolution per min per mg of protein
A157S Fe protein is unable to undergo a MgATP-induced conformational change and does not support MgATP hydrolysis (27)
A157G Fe protein undergoes a delayed conformational change upon MgATP binding, resulting in a reduced substrate-reduction activity (28)
M156C Fe protein undergoes a MgATP-induced conformational change that differs from WT Fe protein, resulting in the loss of substrate-reduction activity (29)
Identical results have been obtained by using 10-, 50-, and 100-fold molar excess of nucleotides relative to Fe protein in the FeMoco-maturation assay
Note that, with the addition of excess MgATP as described in Materials and Methods, these nucleotides do not inhibit substrate-reduction activity of the WT MoFe protein
Discussion
Consistent with what has been proposed (14), we identified a FeMoco precursor synthesized in the presence of nifB gene product through this study. This NifEN-bound precursor contains Fe as the only metal, and Mo and homocitrate are added while the cluster is still bound to the NifEN complex or at a later step. The addition of Mo and homocitrate to the precursor and the final maturation of the holo-MoFe protein require only the participation of Fe protein and MgATP, leading to the following conclusions. (i) FeMoco carrier protein(s) such as γ (18, 19) is not essential for FeMoco maturation; and (ii) Fe protein facilitates Mo and homocitrate insertion into the FeMoco, likely upon MgATP hydolysis. Interestingly, it has been reported that Mo accumulated on the Fe protein during FeMoco biosynthesis (39), an observation in line with our results. Also, the first published x-ray structure of the Fe protein of A. vinelandii contained ADP bound in an unusual location across the subunit–subunit interface and an adjacent Mo located in a position that could correspond to the γ phosphate of ATP (40). It has been speculated that the binding mode in this structure could be involved in the initial entry of the nucleotide into the Fe protein (40). In light of our results, it is also possible that this ADP/Mo binding mode is related to the process of FeMoco maturation.
Although the dual requirement of Fe protein and MgATP for FeMoco maturation has been well documented in the past years (16, 17, 41, 42), previous studies suggest that Fe protein only needs to bind but does not need to hydrolyze MgATP to carry out its function in this process (27–29). Ref. 43 showed that a truncated form of Fe protein was unable to support substrate reduction but was active in FeMoco biosynthesis. This effect could be explained in two ways. (i) Catalytically active Fe protein capable of MgATP hydrolysis may not be required for the FeMoco biosynthesis. (ii) The truncated Fe protein may have lost its ability to interact with the MoFe protein during substrate reduction yet retained its capacity to hydrolyze MgATP, a feature required for FeMoco biosynthesis. Based on our observations that (i) the Fe protein variants defective in MgATP hydrolysis are inactive in FeMoco maturation (Table 3), (ii) nonhydrolyzable analogues of ATP are unable to support FeMoco maturation (Table 3), and (iii) the Fe protein is the only known nucleotide binding protein in our maturation assay, it appears to be likely that the Fe protein carries out its function in FeMoco biosynthesis through MgATP hydrolysis, and the effects of the truncated Fe protein form could be accounted for by the second explanation.
The FeMoco-maturation assay developed in this study is an improvement of earlier FeMoco biosynthesis assays (27–29, 44–46) in that (i) it allows observation of much higher specific activity of reconstituted MoFe protein (≈100-fold) than the reported values of earlier FeMoco biosynthesis assays; (ii) it contains all proteins in purified forms and, therefore, avoids the complication caused by other factors in crude extracts often used in earlier FeMoco biosynthesis assays; and (iii) it uses ΔnifB MoFe protein as the target protein for FeMoco insertion instead of ΔnifH-type MoFe protein used in previous FeMoco biosynthesis assays. Recently, we reported that ΔnifH MoFe protein was not only FeMoco-deficient but also contained a P-cluster precursor (23, 25). Therefore, previous FeMoco biosynthesis assays based on ΔnifH-type MoFe protein were in fact combined assays of FeMoco and P-cluster maturation and consequently, it was difficult to interpret the results accurately. The FeMoco-maturation assay in this study uses ΔnifB MoFe protein (which has fully assembled P-clusters) as the “receptor” for FeMoco synthesized during the assay, and as a result, the sole effect of FeMoco maturation can be observed without interference of P-cluster formation.
Note that despite the fact that our FeMoco-maturation assay yields much higher activity of reconstituted ΔnifB MoFe protein than those of the previously reported FeMoco biosynthesis assays, the maximum activity of reconstitution is only ≈10% of that of the WT holo-MoFe protein (23, 33, 44). This observation indicates that additional factors, albeit not essential for MoFe protein maturation, are required to optimize this process in vivo. Such factors could be GroEL, NifY, or γ, all of which have been implicated in the process of MoFe protein assembly (18, 19, 33, 47, 48).
In summary, we were able to unravel some of the baffling issues regarding FeMoco maturation in this work. However, other key questions such as the structure of the FeMoco precursor on NifEN or the mechanism of Mo insertion into the FeMoco remain unanswered. Also, the fashion by which other factors, such as GroEL, NifY, or γ, facilitate FeMoco insertion and final holo-MoFe protein maturation awaits further investigation.
