Abstract
Previous work has shown that purified capsid protein (CP) of cowpea chlorotic mottle virus (CCMV) is capable of packaging both purified single-stranded RNA molecules of normal composition (comparable numbers of A, U, G, and C nucleobases) and of varying length and sequence, and anionic synthetic polymers such as polystyrene sulfonate. We find that CCMV CP is also capable of packaging polyU RNAs, which—unlike normal-composition RNAs—do not form secondary structures and which act as essentially structureless linear polymers. Following our canonical two-step assembly protocol, polyU RNAs ranging in length from 1000 to 9000 nucleotides (nt) are completely packaged. Surprisingly, negative-stain electron microscopy shows that all lengths of polyU are packaged into 22-nm-diameter particles despite the fact that CCMV CP prefers to form 28-nm-diameter (T = 3) particles when packaging normal-composition RNAs. PolyU RNAs >5000 nt in length are packaged into multiplet capsids, in which a single RNA molecule is shared between two or more 22-nm-diameter capsids, in analogy with the multiplets of 28-nm-diameter particles formed with normal-composition RNAs >5000 nt long. Experiments in which viral RNA competes for viral CP with polyUs of equal length show that polyU, despite its lack of secondary structure, is packaged more efficiently than viral RNA. These findings illustrate that the secondary structure of the RNA molecule—and its absence—plays an essential role in determining capsid structure during the self-assembly of CCMV-like particles.
Introduction
Many viruses use long (thousands of nucleotides (nt)) single-stranded RNAs (ssRNAs) as their genomic material. Such viruses can be as simple as a single RNA molecule encapsidated inside a shell (capsid) composed of many copies of a single capsid protein (CP). The capsid serves to protect the genome from degradation as it moves in and between host cells. Several ssRNA viruses are capable of spontaneous in vitro self-assembly (1, 2), driven by a combination of attractions between the CP and the RNA molecule (CP-RNA) and interactions between neighboring CP subunits (CP-CP) (3).
CCMV, the virus studied throughout this work, is a model ssRNA virus whose virions are composed of 180 identical CPs arranged in 28-nm-diameter icosahedrally symmetric capsids with a T = 3 Caspar-Klug triangulation number (4), and the virions contain either one copy of 3200-nt RNA1, or one copy of 2800-nt RNA2, or one copy each of 2100-nt RNA3 and 700-nt RNA4—hence, ∼3000 nt per capsid. They can be reconstituted in vitro from purified components producing particles that are indistinguishable from the native virus (5). More importantly still, for our purposes, is the fact that CCMV CP is able to package non-viral ssRNA molecules of varying length and sequence (6, 7, 8). It is important to note that these in vitro assembly reactions rely on a two-step protocol in which the components are dialyzed into low-pH (< 6) buffer, which may not be physiologically relevant. Further, a superstoichiometric amount of CP is used in these experiments, as this has been found to be necessary to package all of the RNA in vitro.
Recent experimental and theoretical work has suggested that long ssRNAs exhibit generic properties that enhance their ability to be packaged by viral CP. Specifically, any long RNA molecule with comparable numbers of the bases A, U, G, and C—i.e., “normal” composition—results in secondary structures in which there is extensive Watson-Crick basepairing, typically involving >50% of the nucleotides (9). Because these secondary structures involve a large number of single-stranded loops from which three or more duplexes emanate, they cause these RNAs to behave effectively as branched polymers in solution (10). This branching results in RNA molecules being significantly more compact than structureless linear polymers of similar chain length. The secondary structures associated with viral sequences of RNA involve a larger number of threefold and higher-order “branch points” than do nonviral normal-composition RNAs, and are, as a consequence, more compact (11, 12). Further, ssRNA viral genomes have been shown to have radii of gyration comparable to the radii of the virions into which they are packaged (13).
For RNA molecules of comparable length and similar extent and nature of branching, particular local components of secondary structures, e.g., stem loops with specific sequences in their duplex and single-stranded portions, have been implicated in the preferential packaging, in vivo, of viral RNA over cellular RNA. “Packaging signals” of this kind have been identified in the RNA genomes of several different viruses, and have been established as the origin of nucleation of viral capsids in several in vitro bulk-solution and single-molecule measurements (14, 15, 16, 17, 18).
Competitive self-assembly studies have recently probed the key role of sequence and length in determining the packaging of an RNA (19). In these experiments, equal masses of two different RNA molecules are allowed to compete for an amount of CCMV CP sufficient to completely package only one of them. It was found in such competitive assemblies of branched RNAs of different lengths and sequences that the 3000 nt CCMV RNA1 (C1) outcompeted shorter and longer RNAs. But, surprisingly, the heterologous Brome Mosaic Virus (BMV) RNA1 (B1), essentially identical in length to C1 but with different sequence and, hence, different secondary structure, outcompeted C1 for its own CP.
The role of secondary structure on packaging has been examined in reaction-scheme descriptions of the assembly process (20), theory (21), and computer simulation (22). Erdemci-Tandogan et al. (21) employed mean-field theory to investigate how the secondary structure of an RNA affected the electrostatic interactions with CP and the stability of the assembled virions. They found a deeper minimum for the packaging free energy of a branched polymer compared to that of a linear one with the same number of monomers, and that the optimal packaged length was greater. Perlmutter et al. (22) found similarly that the formation of RNA secondary structure allows longer RNA molecules to be packaged by viral CP. Additionally, Monte Carlo simulations of competition experiments have shown that more compact, branched RNA molecules outcompete less branched RNAs for binding of CP (23).
To provide a direct experimental test of the effect of branching on the self-assembly of virions we have undertaken in vitro packaging studies of polyU, an RNA molecule with no secondary structure. In addition to having no basepairing, the stacking interaction between uridines is very weak, so polyU is an essentially structureless linear polyelectrolyte except at low temperature, where it exhibits some helicity (24). We examine how these unstructured RNAs are packaged in vitro by CCMV CP, and how they compete with viral RNA for CP, to determine whether the presence of secondary structure/branching allows the viral genome to be selectively packaged by its cognate CP. Throughout our discussion, we will refer to polyU molecules alternately/interchangeably as “linear” and structureless polymers, and to normal-composition RNAs as “branched” and structured polymers.
