Abstract
The transcription factor TAL1/SCL is one of the most prevalent oncogenes in T-cell acute lymphoblastic leukemia (T-ALL), a malignant disorder resulting from leukemic transformation of thymus T-cell precursors. TAL1 is normally expressed in hematopoietic stem cells (HSCs) but is silenced in immature thymocytes. We hypothesize that TAL1 contributes to leukemogenesis by activating genes that are normally repressed in immature thymocytes. Herein, we identified a novel TAL1-regulated super-enhancer controlling the GIMAP locus, which resides within an insulated chromosomal locus in T-ALL cells. The GIMAP genes are expressed in HSCs and mature T-cells but are downregulated during the immature stage of thymocyte differentiation. The GIMAP enhancer is activated by TAL1, RUNX1 and GATA3 in human T-ALL cells but is repressed by E-proteins. Overexpression of human GIMAP genes in immature thymocytes alone does not induce tumorigenesis but accelerates leukemia development in zebrafish. Our results demonstrate that aberrant activation of the GIMAP enhancer contributes to T-cell leukemogenesis.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) arises from the clonal expansion of transformed T-lymphoblasts caused by genetic abnormalities that induce differentiation arrest, dysregulated proliferation and aberrant cell survival.1–3 The most frequent molecular abnormality in T-ALL is the dysregulation of transcription factor genes, including overexpression of TAL1/SCL and activating mutations of NOTCH1.1–3 TAL1 is normally expressed in hematopoietic stem cells (HSCs), progenitor cells and erythromegakaryocytic cells.4 In normal HSCs, TAL1 heterodimerizes with E-proteins such as TCF3/E2A and TCF12/HEB and forms a large transcriptional complex with LMO2, LDB1 and GATA2.5–9 TAL1 frequently co-occupies the regulatory elements with other transcription factors, including RUNX1 and the ETS family of proteins.10, 11 Importantly, TAL1 is normally silenced in immature thymocytes,12 whereas E-proteins are upregulated and required for thymocyte development by acting as homo- or heterodimers.12–14 Such stage-specific regulation of TAL1 and E-proteins is essential in normal hematopoiesis.
In contrast, TAL1 is ectopically overexpressed in 40–60% of T-ALL cases as a result of chromosomal translocation, intrachromosomal rearrangement or a somatic mutation in a non-coding intergenic element.15–19 In both human T-ALL and mouse models, TAL1 overexpression leads to a blockage at later stages of differentiation in developing thymocytes.12, 20, 21 We previously reported that in T-ALL cells, TAL1 coordinately regulates gene expression with GATA3, RUNX1 and MYB similar to a mechanism observed in normal HSCs.22 In addition, TAL1 positively regulates the expression of a specific subset of genes that are negatively regulated by E-proteins.22 These results suggested that TAL1 could activate genes that are normally repressed in immature thymocytes by counteracting E-protein function. We hypothesize that such factors would be responsible for the pathogenesis of T-ALL. Interestingly, a recent study showed that TAL1 and its regulatory partners (GATA3, RUNX1 and MYB) are regulated under “super-enhancers”,23 which are clusters of enhancers that exhibit significantly high levels of histone H3 lysine 27 acetylation (H3K27Ac) and mediator bindings. Rapidly accumulating evidence demonstrates that super-enhancers are often enriched at cancer genes in various malignancies.23–27 Hence, we further hypothesized that the critical factors involved in the T-ALL pathogenesis would be regulated by super-enhancers to sustain high expression levels.
Here, we describe a regulatory element differentially controlled by TAL1 and E-proteins in human T-ALL cells. This element is associated with a super-enhancer and is located within a cluster of genes in the same family known as the GTPase of Immunity-Associated protein (GIMAP), which have been implicated in mature T-cell survival.28–32 GIMAP genes and the enhancer are activated in normal HSCs and human T-ALL cells but not in thymocytes in immature stages. Ectopic expression of GIMAP genes in thymocytes accelerates T-cell leukemogenesis in vivo.
Materials and Methods
Cell culture
All T-ALL cell lines were maintained in RPMI-1640 medium supplemented with 10% FBS and L-glutamine (Life Technologies). All cell lines were confirmed by DNA fingerprinting using the PowerPlex 1.2 system (Promega) in January 2013 and were regularly tested for mycoplasma contamination. 293T cells were grown in DMEM medium supplemented with 10% FBS and L-glutamine (Life Technologies).
Analysis of ChIP-Seq, gene expression and ChIA-PET datasets
The ChIP-Seq and microarray gene expression datasets were previously reported by us22 and deposited into the NCBI GEO website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE29181. The ChIP-Seq datasets for NOTCH1 and RBPJ by Wang et al.33, CTCF by Hnisz et al.34 and BRD4 have been deposited under accession numbers GSE29544, GSM1689152, GSM1689151 and GSE83777, respectively. The dataset for H3K27Ac in Jurkat cells23 and normal thymus35 have been deposited under accession numbers GSM1296384 and GSM1013125, respectively. All datasets were analyzed and uploaded into the UCSC genome browser, and sequences were aligned to the human genome sequence (hg19). The microarray gene expression dataset was normalized using dChip software.36 High-confidence chromatin-chromatin interactions were retrieved from the ChIA-PET data34 analysis using the pipeline reported by Dowen et al.37 The interactions were visualized on the WashU Genome Browser.38 Selected genes were included in Supplementary Table 1.
RNA-Seq
Total RNA was extracted from Jurkat cells by the miRNeasy extraction kit (Qiagen). Library construction and RNA-Seq analysis by the Illumina HiSeq 2000 were performed at BGI Tech Solutions (Hong Kong). Paired-end 90 bp long reads were aligned to hg19 using STAR 2.4.0i39 with the outFilterMatchNminOverLread parameter set to 0.80 and outFilterMultimapNmax set to 1. A BEDGRAPH RNA-Seq coverage across the entire genome was calculated by Bedtools genomecov. The data have been deposited to the GEO under accession number GSE81712.
CRISPR/Cas9 knockout
Guide RNAs (gRNAs) targeting either the GIMAP enhancer or the whole GIMAP gene cluster were selected using the CRISPR Design Tool (http://crispr.mit.edu/) (Supplementary Table 2) and cloned into the lentiCRISPRvs2 vector.40 The gRNAs and Cas9 were transduced by lentivirus infection (see Supplementary Method). Genomic DNA was isolated using the QIAamp DNA Blood Mini kit (Qiagen) followed by PCR amplification of targeted loci using specific primers (Supplementary Table 3). PCR products were directly analyzed by Sanger sequencing.
Cloning of constructs
The 6-kb GIMAP enhancer region (hg19, chr7: 150,360,481–150,366,493) was cloned into the pBSII-SK+-I-SceI zebrafish reporter plasmid41 and the pGL4.26 luciferase plasmid (Promega). The RUNX1 enhancer reporter construct41 and the zebrafish rag2 promoter construct42 have been described previously. The cDNA sequence of each of the human GIMAPs was amplified via PCR using primers (Supplementary Table 4) and was cloned into the Rag2-I-SceI zebrafish expression vector. The cDNA of each transcription factor was cloned into the pCS2+ vector.
