Abstract
With the discovery of the prion protein (PrP), immunodiagnostic procedures were applied to diagnose Creutzfeldt–Jakob disease (CJD). Before development of the conformation-dependent immunoassay (CDI), all immunoassays for the disease-causing PrP isoform (PrPSc) used limited proteolysis to digest the precursor cellular PrP (PrPC). Because the CDI is the only immunoassay that measures both the protease-resistant and protease-sensitive forms of PrPSc, we used the CDI to diagnose human prion disease. The CDI gave a positive signal for PrPSc in all 10–24 brain regions (100%) examined from 28 CJD patients. A subset of 18 brain regions from 8 patients with sporadic CJD (sCJD) was examined by histology, immunohistochemistry (IHC), and the CDI. Three of the 18 regions (17%) were consistently positive by histology and 4 of 18 (22%) by IHC for the 8 sCJD patients. In contrast, the CDI was positive in all 18 regions (100%) for all 8 sCJD patients. In both gray and white matter, ≈90% of the total PrPSc was protease-sensitive and, thus, would have been degraded by procedures using proteases to eliminate PrPC. Our findings argue that the CDI should be used to establish or rule out the diagnosis of prion disease when a small number of samples is available as is the case with brain biopsy. Moreover, IHC should not be used as the standard against which all other immunodiagnostic techniques are compared because an immunoassay, such as the CDI, is substantially more sensitive.
Keywords: Creutzfeldt–Jakob disease, detection, endpoint titration, immunoassay, neurodegeneration
Human prion diseases include Creutzfeldt–Jakob disease (CJD), kuru, and Gerstmann–Sträussler–Scheinker disease. Sporadic CJD (sCJD) accounts for 85% of all cases of human prion disease, familial CJD (fCJD) for 10–15%, and infection from exogenous, frequently iatrogenic CJD (iCJD) prions, for <1% (1). Prions consist solely of a disease-causing prion protein (PrPSc) that is derived from the cellular isoform (PrPC) (2). During prion replication, PrPSc stimulates conversion of PrPC into nascent PrPSc.
Human prions from many cases of sCJD, fCJD, and iCJD were transmitted to apes and monkeys, but few titrations were performed, so there is little quantitative data on the levels of prions from these investigations (3). The development of mice expressing human prion protein (HuPrP) and chimeric mouse–human transgenes (MHu2M) (4–7) allowed us to measure the levels of prions in human brains as reported here. The incubation times of these mice were sufficiently abbreviated to allow endpoint titrations. Based on these endpoint titrations, we surmise that each of three cases of sCJD harbors a different strain of prion. We also used the titrations to calibrate PrPSc measurements that were determined by the conformation-dependent immunoassay (CDI). Full-length, protease-resistant PrPSc (rPrPSc) and previously unrecognized protease-sensitive forms of PrPSc (sPrPSc) can be detected by the CDI (8). Most PrPSc accumulating in the frontal cortex and white matter of sCJD cases was sPrPSc. Other immunoassays for PrPSc detect the N-terminally truncated protein PrP 27–30 derived from PrPSc; these include Western blotting, ELISA, and histoblotting. It is unclear what forms of PrPSc are detected by immunohistochemistry (IHC) after hydrolytic autoclaving in the presence of formic acid.
Because the CDI can readily detect PrPSc molecules comprising one ID50 unit, we examined the diagnostic sensitivity of the test by measuring PrPSc in many different brain regions. We performed these measurements on brains of 28 people who died of either sCJD, fCJD(E200K), or iCJD. Whereas the CDI registered a positive signal in every brain region examined in all of the cases, standard histopathology and IHC were much less effective in diagnosing CJD. Indeed, the poor performance of these histological techniques indicates that they should no longer be used to rule out prion disease in a brain biopsy from a single cortical site and must be applied to multiple cortical and subcortical brain samples at autopsy.
Materials and Methods
Preparation of Brain Homogenates. For biochemical analysis only, slices from 24 different anatomical areas of human brains weighing 250–350 mg were homogenized to a final 15% (wt/vol) in 4% (wt/vol) Sarkosyl in PBS, pH 7.4, by three 75-s cycles in a reciprocal homogenizer MiniBeadBeater-8 (BioSpec Products, Bartlesville, OH) as described in refs. 8–10. The resulting homogenate was diluted to a final 5% (wt/vol) by using PBS containing 4% (wt/vol) Sarkosyl. The diluted samples were either treated with a proteinase inhibitor mixture for measurements of PrPSc or digested with 2.5 or 10 μg/ml proteinase K (PK) for 60 min at 37°C on the shaker. After a clarification spin at 500 × g for 5 min at room temperature in a drum rotor (Jouan, Winchester, VA), the samples were mixed with stock solution containing 10% sodium phosphotungstate (NaPTA) and 85 mM MgCl2, pH 7.4, to obtain a final concentration of 0.32% NaPTA. After a 1-h incubation at 37°C on a rocking platform, the samples were centrifuged at 14,000 × g in a Jouan MR23i centrifuge for 30 min at room temperature. The resulting pellets were resuspended in H2O containing protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, and 2 μg/ml leupeptin) and assayed by the CDI.