We find that polyU RNAs are packaged by CCMV CP into capsids with a characteristic size of 22 nm, smaller than the 28-nm diameter of the wild-type (wt) virus, and that their size is independent of the length of the polyU packaged. For lengths of polyU significantly longer than the 3000-nt wt genome, polyU is packaged into multiplets, in which a single RNA is shared between two or more capsids. The packaged polyU is protected against RNase, demonstrating that the particles are closed. Electron microscope images of the particles show that, although they have sizes typical of “T = 2” capsids, a majority of the particles are not icosahedrally symmetric and are significantly polydisperse. A series of experiments in which polyU competes with normal-composition RNAs of equal length demonstrates that, surprisingly, and in contrast to theoretical expectations, the unstructured RNA is preferentially packaged.
Materials and Methods
Buffers
The following buffers were employed: synthesis buffer (100 mM Tris-HCl (pH 9), 1 mM EDTA, and 5 mM MgCl2); buffer B (protein buffer; 20 mM Tris-HCl (pH 7.2), 1 M NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride [PMSF]); RNA assembly buffer (RAB; 50 mM Tris-HCl (pH 7.2), 50 mM NaCl, 10 mM KCl, and 5 mM MgCl2); virus storage buffer (VSB; 50 mM sodium acetate (pH 4.5) and 8 mM magnesium acetate); TE buffer (10 mM Tris-HCl (pH 7.5) and 1 mM EDTA); disassembly buffer (DAB; 50 mM Tris-HCl (pH 7.5), 0.5 M CaCl2, 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF); virus running buffer (VRB; 100 mM sodium acetate (pH 5.5) and 1 mM EDTA); and TAE buffer (40 mM Tris-HCl (pH 8), 20 mM acetic acid, and 1 mM EDTA).
PolyU synthesis and fractionation
PolyU RNAs were prepared in synthesis buffer from a 20-nt-long polyU seed oligo with a 5′ cy5 fluorescent label, using the enzyme polynucleotide phosphorylase (Sigma-Aldrich, St. Louis, MO) isolated from Synechocystis sp. (25). Briefly, 500 pmol of cy5-labeled seed oligo was mixed with 40 mM UDP and 1 unit of PNPase, followed by a 4 h incubation at 37°C. These polymerization reactions produce a polydisperse mixture of fluorescent polyU RNAs ranging in length from 200 to 10,000 nts, with shorter lengths being represented in significantly higher copy number, as determined by fluorescence detection in denaturing 1% TAE agarose gel electrophoresis.
This polydisperse RNA mixture was run on a denaturing agarose gel alongside an ssRNA ladder and fractionated into discrete-length segments using the ssRNA ladder as reference. This fractionation process resulted in the following length fractions of polyU: 500–1500, 1500–2500, 2500–3500, 3500–5000, 5000–7000, and 7000–9000 nts (see Fig. S1). The amount of polyU purified was quantified by ultraviolet-visible absorbance measurements, with the absorbance ratio at 260/280 nm used as a measure of RNA purity (A260/A280 >2.0). These fractionated polyU samples were run on a denaturing agarose gel to demonstrate that single RNA bands were obtained.
In vitro transcription of normal-composition RNA molecules
Normal-composition RNAs were made by in vitro transcription of linearized DNA plasmids with T7 polymerase, rATP, rUTP, rCTP, and rGTP. This work utilized the following plasmids, produced and described in detail by Cadena-Nava et al. (7): pT7B1 for BMV RNA1 and pT7B1-0.5kbp for the 500 nt RNA. Transcription reactions were carried out for 4 h at 37°C, followed by digestion using DNase I (New England Biolabs, Ipswich, MA) for 50 min at 37°C (7). RNA was then purified from transcription mixes by washing through a 100 kDa Amicon filter with TE buffer. The concentration of RNA was determined by ultraviolet-visible absorbance, and the quality of the RNA produced was verified by native 1% agarose gel electrophoresis in TAE buffer.
Purification of CCMV CP
CCMV was purified from infected California cowpea plant (Vigna unguiculata cv black-eye pea) according to the procedure described by Bancroft et al. (1). Virus was dialyzed into disassembly buffer, and CP was isolated as described by Annamalai and Rao (26). An Applied Biosystems Voyager-DE-STR MALDI-TOF was used to assess CP quality, in particular that the N-terminus was not cleaved during purification.
In vitro packaging of RNA by CCMV CP
RNA and CP were mixed in buffer B at a 6:1 CP/RNA mass ratio, unless otherwise stated, to a final RNA concentration of 30 ng/μL, then dialyzed overnight into RAB at 4°C. The assembly mix was then dialyzed against VSB for 6 h at 4°C, at which point the virus-like particles (VLPs) were collected for analysis.
Electrophoretic mobility shift assay
The products of assembly reactions were run on 1% native agarose gels in VRB. Generally, 5 μg of virus was loaded per lane, and run alongside the same mass of wt CCMV, as well as a double-stranded DNA ladder control. Virions containing normal-composition RNAs were visualized using ethidium bromide, whereas polyU VLPs were visualized fluorescently using the cy5 fluorophore at their 5′ end. Virions were also stained using the protein stain Coomassie Blue. Gel bands were visualized on a Pharos FX Plus Molecular Imager, and the quantity of RNA in a particular lane was determined using ImageJ analysis.
Transmission electron microscopy
Negative-stain electron microscopy (EM) grids were prepared by depositing 6 μL of assembly reaction on glow-discharged copper grids (400 mesh), which were previously coated with Parlodion and carbon. After a 1 min deposition, grids were blotted and stained with 6 μL of 2% uranyl acetate for 1 min, then blotted dry. Grids were stored in a desiccator, and imaged on an FEI (Hillsboro, OR) Tecnai TF20 EM (Electron Imaging Center for Nanomachines, California Nanosystems Institute, Los Angeles, CA) at 50,000× magnification. Particles were sized manually from images taken with negative-stain transmission EM. At least 200 particles were analyzed per sample.