Zebrafish studies
Zebrafish studies were conducted in strict adherence to the recommendations of the Institutional Animal Care and Use Committee (IACUC), and all protocols were approved by the Committee at the National University of Singapore (NUS). I-SceI meganuclease-based vectors (pBSII-SK-I-SceI and Rag2-I-SceI) were used in wild-type strain to establish transgenic lines.43 The sample size was determined based on previous similar studies reported by us.43 At least two stable transgenic lines were generated. Each breeding was repeated at least twice. Sample randomization is not required in this study.
Isolation of hematopoietic cells from mice
All mouse experiments followed guidelines set by the National Advisory Committee for Laboratory Animal Research and the NUS IACUC. C57BL/6 mice were maintained, and bone marrow (BM) cells from 8-week-old inbred mice were flushed from the long bones with α-MEM medium supplemented with 10% FBS (Gibco). BM and thymic cells were filtered through a nylon filter (35 µm) to obtain a single-cell suspension. Flow cytometry sorting was performed using FACSAria (BD Biosciences) to isolate hematopoietic cells (see Supplementary Method).
Lentivirus infection
For lentiviral production, either the CRISPR-Cas9 plasmid or pLKO1-puro was co-transfected into 293T cells with the envelope plasmid pMD2.G and packaging plasmids pMDLg/pRRE and pRSV-REV using FuGENE 6 reagent (Roche). Viral supernatants were collected, filtered through a 0.45-µm filter (Millipore) and transduced into Jurkat cells. The infected cells were selected by puromycin (Sigma).
shRNA knockdown analysis
shRNA sequences were cloned into the lentiviral vector pLKO.1-puro (Supplementary Table 6) and transduced by lentiviral infection (see Supplementary Method). Knockdown levels were verified by qRT-PCR.
RNA extraction, cDNA and expression analysis
For analysis of gene expression in human cell lines, zebrafish and mice, total RNA was extracted using a NucleoSpin RNA kit (Macherey-Nagel) and reverse-transcribed using a QuantiTect reverse transcription kit (Qiagen). Quantitative PCR analysis was performed using an Applied Biosystems 7300 Real Time PCR System (Applied Biosystems) in duplicates. The primers used for qPCR are listed in Supplementary Table 5.
Luciferase assay
For Jurkat cells, the GIMAP enhancer reporter construct was co-transfected with the pGL4.74 Renilla reporter plasmid (Promega) using a Neon Transfection System (Invitrogen). Luciferase activity was measured using a microplate reader (Tecan) and a Dual-Luciferase Reporter Assay (Promega). To determine luciferase activity after knockdown, Jurkat cells that stably expressed the GIMAP enhancer reporter construct were selected by hygromycin treatment and were isolated into single clones. The cells were then transduced with lentivirus shRNA. Luciferase activity and cell viability were measured using a ONE-Glo™+Tox Luciferase Reporter and Cell Viability Assay kit (Promega).
Statistical Analysis
Two-tailed student t-tests were used to analyze differences in gene expression, luciferase activity, cell growth rate and apoptosis among groups. The standard deviations, standard error of the mean, mean, error bars and p-values are all indicated. Kaplan-Meier analysis and the Gehan-Breslow-Wilcoxon test were used to compare times to tumor onset in zebrafish study. P-values less than 0.05 were considered statistically significant.
Results
TAL1 binds at the super-enhancer site within the GIMAP gene cluster in T-ALL cells
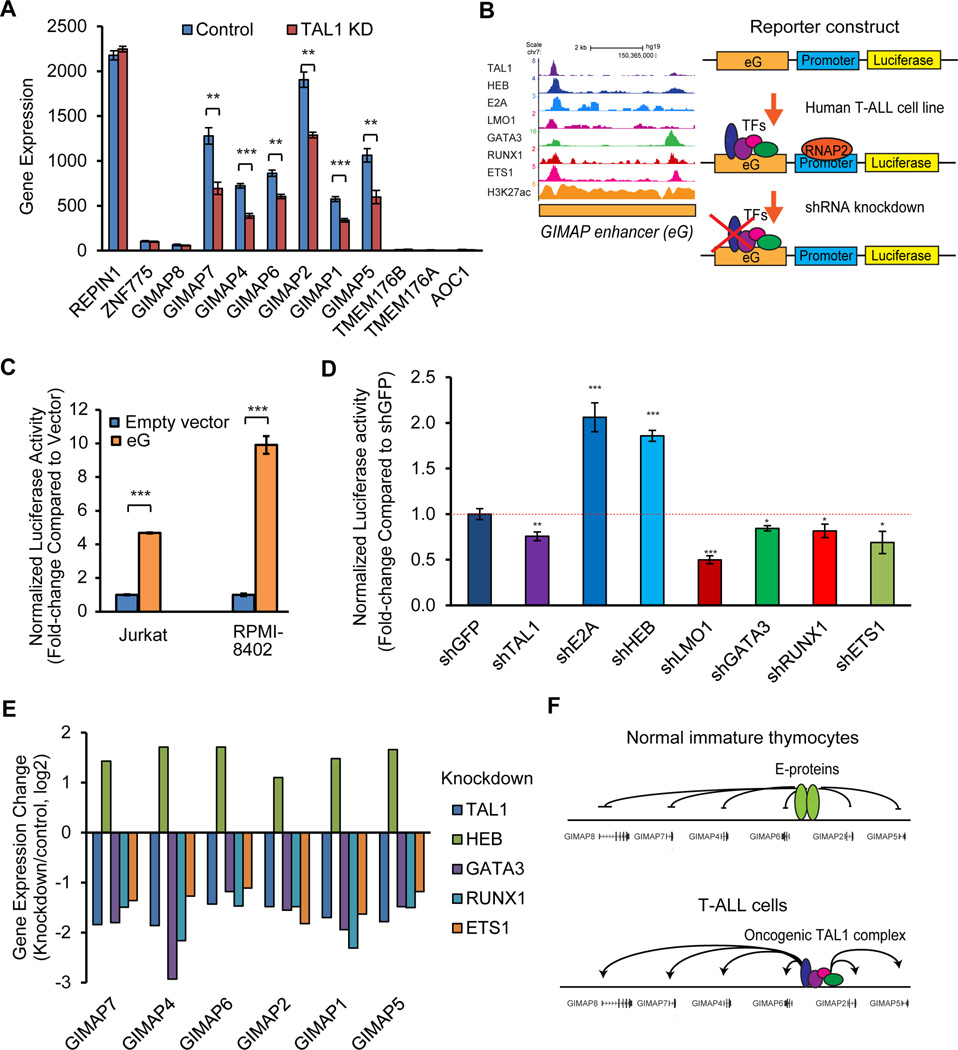
We previously identified transcriptional targets directly regulated by TAL1 and its regulatory partners by chromatin immunoprecipitation-sequencing (ChIP-Seq) and microarray analysis in TAL1-positive T-ALL cell samples.22 In the present study, we employed a targeted approach to identify genes and their regulatory elements that are differentially controlled by TAL1 and E-proteins. Using our previous dataset22, we first filtered genes that were positively regulated by TAL1 and were negatively regulated by E-proteins in a T-ALL cell line (Jurkat) (Supplementary Table 1 and Supplementary Figure 1A). We next explored the Gene Expression Commons database44 to select genes that are expressed in HSCs and are downregulated in double-negative (DN) stages of thymocytes (Supplementary Table 1 and Supplementary Figure 1B). We then selected genes that are associated with super-enhancers in T-ALL cells but not in the normal human thymus using the ChIP-Seq dataset23 (Supplementary Table 1).