Acknowledgments
We thank Prof. Dennis Dean (Virginia Polytechnic Institute and State University, Blacksburg) for kindly providing the A. vinelandii strains DJ1143 and DJ1041. This work was supported by National Institutes of Health Grant GM-67626 (to M.W.R.).
Author contributions: Y.H. and M.W.R. designed research; Y.H., A.W.F., and M.W.R. performed research; Y.H. and M.W.R. analyzed data; and Y.H. and M.W.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FeMoco, FeMo cofactor; IDS, indigo disulfonate.
Footnotes
The C-terminal region of NifB bears significant sequence similarity to NifY, NifX, and γ, all of which are involved in the biosynthesis of the FeMoco (10). The N-terminal region of NifB shows sequence similarity to members of the radical S-adenosylmethionine-dependent enzyme superfamily (10).
Consistent with previous studies (13), in the presence of dithionite, both NifEN and ΔnifB NifEN exhibit broad, nearly featureless UV-visible spectra. However, the intensity of the NifEN spectrum is significantly stronger than that of the ΔnifB NifEN at the same protein concentration (data not shown), indicating the possible presence of an additional metal cluster on NifEN.
It needs to be noted that the conformational rearrangement of the Fe protein induced by such nonhydrolyzable analogues of ATP as ATPγS [adenosine 5′-O-(3-thiotriphosphate)] cannot be distinguished from that induced by ATP based on the properties and/or behavior of the Fe protein in EPR spectroscopy and chelation experiments (data not shown).
The requirement of Fe protein concomitant with ATP hydrolysis suggests that there is/are electron(s) transferred to the emerging FeMoco. Consistent with this observation, preliminary data indicate the requirement of dithionite in the FeMoco-maturation assay (data not shown).
References
- 1.Burgess, B. K. & Lowe, D. J. (1996) Chem. Rev. 96, 2983-3011. [DOI] [PubMed] [Google Scholar]
- 2.Howard, J. B. & Rees, D. C. (1996) Chem. Rev. 96, 2965-2982. [DOI] [PubMed] [Google Scholar]
- 3.Smith, B. E. (1999) Adv. Inorg. Chem. 47, 159-218. [Google Scholar]
- 4.Rees, D. C. & Howard, J. B. (2000) Curr. Opin. Chem. Biol. 4, 559-566. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen, J., Dean, D. R. & Seefeldt, L. C. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 269-295. [DOI] [PubMed] [Google Scholar]
- 6.Igarashi, R. Y. & Seefeldt, L. C. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 351-384. [DOI] [PubMed] [Google Scholar]
- 7.Seefeldt, L. C., Dance, I. G. & Dean, D. R. (2004) Biochemistry 43, 1401-1409. [DOI] [PubMed] [Google Scholar]
- 8.Peters, J. W., Stowell, M. H., Soltis, S. M., Finnegan, M. G., Johnson, M. K. & Rees, D. C. (1997) Biochemistry 36, 1181-1187. [DOI] [PubMed] [Google Scholar]
- 9.Einsle, O., Tezcan, F. A., Andrade, S. L. A., Schmid, B., Yoshida, M., Howard, J. B. & Rees, D. C. (2002) Science 297, 1696-1700. [DOI] [PubMed] [Google Scholar]
- 10.Dos-Santos, P. C., Dean D. R., Hu, Y. & Ribbe, M. W. (2004) Chem. Rev. 104, 1159-1174. [DOI] [PubMed] [Google Scholar]
- 11.Shah, V. K., Allen, J. R., Spangler, N. J. & Ludden, P. W. (1994) J. Biol. Chem. 269, 1154-1158. [PubMed] [Google Scholar]
- 12.Allen, R. M., Chatterjee, R., Ludden, P. W. & Shah, V. K. (1995) J. Biol. Chem. 270, 26890-26896. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin, P. J., Agar, J. N., Roll, J. T., Roberts, G. P., Johnson, M. K. & Dean, D. R. (1998) Biochemistry 37, 10420-10428. [DOI] [PubMed] [Google Scholar]
- 14.Roll, J. T., Shah, V. K., Dean, D. R. & Roberts, G. P. (1995) J. Biol. Chem. 270, 4432-4437. [DOI] [PubMed] [Google Scholar]
- 15.Allen, R. M., Chatterjee, R. Ludden, P. W. & Shah, V. K. (1996) J. Biol. Chem. 271, 4256-4260. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee, R., Allen, R. M., Shah, V. K. & Ludden, P. W. (1994) J. Bacteriol. 176, 2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson, A. C., Dean, D. R. & Burgess, B. K. (1987) J. Biol. Chem. 262, 14327-14332. [PubMed] [Google Scholar]
- 18.Homer, M. J., Dean, D. R. & Roberts, G. P. (1995) J. Biol. Chem. 270, 24745-24752. [DOI] [PubMed] [Google Scholar]