Competition experiments
The competition experiments follow the same basic protocol as the in vitro packaging experiments reported earlier (19), with a few changes. Equal masses of two RNAs were incubated in buffer B at a total RNA concentration of 30 ng/μL and a final volume of 200 μL, with only a 3:1 mass ratio of CP introduced into the system. In other words, there was the same total amount of RNA but half as much CP as in a normal assembly reaction. This RNA-protein mixture was then dialyzed into RAB overnight at 4°C, followed by another 6 h dialysis in VSB. Because of the lower (than 6:1) CP/RNA mass ratio involved, significantly less than all of the RNA was packaged.
RNase A digestion of VLPs
Virions were subjected to RNase digestion to confirm that they were fully closed shells. Briefly, VLPs are incubated with RNase A for 50 min at 4°C at a ratio of 2.5 ng RNase A (PureLink RNase A; Thermo Fisher Scientific, Waltham, MA) to 1 μg VLP. This mild RNase treatment has been shown to have no effect on the wt virus, but is capable of digesting at least 10 μg of unpackaged RNA. After RNase treatment, the mixture was incubated with 40 units of RNase inhibitor (Protector RNase Inhibitor, Sigma-Aldrich) for 15 min at 4°C, then washed through a 100 kDa Amicon filter at least three times with TE buffer to remove the RNase and the RNase inhibitor.
Results
PolyU is completely packaged at the magic ratio
Purified polyU fractions (see Fig. S1) were subjected to the two-step assembly protocol that has been shown to package normal-composition RNAs ranging in length from 140 to 12,000 nt into CCMV VLPs. In this procedure we use a superstoichiometric 6:1 mass ratio of CP to RNA, dubbed the “magic ratio,” to provide the minimum amount of CP required to package completely the RNA (7). It corresponds to an equality between the negative charges on the RNA phosphate backbone and the positive charges in the arginine-rich N-terminus of the CCMV CP.
The RNA and CP are first incubated in neutral-pH high-ionic-strength buffer (buffer B), in which the CP and RNA have little interaction because of the electrostatic screening. This mixture is then dialyzed into neutral-pH low-ionic-strength buffer (RAB), “turning on” RNA:CP interactions involving attraction between the RNA backbone and the CP N-terminus (27, 28). At this point in the assembly protocol, it has been shown for branched RNAs that all of the CP is bound to the RNA in solution. It has also been shown that although all of the protein is bound to the RNA, regular capsid structures do not form under these conditions (3). VLPs are formed after a final dialysis into low-pH low-ionic-strength buffer (VSB), which significantly enhances the lateral attractions between CP subunits.
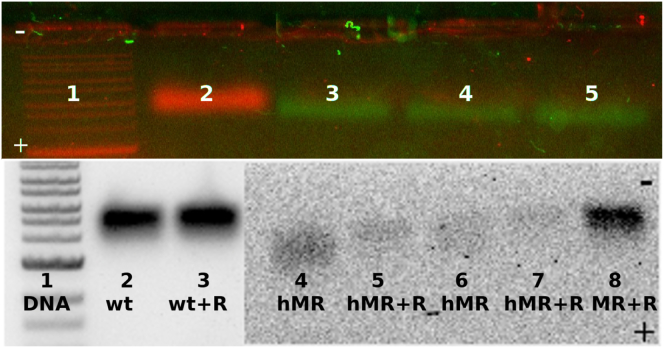
The assembly products were put on a native agarose gel to determine the extent of assembly; the DNA ladders and wild-type CCMV were visualized in red with ethidium bromide staining, while the cy5-labeled-polyU is visualized by its fluorescence, in green. Fig. 1 shows the resulting assembly products for B1 RNA and a variety of polyU lengths packaged at half the magic ratio (where explicitly stated) and at the magic ratio (when not explicitly specified). It is clear to see, for example, the effects of RNase treatment of assembly products shown in lanes 4 and 5 (unfractionated polyU), and in lanes 6 and 7 (B1 RNA and 2500- to 3500-nt polyU), that self-assembly at half the magic ratio yields a large fraction of the RNA being packaged not into RNase-resistant capsids, but rather into a broad range of faster-moving RNA/protein complexes that are susceptible to nuclease digestion. In contrast to this, see lane 8 (1500- to 2500-nt polyU), which shows that self-assembly at the magic ratio results in complete packaging of the RNA into fully closed capsids that are RNase resistant.
Figure 1.
Electrophoretic gel analysis of assembly products. (Top) Samples were as follows: lane 1, double-stranded DNA ladder; lane 2, wt CCMV; lane 3, 1500- to 2500-nt polyU VLPs; lane 4, 2500- to 3500-nt polyU VLPs; and lane 5, 3500- to 5000-nt polyU VLPs. The bands were visualized using two different fluorescent signals: cy5 attached to the polyU RNA (green) and ethidium bromide in structured RNAs (red). (Bottom) Samples were as follows: lane 1, double-stranded DNA ladder; lane 2, wt CCMV; lane 3, wt CCMV + RNase; lane 4, complexes of unfractionated polyU and CP at half the magic ratio (hMR); lane 5, unfractionated polyU complexes at half the magic ratio + RNase; lane 6, 3000-nt polyU and B1 RNA-protein complexes at half the magic ratio; lane 7, 3000-nt polyU and B1 RNA-protein complexes at half the magic ratio + RNase; and lane 8, 1500- to 2500-nt polyU VLPs at the magic ratio (MR) + RNase. Data from the ethidium bromide channel (left) and cy5 fluorophore channel (right) are placed next to each other to create this lower gel image. To see this Figure in color, go online.