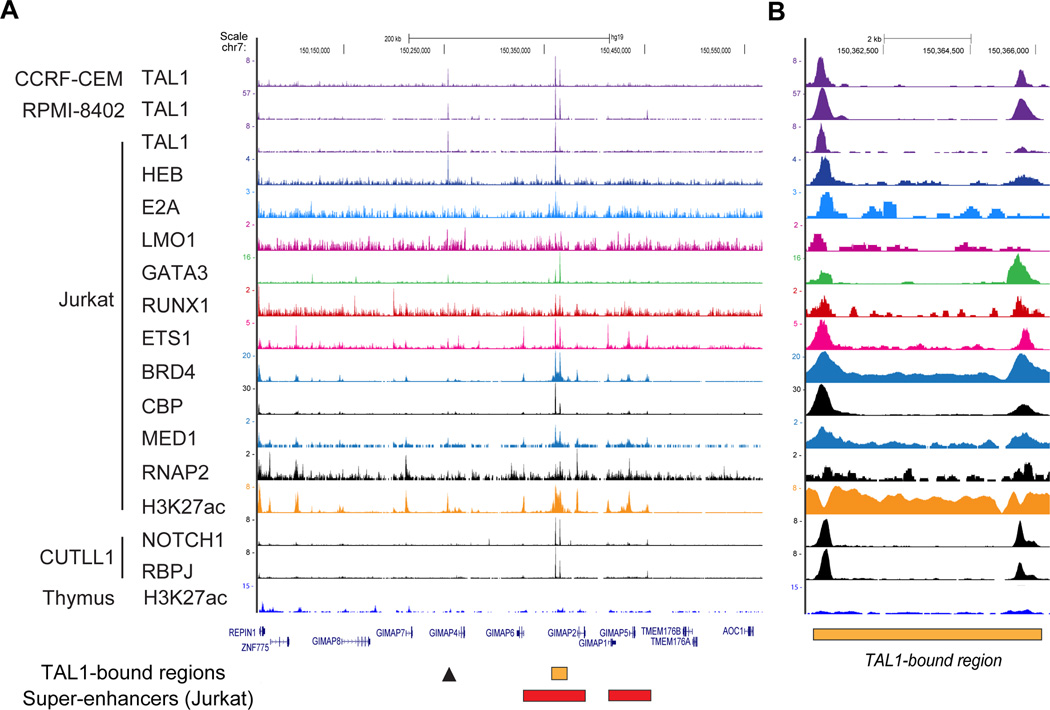
By these criteria, one element located within the gene cluster on chromosome 7q, which contains seven genes that belong to the same family (GIMAP1, GIMAP2, GIMAP4, GIMAP5, GIMAP6, GIMAP7 and GIMAP8), was selected (Figure 1A). DNA binding of the TAL1 complex was observed between the GIMAP2 and GIMAP6 genes across different TAL1-positive T-ALL cell lines (CCRF-CEM, RPMI-8402 and Jurkat) (Figures 1A and 1B, orange box). Notably, this region was also occupied by NOTCH1, which is another prevalent oncogene in T-ALL, with its binding partner RBPJ/CSL. Wang et al. previously reported that activated NOTCH1 regulates the expression of GIMAP genes.33, 45 Importantly, there was substantial binding of RNA polymerase II (RNAP2) and mediator 1 (MED1) as well as extensive acetylation of histone H3 lysine 27 (H3K27Ac), constituting the formation of a super-enhancer in this region (Figure 1A, red box; Supplementary Figure 1C).23, 27 The super-enhancer was not observed in normal thymus (bottom). This result indicates that the GIMAP gene cluster is highly activated in T-ALL cells.
Figure 1. The TAL1-bound super-enhancer in the GIMAP gene cluster.
(A, B) The ChIP-Seq gene tracks represent binding of transcription factors at the GIMAP gene cluster in T-ALL cell samples. Binding of transcription factors (TAL1, HEB, E2A, LMO1, GATA3, RUNX1, ETS1, BRD4, CBP, MED1, NOTCH1, RPBJ and RNA polymerase 2 (RNAP2)) in four T-ALL cell lines (CCRF-CEM, RPMI-8402, Jurkat and CUTLL1) as well as histone H3 acetylation at lysine 27 (H3K27ac) in Jurkat cells and normal thymus are shown. The x-axis indicates the linear sequence of genomic DNA, and the y-axis indicates the total number of mapped reads. The black horizontal bar indicates the genomic scale in kilobases (kb). Blue boxes in the gene map represent exons, and arrows indicate the location and direction of the transcriptional start site. The TAL1-bound region between the GIMAP6 and GIMAP2 genes is indicated as an orange box. The arrowhead shows an additional TAL1 binding site. The super-enhancers are shown as red boxes.
GIMAP genes are controlled by the GIMAP enhancer in T-ALL cells
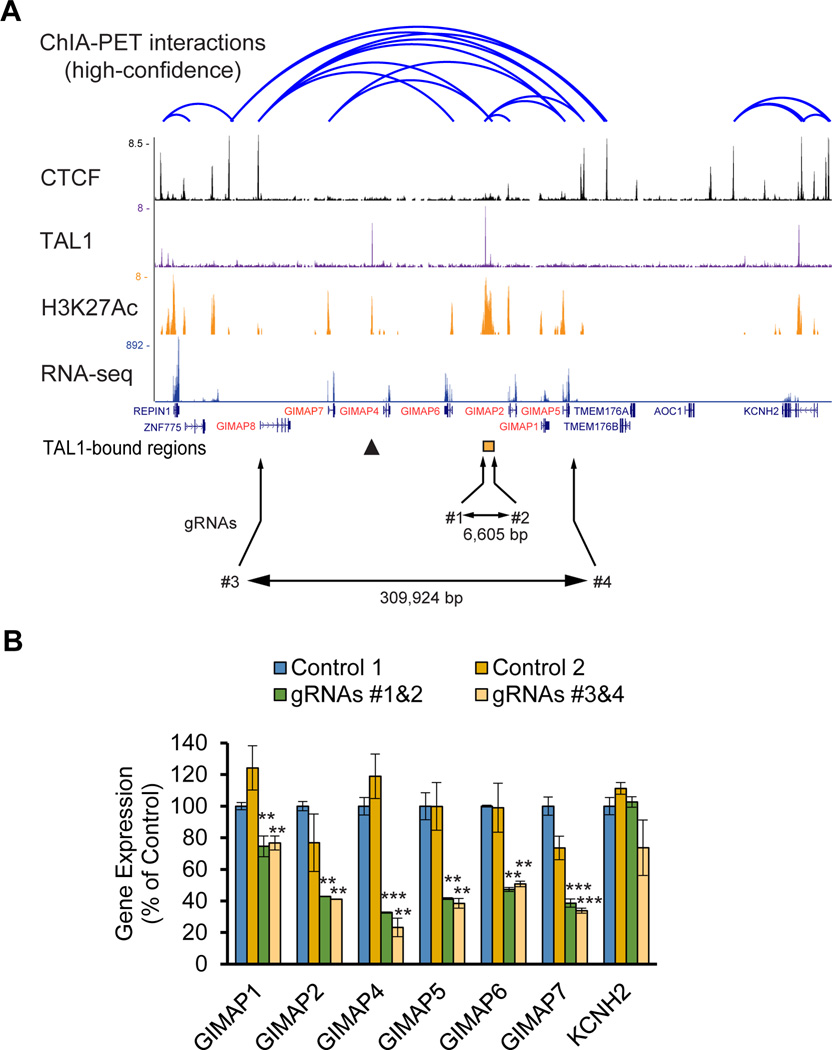
Indeed, GIMAP genes were highly expressed in many T-ALL cell lines (Supplementary Figure 2A). We identified significant positive correlations in expression levels across seven GIMAP genes (Supplementary Figure 2B), suggesting that these genes are coordinately regulated. In fact, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) for the Cohesin protein34 demonstrated multiple chromatin-chromatin interactions within the cluster region in Jurkat cells (Figure 2A). Interestingly, binding of the CTCF insulator protein was observed adjacent to the GIMAP8 and GIMAP5 genes but not within the cluster (Figure 2A), suggesting that these are neighborhood boundaries of the gene cluster.