- 19.Rubio, L. M., Rangaraj, P., Homer, M. J., Roberts, G. P. & Ludden, P. W. (2002) J. Biol. Chem. 277, 14299-14305. [DOI] [PubMed] [Google Scholar]
- 20.Roberts, G. P. & Brill, W. J. (1980) J. Biol. Chem. 144, 210-216. [Google Scholar]
- 21.Brigle, K. E., Weiss, M. C., Newton, W. E. & Dean, D. R. (1987) J. Bacteriol. 169, 1547-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paustian, T. D., Shah, V. K. & Roberts, G. P. (1989) Proc. Natl. Acad. Sci. USA 86, 6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribbe, M. W., Hu, Y., Guo, M., Schmid, B. & Burgess, B. K. (2002) J. Biol. Chem. 277, 23469-23476. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Y., Fay, A. W., Dos Santos, P. C., Naderi, F. & Ribbe, M. W. (2004) J. Biol. Chem., 279, 54963-54971. [DOI] [PubMed] [Google Scholar]
- 25.Corbett, M. C., Hu, Y., Naderi, F., Ribbe, M. W., Hedman, B. & Hodgson, K. O. (2004) J. Biol. Chem. 279, 28276-28282. [DOI] [PubMed] [Google Scholar]
- 26.Christiansen, J., Goodwin, P. J., Lanzilotta, W. N., Seefeldt, L. C. & Dean, D.R. (1998) Biochemistry 37, 12611-12623. [DOI] [PubMed] [Google Scholar]
- 27.Gavini, N. & Burgess, B. K. (1992) J. Biol. Chem. 267, 21179-21186. [PubMed] [Google Scholar]
- 28.Bursey, E. H. & Burgess, B. K. (1998) J. Biol. Chem. 273, 16927-16934. [DOI] [PubMed] [Google Scholar]
- 29.Bursey, E. H. & Burgess, B. K. (1998) J. Biol. Chem. 273, 29678-29685. [DOI] [PubMed] [Google Scholar]
- 30.Burgess, B. K., Jacobs, D. B. & Stiefel, E. I. (1980) Biochim. Biophys. Acta 614, 196-209. [DOI] [PubMed] [Google Scholar]
- 31.Rangaraj, P., Shah, V. K. & Ludden, P. W. (1997) Proc. Natl. Acad. Sci. USA 94, 11250-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah, V. K., Ugalde, R. A., Imperial, J. & Brill, W. J. (1985) J. Biol. Chem. 260, 3891-3894. [PubMed] [Google Scholar]
- 33.Ribbe, M. W. & Burgess, B. K. (2001) Proc. Natl. Acad. Sci. USA 98, 5521-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madden, M. S., Paustian, T. D., Ludden, P. W. & Shah, V. K. (1991) J. Bacteriol. 173, 5403-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbin, J. L. (1984) Appl. Environ. Microbiol. 47, 1027-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark, L. J. & Axley, J. H. (1955) Anal. Biochem. 27, 2000. [Google Scholar]
- 37.Van de Bogart, M. & Beinert, H. (1967) Anal. Biochem. 20, 325-334. [DOI] [PubMed] [Google Scholar]
- 38.Schmid, B., Ribbe, M. W., Einsle, O., Yoshida, M., Thomas, L. M., Dean, D. R., Rees, D. C. & Burgess, B. K. (2002) Science 296, 352-356. [DOI] [PubMed] [Google Scholar]
- 39.Rangaraj, P. & Ludden, P. W. (2002) J. Biol. Chem. 277, 40106-40111. [DOI] [PubMed] [Google Scholar]
- 40.Georgiadis, M. M., Komiya, H., Chakrabarti, P., Woo, D., Kornuc, J. J. & Rees, D. C. (1992) Science 257, 1653-1659. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, A. C., Chun, T. W., Li, J.-G. & Burgess, B. K. (1989) J. Biol. Chem. 264, 10088-10095. [PubMed] [Google Scholar]
- 42.Rangaraj, P., Ryle, M. J., Lanzilotta, W. N., Ludden, P. W. & Shah, V. K. (1999) J. Bio. Chem. 274, 19778-19784. [DOI] [PubMed] [Google Scholar]
- 43.Filler, W. A., Kemp, R. M., NG, J. C., Hawkes, T. R., Dixon, R. A. & Smith, B. E. (1986) Eur. J. Biochem. 160, 371-377. [DOI] [PubMed] [Google Scholar]
- 44.Ribbe, M. W., Bursey, E. H. & Burgess, B. K. (2000) J. Biol. Chem. 275, 17631-17638. [DOI] [PubMed] [Google Scholar]
- 45.Shah, V. K., Imperial, J., Ugalde, R. A., Ludden, P. W. & Brill, W. J. (1986) Proc. Natl. Acad. Sci. USA 83, 1636-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imperial, J., Shah, V. K., Ugalde, R. A., Ludden, P. W. & Brill, W. J. (1987) J. Bacteriol. 169, 1784-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rangaraj, P., Ruttimann-Johnson, C., Shah, V. K. & Ludden, P. W. (2001) J. Biol. Chem. 276, 15968-15974. [DOI] [PubMed] [Google Scholar]
- 48.Shah, V. K., Rangaraj, P., Chatterjee, R., Allen, R. M., Roll, J. T., Roberts, G. P. & Ludden, P. W. (1999) J. Bacteriol. 181, 2797-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]