PolyU RNA is packaged into 22-nm capsids
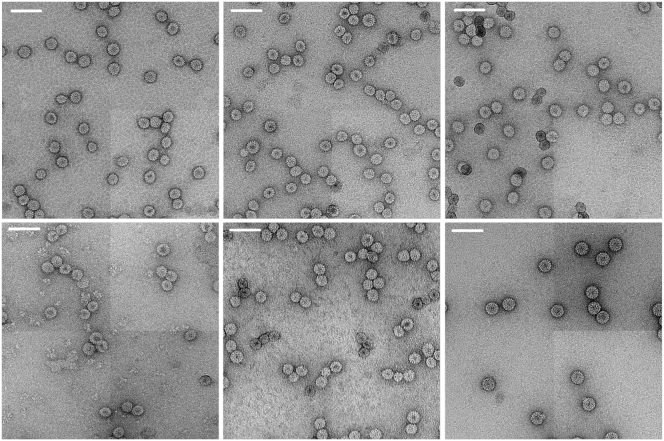
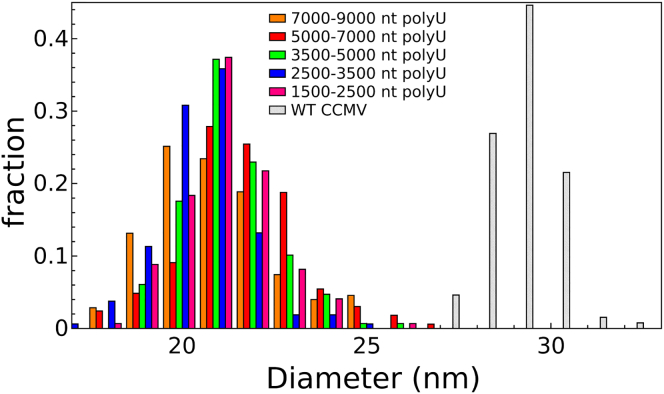
The assembly products were visualized before and after RNase A digestion using negative-stain EM (Fig. 2). Analysis of the micrographs allows for size determination of the particles, measured by taking the geometric mean of their diameters along the horizontal and vertical axes of the image. Surprisingly, as shown in Fig. 3, all polyU VLPs have a characteristic size of 22 nm, smaller than the 28-nm wt particles of CCMV. Similarly small particles have previously been found for VLPs containing branched RNAs <2300 nt long (7), whereas longer RNAs are packaged into 28-nm-diameter capsids.
Figure 2.
Negative-stain EM images of polyU packaged by CCMV CP and treated with RNase. (Top row, left to right) 1500–2500, 2500–3500, and 3500–5000 nt. (Bottom row, left to right) 5000–7000 nt, 7000–9000 nt, and wt CCMV. All lengths of polyU assembled (ranging between 1500 and 9000 nt) have resulted in the formation of 22 nm diameter particles, despite the fact that wild-type virions are 28 nm in diameter. Scale bar represents 50 nm.
Figure 3.
Histogram of particle sizes formed from assemblies using various lengths of polyU compared to particle sizes from wt CCMV virions purified from plants. All polyU VLPs were treated with RNase before imaging, although the plot for VLPs that have not been subject to RNase treatment is essentially identical. To see this figure in color, go online.
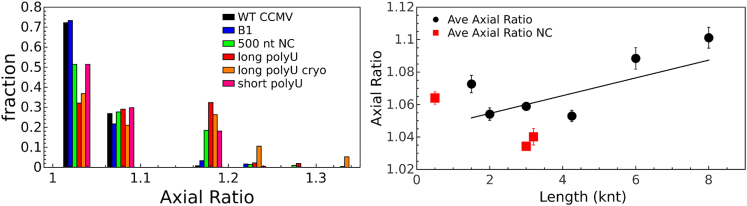
To quantify their asymmetry, the axial ratio of the VLPs as a function of length of polyU packaged was determined from negative-stain electron micrographs. Fig. 4 (left) shows the axial ratios of VLPs containing various lengths of polyU in comparison to particles containing normal-composition RNAs and (right) distributions of axial ratios for those same samples. The average axial ratios of the polyU VLPs are larger than the ratio determined for both wt virus and VLPs containing the viral RNA 1 (3000- and 3200-nt data points, respectively, shown in red). Interestingly, VLPs containing 500 nt normal-composition RNAs, which were previously reported to be T = 2 in size (29), have average axial ratios similar to those for VLPs formed around shorter polyU fractions. It is also clear that, for all samples, only certain ranges of axial ratios are populated, indicating that the assembly reactions result in specific structures. Specifically, axial ratios between 1 and 1.1 (these are considered to be spherical given the fact that in negative stain, even wt CCMV exhibits some asymmetry) and 1.15–1.25 have majority populations, whereas axial ratios around 1.3, 1.4, and 1.5 show minority populations. VLPs containing 3000 nt polyU imaged using cryo-electron microscopy, in which the particles are suspended in vitreous buffer, show that the T = 2-sized particles are indeed asymmetric in solution, and that this asymmetry does not occur due to adsorption onto the negative-stain EM grid. Additionally, the fact that discrete axial ratios are seen for all samples suggests that the particles are asymmetric (see Discussion), and that this asymmetry is not a result of particles being distorted by adhesion to a substrate.
Figure 4.
(Left) Distributions of axial ratios for VLPs containing polyUs of various length, for normal-composition (NC) RNA VLPs (500 nt normal composition and B1), and for wt CCMV. For clarity, average distributions for short-polyU VLPs (polyUs of <5000 nt) and long-polyU VLPs (polyUs of >5000 nt) are plotted, as the individual samples within each set exhibited similar distributions. Additionally, the axial ratios of long-polyU VLPs were inspected in cryo-EM to rule out the effect of drying the samples on EM grids. (Right) Axial ratios of polyU VLPs as a function of length, measured from negative-stain electron micrographs. The black data points represent the average axial ratio of polyU VLPs of a given length. Linear regression (black line) shows the increase in the axial ratio as a function of polyU length. The red data points represent the average axial ratio of several normal-composition RNA VLPs (29). The normal-composition RNA data point at 3000 nt corresponds to the wt virus. Error bars indicate the mean ± SD. To see this figure in color, go online.
Multiplets form for longer lengths of polyU
Although polyU is packaged into 22-nm sized particles regardless of its length, longer polyUs begin to be packaged into “multiplet” capsids in which RNA is shared between two or more capsids. Fig. 5 depicts the various types of capsids formed for polyU: a singlet has at least one RNA molecule in a single capsid (for RNAs <1000 nt, several RNAs are copackaged into the same capsid), a doublet contains RNA shared between two capsids, whereas triplets have RNA shared between three capsids. Similar multiplets have been found previously for branched RNAs that are significantly longer than the genome of CCMV (i.e., >4500 nt) (7).