Figure 2. GIMAP genes are controlled by the GIMAP enhancer in T-ALL cells.
(A) ChIA-PET chromatin-chromatin interactions and CTCF bindings in Jurkat cells were analyzed using the dataset reported by Hnisz et al. (Science, 2016). The ChIP-Seq gene tracks represent bindings of TAL1 and CTCF as well as H3K27Ac in Jurkat cells. The mRNA expressions were analyzed by RNA-Seq. Combinations of gRNAs (#1 and #2; #3 and #4) were used to knock out the TAL1-bound region (orange box) and the entire GIMAP gene cluster, respectively. The sizes of the deleted regions are shown. The TAL1-bound region between the GIMAP6 and GIMAP2 genes is indicated as an orange box. The arrowhead shows an additional TAL1 binding site. (B) Expression levels of each GIMAP gene in the knockout clones were measured by quantitative RT-qPCR (qRT-PCR) analysis. Expression values were normalized to that of GAPDH and are shown as the percentage of control values, which were transduced by gRNA targeting of the eGFP gene. The values are presented as the means ± SD of two samples. The KCNH2 gene resides to the right of the GIMAP gene cluster and serves as a negative control. *p<0.05, **p<0.01, ***p<0.001 by a two-sample, two-tailed t-test compared to the expression in control 1.
We next examined whether the region bound by the TAL1 complex (Figures 1 and 2A, orange box) can control the expression of GIMAP genes in T-ALL cells. We designed guide RNAs (gRNAs) targeting either this locus or the whole GIMAP gene cluster (Figure 2B, bottom) and transduced a pair of gRNAs with the Cas9 endonuclease to enable removal of these genomic regions using CRISPR/Cas9 technology. PCR analysis showed successful deletions of the target locus (Supplementary Figure 2C). The chromatogram demonstrated a successful recombination after the cleavage (Supplementary Figure 2D). Importantly, compared to cells transduced with control gRNAs, gene expression of the GIMAP genes was concomitantly downregulated in samples where either the TAL1-bound region or the whole cluster was deleted (Figure 2B). Expression of the KCNH2 gene, which is located outside of the cluster and does not interact with the TAL1-bound region (Figure 2A, right), was not affected. GIMAP8 expression was not shown because it is not highly expressed in Jurkat cells (Figure 2A, see RNA-seq). These results demonstrate that the locus bound by the TAL1 complex possesses enhancer activity to induce the expression of the GIMAP genes (GIMAP enhancer). Of note, an additional TAL1 binding site was identified between the GIMAP4 and GIMAP7 genes (Figures 1A and 2A, arrowhead). However, we did not observe any chromatin-chromatin interactions between this region and GIMAP genes by ChIP-PET analysis (Figure 2A).
The GIMAP enhancer can be activated in normal HSCs in vivo
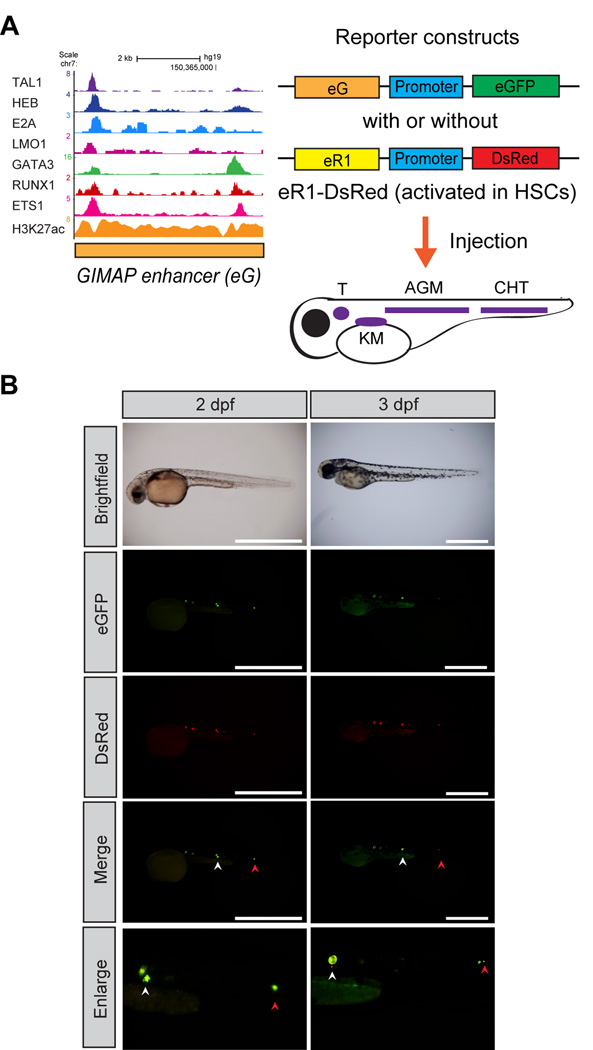
We next examined the activity of the GIMAP enhancer in normal hematopoietic cells using an in vivo reporter system. We cloned the enhancer sequence into a reporter construct that encodes the eGFP fluorescent protein with a minimal promoter sequence (Figure 3A). To analyze specific cells where the element is activated in vivo, we injected the construct into zebrafish embryos and analyzed the resulting eGFP expression patterns during embryogenesis. Interestingly, eGFP-positive cells were specifically detected along the posterior dorsal aorta at 2–3 days post fertilization (dpf) (Figure 3B). This region corresponded with the aorta-gonad-mesonephros (AGM), where zebrafish HSCs arise. To confirm the origin of fluorescence-positive cells, we co-introduced the RUNX1 +23/24 enhancer (eR1) reporter construct encoding DsRed, which has been reported to be activated in normal HSCs and progenitor cells both in zebrafish and mice.11, 41 This analysis revealed co-localization of eGFP and DsRed in the same cells (Figure 3B, middle and bottom panels). The same findings were reproducibly observed (Supplementary Figure 3A). These results show that the TAL1-bound region near the GIMAP2 gene locus can drive expression in HSC compartments in zebrafish. Of note, injection of the construct encoding the TAL1-bound region between the GIMAP4 and GIMAP7 genes showed eGFP-positive cells in circulating cells and muscles (Supplementary Figure 3B) and were not specific to HSCs.
