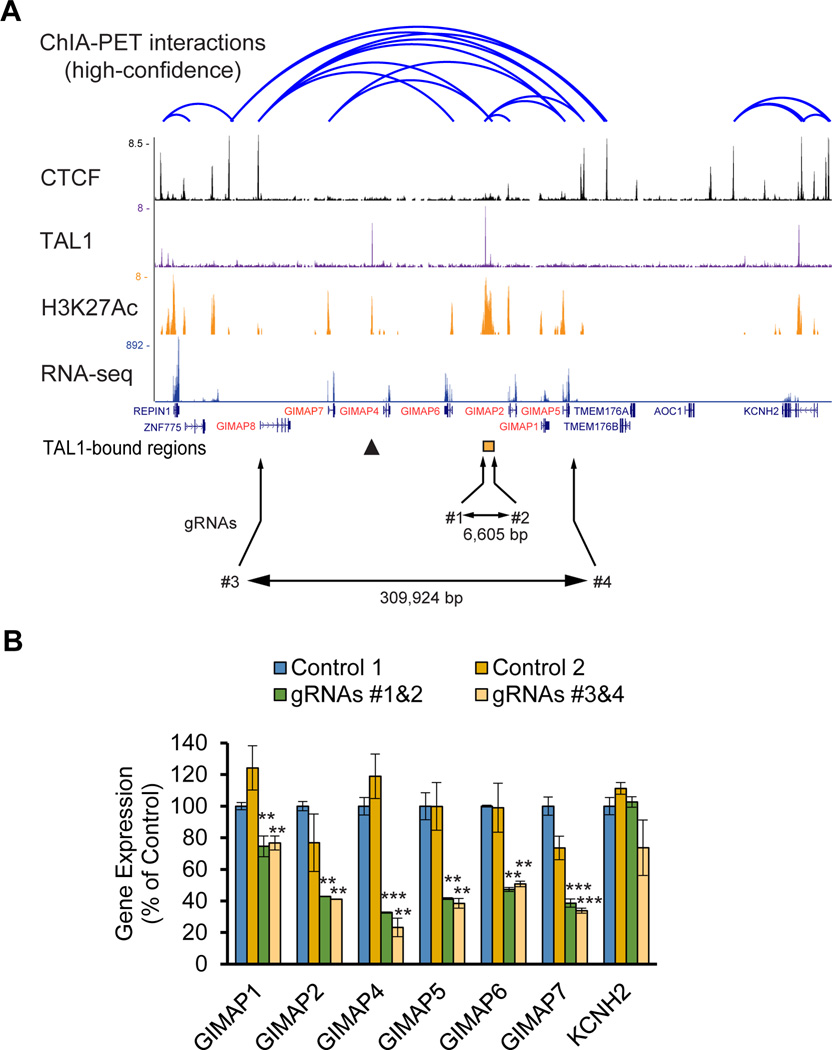
Figure 2. GIMAP genes are controlled by the GIMAP enhancer in T-ALL cells.
(A) ChIA-PET chromatin-chromatin interactions and CTCF bindings in Jurkat cells were analyzed using the dataset reported by Hnisz et al. (Science, 2016). The ChIP-Seq gene tracks represent bindings of TAL1 and CTCF as well as H3K27Ac in Jurkat cells. The mRNA expressions were analyzed by RNA-Seq. Combinations of gRNAs (#1 and #2; #3 and #4) were used to knock out the TAL1-bound region (orange box) and the entire GIMAP gene cluster, respectively. The sizes of the deleted regions are shown. The TAL1-bound region between the GIMAP6 and GIMAP2 genes is indicated as an orange box. The arrowhead shows an additional TAL1 binding site. (B) Expression levels of each GIMAP gene in the knockout clones were measured by quantitative RT-qPCR (qRT-PCR) analysis. Expression values were normalized to that of GAPDH and are shown as the percentage of control values, which were transduced by gRNA targeting of the eGFP gene. The values are presented as the means ± SD of two samples. The KCNH2 gene resides to the right of the GIMAP gene cluster and serves as a negative control. *p<0.05, **p<0.01, ***p<0.001 by a two-sample, two-tailed t-test compared to the expression in control 1.