Figure 5.
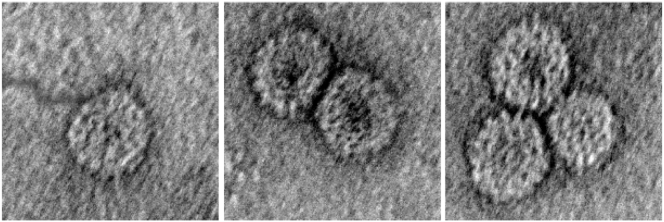
Representative negative-stain electron micrographs of singlet, doublet, and triplet capsids formed by in vitro self-assembly of a 7000- to 9000-nt polyU. For this length of polyU, predominantly doublets are formed.
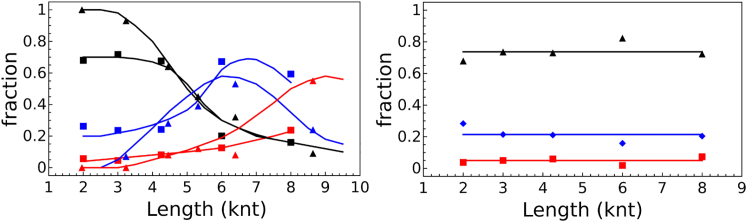
As Fig. 6 (left) shows, both normal-composition and polyU RNAs <4000 nt are packaged exclusively into singlet particles. RNAs with lengths between 4000 and 7000 nt are packaged predominantly into doublets; triplets begin to form at lengths >7000 nt. Despite multiplet formation, all the capsids containing polyU are still the smaller 22-nm diameter capsids, whereas the capsids containing normal-composition RNAs are 28 nm in diameter. It is seen further (Fig. 6, right) that after RNase digestion, the RNA shared between the capsids has been digested, resulting in only singlet VLPs. The right-hand plot also shows that the apparent fraction of doublets seen at the left for short polyU lengths is due simply to the density of particles on the EM grid. The normal-composition RNA plots refer to data from significantly higher dilution, and therefore, doublets are not seen before RNase digestion for RNAs <4000 nt, as particles are far less likely to be near one another under these conditions. PolyU VLPs imaged at higher dilutions show that multiplets disappear for shorter polyU assemblies, but that for longer polyU assemblies (>5000 nt), the number of multiplets stays constant upon dilution.
Figure 6.
(Left) Fractions of singlet (black), doublet (blue), and triplet (red) capsids as a function of increasing RNA length for polyU (squares) and for normal-composition RNAs (triangles). Note that the onsets of doublets (at ∼4000 nt) and triplets (at ∼7000 nt) occur at similar lengths for both polyU and normal-composition RNA assemblies. (Right) Effect of RNase on the fractions of singlet (black), doublet (blue), and triplet (red) capsids formed from polyUs of increasing length. RNase digestion converts multiplets into singlet capsids, demonstrating that the polyU RNA is threaded between the capsids. The 20% baseline of doublets is very probably due to particle crowding upon drying on the EM grid, as demonstrated by imaging solutions at higher dilution, resulting in a decrease in the number of doublets for VLPs containing short polyUs, whereas doublets survive dilution for VLPs containing longer polyUs. To see this figure in color, go online.
PolyU outcompetes viral RNA for CP
Competition experiments—self-assembly reactions in which equal masses of two different RNAs compete for CP that is sufficient to package only one of them—were carried out between viral RNA and polyU of the same length (2500–3500 nt). As these competition experiments are performed in vitro they necessarily lack the host-cell machinery that could influence RNA packaging. Previous competitions between structured RNAs have shown that RNA1 of BMV, a sibling virus of CCMV, outcompetes other RNAs (including RNA1 of CCMV) for viral CP in an in vitro assembly reaction (19). In addition, these experiments showed that the order in which components are mixed at neutral pH does not affect the outcome of the experiment. In other words, if RNA1 outcompetes RNA2, it will do so even if RNA2 is first equilibrated with CP at neutral pH before the addition of RNA1. This result suggests that the battle for CP is fought at neutral pH, and that whichever RNA binds CP more readily at neutral pH will be packaged upon acidification.
Three distinct competitions were carried out between linear and branched RNAs of equal length: the viral RNA and polyU were incubated at neutral pH, followed by the addition of CP (called Both First), viral RNA was first equilibrated with CP followed by the addition of polyU (B1 First), and polyU was first equilibrated with CP followed by addition of B1 (polyU First). It is important to note that all of these steps were taken before the pH was lowered; acidification results in complete capsid formation, which is an irreversible process.
The relative amounts of each RNA packaged were analyzed after RNase treatment using two methods. Negative-stain electron micrographs (Fig. 7) were taken of the mixture, and the numbers of 22- and 28-nm-diameter particles were compared. The relative numbers of each type of RNA packaged could then be determined under the assumption that polyU was packaged in the smaller capsids and B1 in the larger. The amounts of each RNA packaged were also determined by gel electrophoresis of the assembly mixtures alongside calibration curves of viral RNA VLPs and fluorescently labeled polyU VLPs. The molecules can be distinguished because the polyU is labeled with cy5 and the viral RNA is stained by ethidium bromide. The intensity attributed to each RNA molecule was then compared to its respective calibration curve, allowing a quantitative method of determining the amount of each type of VLP formed (data not shown).
Figure 7.
Negative-stain electron micrograph of the assembly products of the Both First competition reaction between 3000 nt polyU (red circles) and B1 RNA (blue circles) after RNase treatment. The smaller, 22-nm-diameter VLPs contain polyU, whereas the larger, 28-nm-diameter particles contain B1. Scale bar represents 50 nm. To see this figure in color, go online.