Figure 3. The GIMAP enhancer is activated in normal zebrafish HSCs.
(A) Schematic representation of the experimental design. The GIMAP enhancer (eG) was introduced into the reporter construct carrying a minimal promoter sequence (promoter) and an eGFP gene. The RUNX1 stem cell enhancer (eR1) was cloned into the reporter construct encoding the DsRed gene. The reporter constructs were injected into a single-cell zebrafish embryo. The cartoon displays regions of zebrafish hematopoietic tissues. The kidney marrow (KM), aorta-gonad-mesonephros (AGM), caudal hematopoietic tissue (CHT), and thymus (T) are indicated. (B) Activation of the reporter constructs in early hematopoiesis in zebrafish. Fluorescent images show one representative zebrafish embryo (n=16) injected with reporter constructs at 2 and 3 dpf. The AGM and CHT are indicated by white and red arrowheads, respectively. Scale bar, 1 mm.
The GIMAP genes are repressed in immature thymocytes
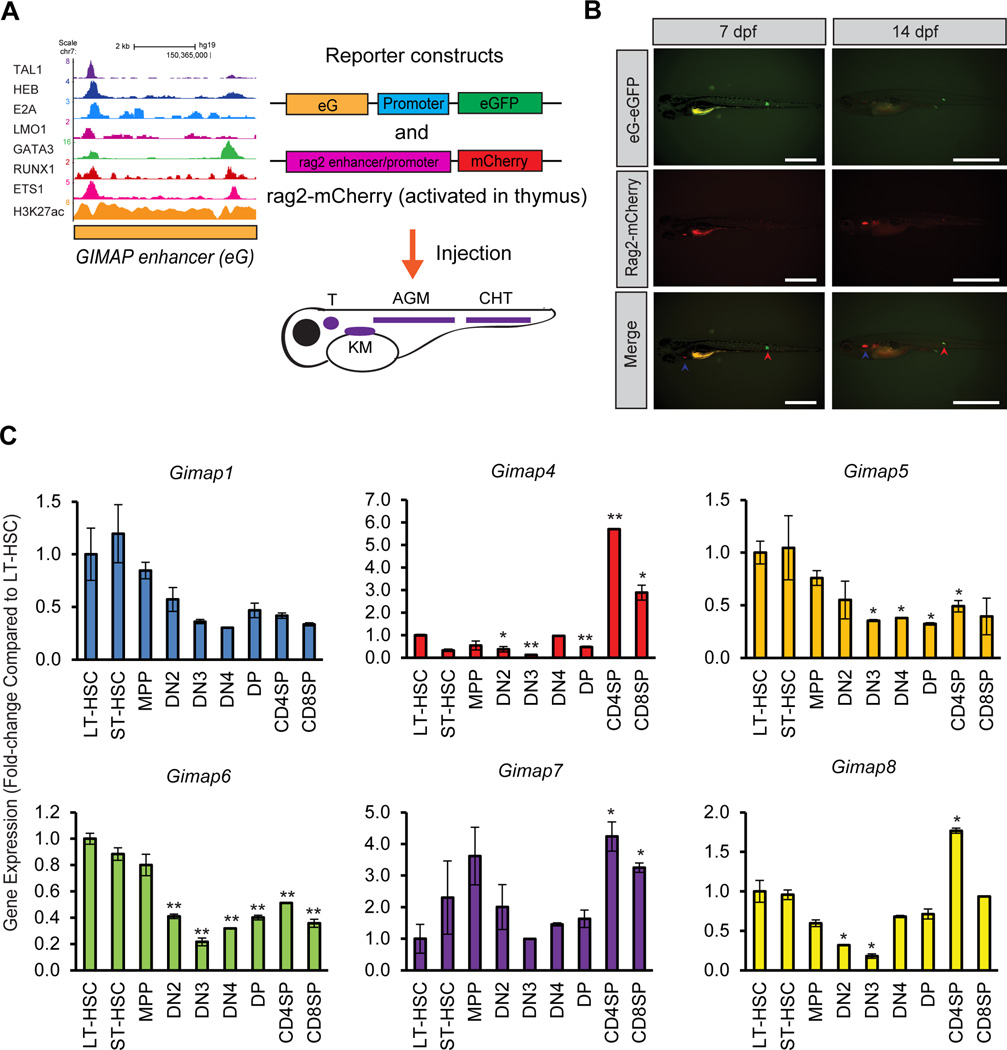
We next analyzed the activity of the GIMAP enhancer in the normal thymus. We co-injected a GIMAP enhancer reporter plasmid with the zebrafish rag2 reporter construct, which can be activated during the immature stage of thymocyte differentiation (Figure 4A).42 In this setting, we detected mCherry signals (by the rag2 promoter) but not eGFP signals (by the GIMAP enhancer) in the thymus (Figure 4B). This result indicates that the GIMAP enhancer cannot be activated in immature thymocytes.
Figure 4. The GIMAP enhancer is repressed in immature thymocytes.
(A) Schematic representation of the experimental design. The GIMAP enhancer (eG) was introduced into the reporter construct carrying a minimal promoter sequence (promoter) and an eGFP gene. The zebrafish rag2 promoter was cloned into the reporter construct encoding a mCherry gene. The reporter constructs were injected into a single-cell zebrafish embryo. See Figure 3A for details. (B) The GIMAP enhancer is not activated in immature zebrafish thymocytes. Fluorescent images show one representative zebrafish (n=4) injected with reporter constructs at 7 and 14 dpf. The CHT and thymus are indicated by red and blue arrowheads, respectively. Scale bar, 1 mm. (C) mRNA expression levels of Gimap genes in mouse hematopoietic cells. Total RNA was harvested from mouse hematopoietic cells at different stages: long-term HSC (LT-HSC), short-term HSC (ST-HSC), multi-potent progenitor (MPP), double-negative 2 (DN2), DN3, DN4, double-positive (DP), CD4-single-positive (CD4SP) and CD8-single-positive (CD8SP). Expression levels in DN1 cells could not be analyzed due to the lack of a sufficient sample size. The mRNA expression levels of six Gimap genes (Gimap1, Gimap4, Gimap5, Gimap6, Gimap7, and Gimap8) as well as β-actin were measured by quantitative RT-PCR analysis. The expression values were normalized to that of β-actin and were shown as fold-changes compared to LT-HSCs. The values are presented as the means ± SD of two samples. *p<0.05, **p<0.01 by a two-sample, two-tailed t-test compared to the expression in LT-HSC.
We next examined expression patterns of the orthologs of human GIMAP genes during mouse hematopoiesis. We analyzed the mRNA expression levels of six mouse Gimap genes during different stages of mouse hematopoietic cell differentiation. No ortholog of the human GIMAP2 gene was found in mice. This analysis revealed that Gimap1, Gimap5, Gimap6 and Gimap8 were highly expressed in both long-term (LT) and short-term (ST)-HSCs (Figure 4C). Gimap4, Gimap7 and Gimap8 were also highly expressed in CD4+ or CD8+ single-positive (SP) T-cells. Importantly, the expression of all Gimap genes was downregulated during the DN stage in thymocytes, which is consistent with the result by Gene Expression Commons (Supplementary Figure 1B). Similarly, the mouse Tal1 gene was downregulated at the DN stage (Supplementary Figure 4). In contrast, the expression of an E-protein Tcf12/Heb and Ets1 was upregulated in the same cells. These results indicate that Gimap genes are expressed in normal HSCs and progenitor cells but are repressed in immature thymocytes where Tal1 is silenced.