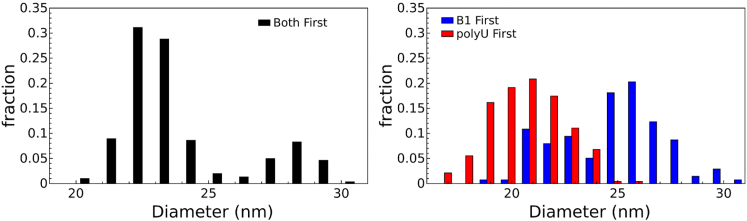
The results of the EM analysis are shown in Fig. 8. Surprisingly, polyU outcompetes the viral RNA in head-to-head competition for CP. Additionally, unlike the case of the competition between B1 and other branched RNAs, the order in which the polyU and B1 RNAs are incubated with CP has a significant effect on the outcome of the competition. Neither of these results was expected.
Figure 8.
The size distribution of the VLPs formed from competitive self-assembly reactions after RNase treatment. The peak centered around 22 nm corresponds to polyU VLPs, whereas the peak centered at 28 nm represents B1 VLPs. The separate colors represent distinct self-assembly reactions, in which the order of mixing of the assembly components was altered. In the Both First reaction (left, black,), the two molecules were mixed and CP was then added, whereas in the B1 First (right, blue) and polyU First (right, red) assemblies, the respective RNA was first mixed with CP and allowed to equilibrate before the other RNA was introduced. The figure shows that polyU wins the competition, and that the order of mixing is important. To see this figure in color, go online.
Table 1 shows the percentages of 22- and 28-nm-diameter capsids produced for each competition reaction, as analyzed by negative-stain EM or by fluorescence gel electrophoresis (in parentheses). The table shows good agreement between the two forms of analysis, and that polyU outcompetes B1 RNA for CP. Additionally, the data show that the order in which the RNAs sees the CP alters the results of the competition experiment: if polyU is equilibrated with CP first, it wins the competition outright; if B1 is equilibrated with CP first, B1 is preferentially packaged; and if both RNAs interact with CP simultaneously, polyU is preferentially packaged. This suggests that polyU does not bind CP reversibly, as the order of mixing changes the outcome of the competition. In contrast, for normal-composition RNAs, the result obtained for a competition between two RNAs was independent of the order of mixing.
Table 1.
EM Results and Agarose-Gel Fluorescence Intensity Analyses, in Parentheses, Showing the Percentages of polyU and B1 VLPs Formed during the Competition Reactions
| Particle Diameter (nm) | Both First | polyU First | B1 First |
|---|---|---|---|
| 22 | 78 (76) | 99 (91) | 35 (30) |
| 28 | 22 (24) | 1 (9) | 65 (70) |
Discussion
As mentioned in the Introduction, it is significant that although RNA molecules of the same nucleotide length and composition have comparable amounts of intramolecular self-complementarity (Watson-Crick basepairing), non-viral sequences result in significantly less compact 3D structures. This is because even though secondary structure formation leads in all cases to an effective branching of RNA, associated with the presence of many single-stranded loops, from which three or more duplexes emanate, viral sequences involve more “branch points” of this kind, as well as a concentration of them in the center of the overall secondary structure rather than on its periphery (where they do not contribute much to the “gathering-in”—compaction—of the molecule). In addition, recent theoretical work has shown that this branching and compactness of RNA results in a more efficient binding of CP (23).
Accordingly, in choosing to work with polyU—for which neither self-complementarity nor base stacking plays a role—we expected that it would be both less compact than viral RNA of the same length and less efficiently packaged. Indeed, DLS measurements (see Fig. S2) indicate that in assembly buffer at 25°C in the absence of CP, polyU molecules are ∼25% larger in average diameter than branched RNAs of the same length. And yet we find that polyU outcompetes viral RNA for binding CP and for being packaged into RNase-resistant virus-like particles. Further, independent of its length, even when it is three times longer than viral RNA, the VLPs formed are always of a size (22 nm) typical of T = 2, rather than the wt 28-nm T = 3 nucleocapsids formed in the case of viral (or normal-composition) RNAs.
This work therefore raises many more questions than it answers. Basically, there are two sets of issues. One involves the binding of RNA by CP with subsequent formation of a VLP, and its dependence on RNA secondary structure (or the lack thereof). The other involves the spontaneous selection of a preferred size of capsid, and its dependence on RNA secondary structure and on overall length/size. These two issues are of course interconnected, in particular because the preferred capsid size depends not just on the preferred curvature of protein-protein interactions, but also on the sequence-dependent local secondary structure of the RNA molecule to which the CP binds. One way to disentangle these effects is to keep the secondary structure fixed and change only the overall length/size of the RNA, and this can be done most simply by working with synthetic linear anionic polymers (e.g., poly(styrenesulfonate) (PSS)) or with RNA that has no secondary structure (e.g., polyU).
In the case of PSS, earlier work with in vitro self-assembly with CCMV CP has shown that the spontaneously formed VLPs change from T = 1-size to wild-type T = 3 nucleocapsids as the size of PSS is increased from hundreds of kDa to several MDa (8, 30). This result was understood in terms of there being three curvatures that are intrinsically preferred by interacting CP, corresponding to T = 1, T = 2, and T = 3 sizes, with the final result being determined by the size of the polyanionic “template” on which the protein binds.
A similar result was found for normal-composition RNA: when the length of RNA was increased from hundreds to thousands of nucleotides, the dominant VLP product switched from T = 2 to T = 3 sizes (7). However, as found in this work, in the case of polyU, the only VLP formed is of T = 2 size, even when the polyanion length is increased by a factor of 10, up to 10,000 nt. In both (PSS and normal-composition RNA) cases, the polyanion is shared by two or more T = 3 capsids when it becomes sufficiently long; with polyU, the increasingly long polyanion is shared by two or more T = 2 capsids.
However, the increase in size of the polyU could also be accommodated by a switch from a T = 2 size to T = 3. Why does this not happen? It is here that the absence of secondary structure in polyU likely plays a role. Structural determinations of T = 3 capsids show that the protein residues are essentially coplanar, with the 180° dihedral angle between the residues in the hexamer the result of double-stranded RNA that runs beneath it, a mechanism clearly not operative for the necessarily single-stranded polyU (5, 31).
In this context, it is important to mention the example of MS2 capsids, for which it has been shown that interaction of the CP with specific local components of RNA secondary structure—the “packaging sequences/signals”—are essential for the formation of appropriate size (T = 3) shells for efficient packaging of the viral genome (14). In fact, Krol et al. (32) showed that in the presence of replication-incompetent genomic RNAs, BMV CP forms T = 2 particles in vivo. However, these particles were polydisperse and only the most spherical particles were chosen for the structure determination. In reviewing several previous works that involved the formation of T = 2-sized particles made from CCMV or BMV CPs, it appears that all of these assemblies have resulted in heterogeneous particles (8, 29, 30, 32, 33, 34). A mutated CCMV CP lacking most of its cationic N-terminal domain has been shown to produce, in vitro, heterogenous, T = 2-sized particles, in addition to producing T = 1 and T = 3 particles. Additionally, Tresset et al. (35) found that polydisperse, T = 2-sized, VLPs containing PSS were less thermally stable than wt CCMV.
Small nonisometric closed capsids have been described in two theoretical papers treating polymorphism of empty capsids formed from CP alone (36, 37). They both find a family of closed-shell structures based on icosahedral caps of pentamers separated by belts of hexamers. Similar structures have been inferred for the in vivo virions of Alfalfa Mosaic Virus (AMV) (38) and Flock House Virus (FHV) (39). In both these cases, the virus particles contain RNAs shorter than the wild-type genome and have characteristic sizes that lie between T = 1 and T = 3 structures. The aspect ratios observed in electron micrographs for FHV are similar to those expected for one or two belts of hexamers. More explicitly, Dong et al. (39) have made estimates of the maximum number of nucleotides that could be packaged within different polymorphs. They assume that one nucleotide occupies 0.655 nm3 and calculate that a T = 1 FHV particle could accommodate 600 nt and that bacilliform particles with one belt and two belts of hexamers could accommodate 1740 and 2700 nt, respectively. From sucrose gradient sedimentation studies, however, Martin and Ames inferred a value of 0.57 cm3/g for the partial specific volume of polyU (40); this corresponds to a nucleotide volume of 0.31 nm3, about half that of the volume used in the computation in the Dong article, and suggests that single-belt and two-belt capsids could accommodate ∼3500 and 5400 nt of polyU. These values are consistent with the onset of multiplet structures, the point at which the capacity of a single capsid is exceeded. Additionally, axial ratios of polyU VLPs are discretized, suggesting “hexamer-belted” structures similar to those shown for AMV and FHV.
This polymorphism suggests that T = 2 sized particles have a high probability of forming kinetically-trapped, non-icosahedral, structures. In fact, it has been suggested that T = 2 particles form following a kinetically-trapped pathway en route to T = 3 (41, 42). The idea that kinetics plays a role in the assembly of T = 2 sized polyU VLPs is further supported by the results of the competition experiments between polyU and BMV RNA1, wherein the order of mixing the RNAs resulted in different amounts of each RNA encapsidated. This is unique to polyU RNA, in contradistinction to competitions between normal-composition RNAs where the order of mixing is irrelevant (19). We have not investigated whether CP prefers to bind polyU or B1 at neutral pH, but we expect that polyU is preferred, as previous work has shown that the RNA that binds CP more readily at neutral pH is the one that is subsequently packaged upon acidification (3, 19).
This work highlights the importance of RNA secondary structure in capsid formation, as it affects both the ability of an RNA to bind CP and the size and structure of the capsid formed. Although polyU both outcompetes viral RNA for CP and is packaged into RNase-resistant nucleocapsids, the capsids formed appear to involve kinetically trapped assembly pathways, resulting in structural heterogeneity. It is also clear that models of capsid assembly need to be refined to include the effect of secondary structure on capsid assembly, specifically addressing the impact of particular secondary structures on resultant capsid morphology.
Author Contributions
All authors were involved in designing the experiments. C.B., L.L.D., and K.N.L. performed the experiments and analyzed the data. C.B., W.M.G., and C.M.K. wrote the manuscript together.
Acknowledgments
We thank Dr. Feng Guo for providing the T7 RNA polymerase, Dr. A. L. N. Rao for providing the cDNA used to generate BMV RNA1, and Dr. Hong Zhou, Dr. Ivo Atanasov, and Dr. Yanxiang Cui for helpful discussions related to EM. EM images were acquired at the California NanoSystems Institute (CNSI) Electron Imaging Center for NanoMachines.
This research was supported by grant CHE1051507 from the National Science Foundation. K.N.L. was supported by the Amgen Scholars Program over the course of this work.
Editor: Karin Musier-Forsyth.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30688-4.