The GIMAP enhancer is activated by TAL1 and repressed by E-proteins in T-ALL cells
TAL1 is normally expressed together with GATA2, RUNX1 and ETS1 in HSCs and progenitor cells but is silenced during early thymocyte development.9, 10 TAL1 expression is undetectable in normal DN4 and DP cells12, a finding that we also confirmed. In contrast, TAL1 is overexpressed in T-ALL cells,15–19 resulting in differentiation arrest of immature thymocytes.18, 20 Thus, we expect that ectopic expression of TAL1 leads to aberrant expression of GIMAP genes via activation of the GIMAP enhancer. In fact, when we analyzed changes in GIMAP gene expression after knockdown of TAL1 in Jurkat cells, the mRNA expression of six GIMAP genes was concomitantly downregulated (Figure 5A). Notably, the expression of genes outside the GIMAP cluster (REPIN1, ZNF775, TMEM176B, TMEM176A and AOC1; see Figures 1A and 2A) was not affected by TAL1 knockdown. These results indicate that GIMAP genes located within the cluster are specifically controlled by TAL1 in T-ALL cells.
Figure 5. The GIMAP enhancer is activated by TAL1 and is repressed by E-proteins in T-ALL cells.
(A) Expression changes for GIMAP genes after TAL1 knockdown in Jurkat cells. Global gene expression was analyzed using microarrays in four control samples and four TAL1 knockdown samples as previously described.22 The expression values were normalized by dChip software and are shown as the means ± standard deviations (SDs) of four samples; **p<0.01, ***p<0.001 by a two-sample, two-tailed t-test. (B) Schematic representation of the experimental design. The GIMAP enhancer (eG) was introduced into the reporter construct carrying a minimal promoter sequence (promoter) and a luciferase gene. The luciferase reporter construct was transfected into human T-ALL cells. Luciferase activity was measured in T-ALL cells after knockdown of the transcription factor (TF). (C) Induction of GIMAP enhancer activity in TAL1-positive T-ALL cells. The GIMAP enhancer reporter construct was co-transfected with the renilla plasmid (internal control for transfection efficiency) into two TAL1-positive T-ALL cell lines (Jurkat and RPMI-8402). An empty luciferase vector was used as a control. Luciferase activity values were normalized to renilla activity and are shown as fold-changes compared to the empty vector. The values are presented as the means ± SDs of biological triplicates. ***p<0.001 by a two-sample, two-tailed t-test. (D) Inhibition of GIMAP enhancer activity by knockdown of transcription factors in T-ALL cells. Jurkat cells that stably expressed the GIMAP enhancer reporter construct were established and were transduced with shRNA targeting a transcription factor using lentivirus infection. Luciferase activity and cell viability were measured after 5 days of virus infection. Luciferase activity values were normalized to cell viability and are shown as fold-changes compared to the control sample, which was transduced with shGFP. The values are presented as the means ± SDs of biological triplicates; *p<0.05, **p<0.01, ***p<0.001 by a two-sample, two-tailed t-test. (E) Expression changes of GIMAP genes after knockdown of transcription factors in Jurkat cells (see the Figure 5A legend for details). The mean fold-change is shown as log 2 of knockdown over control cells. (F) Proposed model of GIMAP gene activation in normal and T-ALL cells. E-protein dimers negatively regulate GIMAP gene expression in normal immature thymocytes. The oncogenic TAL1 complex positively regulates GIMAP gene expression in T-ALL cells. Arrows indicate cases of gene activation.
Subsequently, we examined the functional contribution of each transcription factor involved in the TAL1 complex in regulating the GIMAP enhancer. Analysis of associated DNA-binding sequences revealed that E-box, GATA, RUNX and ETS motifs were observed within this region (Supplementary Figure 5A). To determine whether the GIMAP enhancer can be activated in human T-ALL cells, we introduced the GIMAP enhancer element into the luciferase reporter construct and transfected the construct into human T-ALL cells (Figure 5B). Compared with empty vector-transfected cells, significant inductions of luciferase activity were observed in TAL1-positive T-ALL cell lines (Jurkat and RPMI-8402) (Figure 5C), indicating that the GIMAP enhancer can be activated by endogenous factors in TAL1-positive T-ALL cells. Next, we measured luciferase activity after depletion of either TAL1 or each of the regulatory partners in Jurkat cells that stably expressed the reporter construct. We transduced short-hairpin RNA (shRNA) by lentiviral infection to specifically knock down each of the target proteins (Supplementary Figure 5B). In this analysis, depletion of TAL1, LMO1, GATA3, RUNX1 or ETS1 reduced luciferase activity (Figure 5D) compared to control samples in which control GFP shRNA was transduced. This result indicates that the TAL1 complex can activate the GIMAP enhancer in T-ALL cells.
Notably, we found that knockdown of the E-proteins E2A and HEB increased luciferase activity (Figure 5D). When we examined changes in the expression of GIMAP genes after knockdown of each member of the TAL1 complex in Jurkat cells, the mRNA expression of all GIMAP genes was downregulated after depletion of TAL1, GATA3, RUNX1 and ETS1 but was upregulated after depletion of the E-protein HEB (Figure 5E). Our results indicate that the GIMAP genes are positively regulated by the TAL1 complex and negatively regulated by E-proteins (Figure 5F).
Overexpression of human GIMAP genes in immature thymocytes promotes T-cell leukemogenesis
Because GIMAP genes are normally repressed during the DN stage in thymocytes (Figure 4C) but are overexpressed in human T-ALL cells, we postulated that ectopic expression of GIMAP genes in immature thymocytes could contribute to leukemogenesis. Hence, we examined the tumorigenic ability of GIMAP genes in vivo. Prior to this analysis, we first confirmed cell growth in a human T-ALL cell line where either the GIMAP enhancer or the whole GIMAP gene cluster had been deleted (Figure 2). These conditions slightly reduced the growth of T-ALL cells (Supplementary Figure 6A). Individual gene knockdown using validated shRNAs (Supplementary Figure 6B) demonstrated that inhibition of GIMAP5 and GIMAP7 reduced cell growth (Supplementary Figure 6C). Analysis with annexin staining also revealed that knockdown of GIMAP5 and GIMAP7 induced apoptotic cell death (Supplementary Figure 6D). Although the inhibitory effect on growth after the loss of GIMAP proteins is relatively weak in vitro, these results supported previous findings that demonstrated a protective role of Gimap5 against cell death.46, 47 Based on these results, we selected GIMAP5 and GIMAP7 genes for the transgenic study.
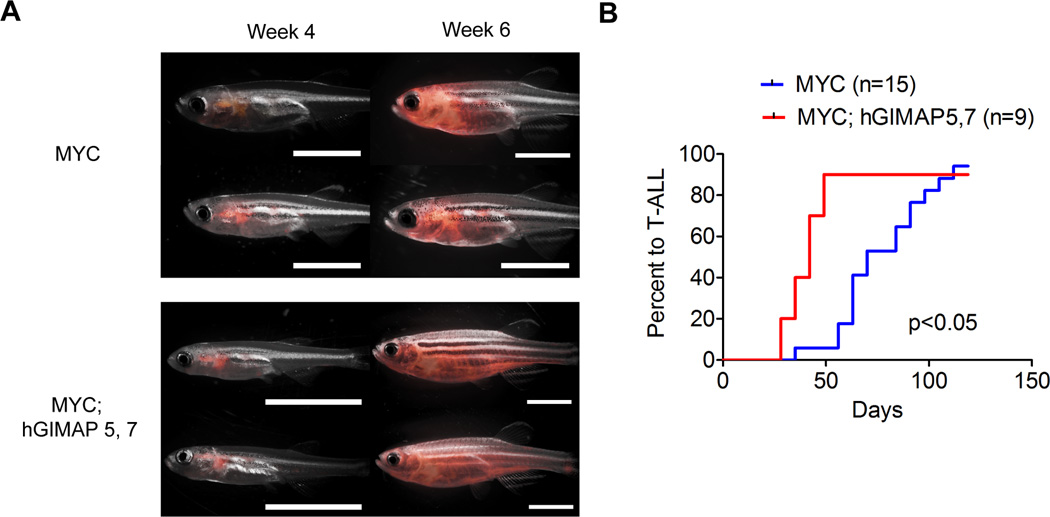
We used the zebrafish rag2 promoter to specifically overexpress multiple human GIMAP genes in immature thymocytes to recapitulate human T-ALL. In this setting, overexpression of GIMAP5 and GIMAP7 either individually or in combination did not induce T-ALL in zebrafish (data not shown), suggesting that GIMAP genes alone do not possess sufficient oncogenic potential to initiate leukemia. Hence, we next overexpressed the GIMAP5 and GIMAP7 genes in immature thymocytes together with the MYC T-ALL oncogene. Strikingly, transgenic zebrafish that showed concomitant overexpression of the GIMAP5 and GIMAP7 genes significantly accelerated tumor onset compared to the MYC single transgenic fish (Figures 6A and B). These results indicate that ectopic expression of GIMAP genes promotes T-cell leukemogenesis in vivo when overexpressed with other oncogenes.
Figure 6. Overexpression of GIMAP genes accelerates T-ALL onset in vivo.
(A) Tumor development in transgenic zebrafish. Tumor cells expressing fluorescent proteins (eGFP and mCherry) in rag2::Myc-EGFP; rag2::mCherry (MYC) (n=15) and rag2::Myc-EGFP; rag2::hGIMAP5; rag2::hGIMAP7; rag2::mCherry (MYC; hGIMAP5,7) (n=9) transgenic fish were analyzed at 4 and 6 weeks using epi-fluorescence stereomicroscopy. Two representative zebrafish for each group are shown. Panels show the merged fluorescent and bright-field images. Scale bar, 5 mm. (B) The tumor onset of T-ALL in MYC-transgenic fish (n=15) and MYC; GIMAP5, 7 transgenic fish (n=9) were analyzed via the Kaplan-Meier method. P<0.05 by the Gehan-Breslow-Wilcoxon test.
Discussion
In the present study, we report that GIMAP genes are highly expressed in human T-ALL cells as well as in normal mouse HSCs. Expression of the GIMAP genes is differentially controlled by TAL1 and E-proteins via the GIMAP enhancer. Ectopic overexpression of human GIMAP5 and GIMAP7 genes in the zebrafish thymus accelerated tumor development in the presence of MYC. Our results demonstrate that aberrant activation of the GIMAP enhancer contributes to T-cell leukemogenesis.
The GIMAP family of proteins has been implicated in lymphocyte development. Studies with knockout mice show that these proteins are involved in the development and survival of mature T-lymphocytes.28, 32, 48 One research group reported that mouse Gimap1, Gimap4, Gimap5 and Gimap6 are downregulated in DP cells and that their expression is dramatically increased in CD4+ or CD8+ SP T-cells,28, 48, 49 which was consistent with our results. In general, the vast majority of thymocytes undergo apoptosis by positive selection during the transition from DP to SP cells, and only selected T-cells acquire the ability to survive.50 These findings suggest a possible role of GIMAPs in the positive selection of T-cells. In addition, we have now shown that GIMAP genes are highly expressed in HSCs and progenitor cells. This suggests that GIMAP genes could be physiological targets of TAL1 in normal HSCs.
Notably, the roles of GIMAPs in survival-or-death determinations vary among the family proteins. For example, GIMAP4 has been reported to accelerate T-cell death, whereas GIMAP5 exhibits a protective role against cell death through association with the Bcl-2 protein family.28, 30, 46, 51 Gimap5 knockout mice show a dramatic decrease in T-cell numbers.52 GIMAP1 is also vital for the development/survival of mature T-lymphocytes.49 Additionally, GIMAP2 can heterodimerize with GIMAP7 protein to activate GIMAP7 function in lymphocytes.53, 54 These studies indicate that multiple GIMAP proteins cooperate to maintain T-cell survival. Further investigation is necessary to elucidate the roles of GIMAP proteins during T-cell leukemogenesis.
Interestingly, the GIMAP genes have also been reported to be downstream targets of the NOTCH1 oncogene in T-ALL.31, 33 ChIP-Seq data demonstrate binding of NOTCH1 and its partner RBPJ/CSL at the GIMAP enhancer in T-ALL cells.33 Notch1 upregulates the GIMAP family genes, and pharmacological inhibition of the Notch signaling pathway perturbs T-ALL cell proliferation.31 Hence, GIMAP can be positively regulated by two major T-ALL oncogenes, namely, TAL1 and NOTCH1. In normal T-cell development, NOTCH1 is downregulated during the transition from the DN3 to the DN4 stage,55 when GIMAP genes are silenced. Activating mutations of NOTCH1 are frequently found in TAL1-positive human T-ALL as well as in transgenic mouse models, leading to increased MYC expression.56, 57 Hence, progressive increases in GIMAP gene expression may contribute to the oncogenic collaboration among TAL1, NOTCH1 and MYC in T-ALL.
Importantly, we showed that E-proteins (E2A and HEB) negatively regulate GIMAP enhancer activity in T-ALL cells. E-proteins form a heterodimer with TAL1 but can also form homodimers by themselves to regulate gene expression.14 E-proteins are normally required for thymocyte development in a stage-specific manner.12, 13 E-proteins are upregulated during thymocyte development, and their expression levels are highest during the DN and DP stages in thymocytes as reported by other researchers12 and by us (Supplementary Figure 4), in which TAL1 is silenced and GIMAP genes are downregulated (Figure 4). These findings indicate that GIMAP genes are repressed by E-proteins in normal immature thymocytes but can be reactivated by ectopic expression of TAL1 and/or by inhibition of the E-protein dimers. E-proteins have been implicated as tumor suppressors in T-ALL, and a heterozygous loss of either E2a or Heb markedly accelerates the onset of T-ALL in Tal1-transgenic mice.21 Therefore, we hypothesize that overexpression of TAL1 and loss of E-proteins synergistically upregulate GIMAPs (Figure 5F), thereby accelerating T-cell leukemogenesis. Taken together, our study provides a novel mechanism by which TAL1 contributes to T-cell leukemogenesis through induction of the GIMAP enhancer activity in developing thymocytes.