Supporting Material
References
- 1.Bancroft J.B., Hiebert E. Formation of an infectious nucleoprotein from protein and nucleic acid isolated from a small spherical virus. Virology. 1967;32:354–356. doi: 10.1016/0042-6822(67)90284-x. [DOI] [PubMed] [Google Scholar]
- 2.Fraenkel-Conrat H., Williams R.C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc. Natl. Acad. Sci. USA. 1955;41:690–698. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garmann R.F., Comas-Garcia M., Gelbart W.M. The assembly pathway of an icosahedral single-stranded RNA virus depends on the strength of inter-subunit attractions. J. Mol. Biol. 2014;426:1050–1060. doi: 10.1016/j.jmb.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caspar D.L., Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 5.Fox J.M., Wang G., Young M.J. Comparison of the native CCMV virion with in vitro assembled CCMV virions by cryoelectron microscopy and image reconstruction. Virology. 1998;244:212–218. doi: 10.1006/viro.1998.9107. [DOI] [PubMed] [Google Scholar]
- 6.Hiebert E., Bancroft J.B. Factors affecting the assembly of some spherical viruses. Virology. 1969;39:296–311. doi: 10.1016/0042-6822(69)90050-6. [DOI] [PubMed] [Google Scholar]
- 7.Cadena-Nava R.D., Comas-Garcia M., Gelbart W.M. Self-assembly of viral capsid protein and RNA molecules of different sizes: requirement for a specific high protein/RNA mass ratio. J. Virol. 2012;86:3318–3326. doi: 10.1128/JVI.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y., Zandi R., Gelbart W.M. Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size. Biophys. J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang L.T., Yoffe A.M., Ben-Shaul A. A sequential folding model predicts length-independent secondary structure properties of long ssRNA. J. Phys. Chem. B. 2011;115:3193–3199. doi: 10.1021/jp110680e. [DOI] [PubMed] [Google Scholar]
- 10.Fang L.T., Gelbart W.M., Ben-Shaul A. The size of RNA as an ideal branched polymer. J. Chem. Phys. 2011;135:155105. doi: 10.1063/1.3652763. [DOI] [PubMed] [Google Scholar]
- 11.Garmann R.F., Gopal A., Harvey S.C. Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy. RNA. 2015;21:877–886. doi: 10.1261/rna.047506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoffe A.M., Prinsen P., Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc. Natl. Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal A., Zhou Z.H., Gelbart W.M. Visualizing large RNA molecules in solution. RNA. 2012;18:284–299. doi: 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basnak G., Morton V.L., Stockley P.G. Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid. J. Mol. Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner D.R., Butler P.J. Essential features of the assembly origin of tobacco mosaic virus RNA as studied by directed mutagenesis. Nucleic Acids Res. 1986;14:9229–9242. doi: 10.1093/nar/14.23.9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolfsson Ó., Middleton S., Stockley P.G. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016;428(2 Pt B):431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stockley P.G., White S.J., Twarock R. Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage. 2016;6:e1157666. doi: 10.1080/21597081.2016.1157666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel N., Dykeman E.C., Stockley P.G. Revealing the density of encoded functions in a viral RNA. Proc. Natl. Acad. Sci. USA. 2015;112:2227–2232. doi: 10.1073/pnas.1420812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comas-Garcia M., Cadena-Nava R.D., Gelbart W.M. In vitro quantification of the relative packaging efficiencies of single-stranded RNA molecules by viral capsid protein. J. Virol. 2012;86:12271–12282. doi: 10.1128/JVI.01695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zlotnick A., Porterfield J.Z., Wang J.C. To build a virus on a nucleic acid substrate. Biophys. J. 2013;104:1595–1604. doi: 10.1016/j.bpj.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erdemci-Tandogan G., Wagner J., Zandi R. Effects of RNA branching on the electrostatic stabilization of viruses. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2016;94:022408. doi: 10.1103/PhysRevE.94.022408. [DOI] [PubMed] [Google Scholar]
- 22.Perlmutter J.D., Qiao C., Hagan M.F. Viral genome structures are optimal for capsid assembly. eLife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singaram S.W., Garmann R.F., Ben-Shaul A. Role of RNA branchedness in the competition for viral capsid proteins. J. Phys. Chem. B. 2015;119:13991–14002. doi: 10.1021/acs.jpcb.5b06445. [DOI] [PubMed] [Google Scholar]
- 24.Richards E.G., Flessel C.P., Fresco J.R. Polynucleotides. VI. Molecular properties and conformation of polyribouridylic acid. Biopolymers. 1963;1:431–446. [Google Scholar]
- 25.Vanzi F., Vladimirov S., Cooperman B.S. Protein synthesis by single ribosomes. RNA. 2003;9:1174–1179. doi: 10.1261/rna.5800303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Annamalai P., Rao A.L. Dispensability of 3′ tRNA-like sequence for packaging cowpea chlorotic mottle virus genomic RNAs. Virology. 2005;332:650–658. doi: 10.1016/j.virol.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Bayer T.S., Booth L.N., Ellington A.D. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA. 2005;11:1848–1857. doi: 10.1261/rna.2167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni P., Wang Z., Kao C.C. An examination of the electrostatic interactions between the N-terminal tail of the Brome Mosaic Virus coat protein and encapsidated RNAs. J. Mol. Biol. 2012;419:284–300. doi: 10.1016/j.jmb.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comas-Garcia M., Garmann R.F., Gelbart W.M. Characterization of viral capsid protein self-assembly around short single-stranded RNA. J. Phys. Chem. B. 2014;118:7510–7519. doi: 10.1021/jp503050z. [DOI] [PubMed] [Google Scholar]
- 30.Cadena-Nava R.D., Hu Y., Gelbart W.M. Exploiting fluorescent polymers to probe the self-assembly of virus-like particles. J. Phys. Chem. B. 2011;115:2386–2391. doi: 10.1021/jp1094118. [DOI] [PubMed] [Google Scholar]
- 31.Speir J.A., Munshi S., Johnson J.E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by x-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krol M.A., Olson N.H., Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tresset G., Tatou M., Porcar L. Weighing polyelectrolytes packaged in viruslike particles. Phys. Rev. Lett. 2014;113:128305. doi: 10.1103/PhysRevLett.113.128305. [DOI] [PubMed] [Google Scholar]
- 34.Tang J., Johnson J.M., Johnson J.E. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J. Struct. Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Tresset G., Chen J., Lansac Y. Two-dimensional phase transition of viral capsid gives insights into subunit interactions. Phys. Rev. Appl. 2017;7:014005. [Google Scholar]
- 36.Nguyen H.D., Reddy V.S., Brooks C.L., 3rd Invariant polymorphism in virus capsid assembly. J. Am. Chem. Soc. 2009;131:2606–2614. doi: 10.1021/ja807730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner J., Zandi R. The robust assembly of small symmetric nanoshells. Biophys. J. 2015;109:956–965. doi: 10.1016/j.bpj.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cusack S., Oostergetel G.T., Mellema J.E. Structure of the Top a-t component of alfalfa mosaic virus. A non-icosahedral virion. J. Mol. Biol. 1983;171:139–155. doi: 10.1016/s0022-2836(83)80350-7. [DOI] [PubMed] [Google Scholar]
- 39.Dong X.F., Natarajan P., Schneemann A. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 1998;72:6024–6033. doi: 10.1128/jvi.72.7.6024-6033.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin R.G., Ames B.N. The effect of polyamines and of poly U size on phenylalanine incorporation. Proc. Natl. Acad. Sci. USA. 1962;48:2171–2178. doi: 10.1073/pnas.48.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J.C., Chen C., Zlotnick A. Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano. 2015;9:8898–8906. doi: 10.1021/acsnano.5b02632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zlotnick A., Aldrich R., Young M.J. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.