Supplementary Material
Acknowledgments
We thank Rick Young, Lee Lawton and members of the Sanda laboratory for their discussions and critical reviews. We thank Wei Zhong Leong and Stella Amanda for their help with the zebrafish experiments. We are grateful to the NUS zebrafish facility for zebrafish analyses. This research is supported by the National Research Foundation, Prime Minister’s Office, Singapore, under its Competitive Research Programme (award no. NRF-NRFF2013-02) and the U.S. National Cancer Institute (1K99CA157951).
Footnotes
Authorship contributions
W-S.L., S.H.T. and T.S. performed the experiments; C.Q.W., Z.G. and M.O. provided materials; W-S.L., P.C.T.N. and T.S. analyzed the results; V.T., M.O., and A.T.L. supervised this study; and W-S.L. and T.S. designed the research and wrote the paper.
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008 May;8(5):380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005 Sep 10;23(26):6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 3.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997 Nov 7;278(5340):1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 4.Lecuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004 Jan;32(1):11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Hsu HL, Cheng JT, Chen Q, Baer R. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol Cell Biol. 1991 Jun;11(6):3037–3042. doi: 10.1128/mcb.11.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu HL, Wadman I, Baer R. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3181–3185. doi: 10.1073/pnas.91.8.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, et al. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002 Oct 1;100(7):2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- 8.Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997 Jun 2;16(11):3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003 Nov;23(21):7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry JR, Kinston S, Knezevic K, de Bruijn MF, Wilson N, Nottingham WT, et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood. 2008 Mar 15;111(6):3005–3014. doi: 10.1182/blood-2007-07-098830. [DOI] [PubMed] [Google Scholar]
- 11.Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007 Dec 15;110(13):4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol. 2000 Aug;1(2):138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 13.Bernard M, Delabesse E, Smit L, Millien C, Kirsch IR, Strominger JL, et al. Helix-loop-helix (E2–5, HEB, TAL1 and Id1) protein interaction with the TCRalphadelta enhancers. Int Immunol. 1998 Oct;10(10):1539–1549. doi: 10.1093/intimm/10.10.1539. [DOI] [PubMed] [Google Scholar]
- 14.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009 Mar;9(3):175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 15.Begley CG, Aplan PD, Davey MP, Nakahara K, Tchorz K, Kurtzberg J, et al. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor delta-chain diversity region and results in a previously unreported fusion transcript. Proc Natl Acad Sci U S A. 1989 Mar;86(6):2031–2035. doi: 10.1073/pnas.86.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown L, Cheng JT, Chen Q, Siciliano MJ, Crist W, Buchanan G, et al. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990 Oct;9(10):3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrando AA, Look AT. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin Hematol. 2000 Oct;37(4):381–395. doi: 10.1016/s0037-1963(00)90018-0. [DOI] [PubMed] [Google Scholar]
- 18.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002 Feb;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 19.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 Dec 12;346(6215):1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, et al. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010 Jun 1;24(11):1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004 Jun;5(6):587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 22.Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012 Aug 14;22(2):209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014 Jul 31;511(7511):616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pott S, Lieb JD. What are super-enhancers? Nature genetics. 2015 Jan;47(1):8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 25.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013 Nov 7;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013 Apr 11;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013 Apr 11;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006 Apr;4(4):e103. doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cambot M, Aresta S, Kahn-Perles B, de Gunzburg J, Romeo PH. Human immune associated nucleotide 1: a member of a new guanosine triphosphatase family expressed in resting T and B cells. Blood. 2002 May 1;99(9):3293–3301. doi: 10.1182/blood.v99.9.3293. [DOI] [PubMed] [Google Scholar]
- 30.Chadwick N, Zeef L, Portillo V, Boros J, Hoyle S, van Doesburg JC, et al. Notch protection against apoptosis in T-ALL cells mediated by GIMAP5. Blood Cells Mol Dis. 2010 Oct 15;45(3):201–209. doi: 10.1016/j.bcmd.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Chadwick N, Zeef L, Portillo V, Fennessy C, Warrander F, Hoyle S, et al. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer. 2009;8:35. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krucken J, Epe M, Benten WP, Falkenroth N, Wunderlich F. Malaria-suppressible expression of the anti-apoptotic triple GTPase mGIMAP8. J Cell Biochem. 2005 Oct 1;96(2):339–348. doi: 10.1002/jcb.20552. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011 Sep 6;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016 Mar 25;351(6280):1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010 Oct;28(10):1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004 May 22;20(8):1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 37.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014 Oct 9;159(2):374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Lowdon RF, Li D, Lawson HA, Madden PA, Costello JF, et al. Exploring long-range genome interactions using the WashU Epigenome Browser. Nature methods. 2013 May;10(5):375–376. doi: 10.1038/nmeth.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 Jan 1;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014 Aug;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng CE, Yokomizo T, Yamashita N, Cirovic B, Jin H, Wen Z, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells. 2010 Oct;28(10):1869–1881. doi: 10.1002/stem.507. [DOI] [PubMed] [Google Scholar]
- 42.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003 Feb 7;299(5608):887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 43.Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010 Oct 19;18(4):353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One. 2012;7(7):e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnell S, Demolliere C, van den Berk P, Jacobs H. Gimap4 accelerates T-cell death. Blood. 2006 Jul 15;108(2):591–599. doi: 10.1182/blood-2005-11-4616. [DOI] [PubMed] [Google Scholar]
- 47.Schulteis RD, Chu H, Dai X, Chen Y, Edwards B, Haribhai D, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood. 2008 Dec 15;112(13):4905–4914. doi: 10.1182/blood-2008-03-146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. 2007 Feb;28(2):58–65. doi: 10.1016/j.it.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Saunders A, Webb LM, Janas ML, Hutchings A, Pascall J, Carter C, et al. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 2010 Apr 22;115(16):3249–3257. doi: 10.1182/blood-2009-08-237586. [DOI] [PubMed] [Google Scholar]
- 50.Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate 'decisions' and effector function. Nat Immunol. 2014 Sep;15(9):815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moralejo DH, Park HA, Speros SJ, MacMurray AJ, Kwitek AE, Jacob HJ, et al. Genetic dissection of lymphopenia from autoimmunity by introgression of mutated Ian5 gene onto the F344 rat. J Autoimmun. 2003 Dec;21(4):315–324. doi: 10.1016/S0896-8411(03)00138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barnes MJ, Aksoylar H, Krebs P, Bourdeau T, Arnold CN, Xia Y, et al. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J Immunol. 2010 Apr 1;184(7):3743–3754. doi: 10.4049/jimmunol.0903164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwefel D, Arasu BS, Marino SF, Lamprecht B, Kochert K, Rosenbaum E, et al. Structural insights into the mechanism of GTPase activation in the GIMAP family. Structure. 2013 Apr 2;21(4):550–559. doi: 10.1016/j.str.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Yano K, Carter C, Yoshida N, Abe T, Yamada A, Nitta T, et al. Gimap3 and Gimap5 cooperate to maintain T-cell numbers in the mouse. Eur J Immunol. 2014 Feb;44(2):561–572. doi: 10.1002/eji.201343750. [DOI] [PubMed] [Google Scholar]
- 55.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014 Aug;14(8):529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 57.O'Neil J, Calvo J, McKenna K, Krishnamoorthy V, Aster JC, Bassing CH, et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006 Jan 15;107(2):781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.