Sandwich CDI for PrPSc. For capture of HuPrP, the mAb MAR1 was used (11), and detection was accomplished with mAb 3F4 (12) labeled with Eu-chelate of N-(p-isothiocyanatobenzyl)-diethylenetriamine-N1,N2,N3,N3-tetraacetic acid at pH 9.6 for 16 h at room temperature according to the manufacturer's protocols (Wallac, Turku, Finland), as described in ref. 8. The principle, development, calibration, and calculation of PrPSc concentration from CDI data have been described in refs. 8–10. The results were expressed as the difference in Ab-binding between native and denatured samples [(D - N)] of the time-resolved fluorescence of aliquots measured in cpm. In some cases, the concentration of PrPSc is directly proportional to (D - N) value and was calculated from the formula described in refs. 8–10.
Histopathologic Procedures. Autopsies were performed shortly after death, and brain tissue was either immediately frozen or immersion-fixed in 10% buffered formalin for embedding in paraffin. We stained 8-μm-thick sections with hematoxylin and eosin (H&E) to evaluate vacuolation. Vacuolation scores, visual estimates of the percentage of gray matter area in a slide occupied by vacuoles, were determined by a single observer (S.J.D.). Reactive astrocytic gliosis was evaluated by glial fibrillary acidic protein immunostaining by using a rabbit antiserum (DAKO). Hydrolytic autoclaving pretreatment of the formalin-fixed tissue sections was used to detect PrPSc, as described in ref. 13. The Bielschowsky silver stain and IHC for α-synuclein, tau, and ubiquitin were used as needed to test for Alzheimer's disease, synucleinopathies, tauopathies, and other neurodegenerative processes.
Additional methods describing the diagnosis of prion disease, human samples acquisition, construction of transgenic (Tg) mice, endpoint titrations in Tg mice, sample tracking and data processing, and sandwich CDI for PrPSc are described in Supporting Text, which is published as supporting information on the PNAS web site.
Results
Patient Groups, Clinical Diagnosis, and Codon 129 Polymorphism. Human brain specimens were obtained from 46 patients who underwent pathologic evaluation. Of the 28 cases in the prion disease group, 24 were diagnosed with sCJD, three with fCJD(E200K), and one with iCJD (see Table 3, which is published as supporting information on the PNAS web site). The PrP polymorphism at codon 129 [methionine (M) or valine (V)] was determined by DNA sequencing for all 28 patients in the prion disease group. As shown for sCJD, 13 patients were MM, 7 were MV, and 4 were VV (Table 3).
Analytical Sensitivity of the CDI for Detection of Human Prions. We used Eu-labeled 3F4 mAb (12) for detection and MAR1 mAb (11) to capture HuPrPSc in a sandwich CDI format (10). For a normal human brain homogenate that contains only PrPC, the (D - N) was ≤1,789 cpm.
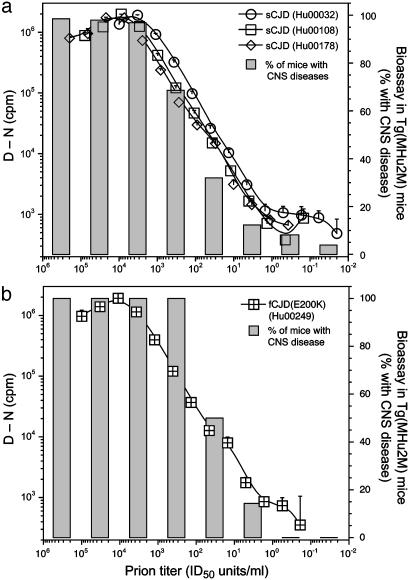
Brain homogenates from three cases of sCJD and one case of fCJD were serially diluted in 3-fold increments into normal human plasma and assayed by the CDI (Fig. 1). The sensitivity of the CDI in detecting both sCJD and fCJD prions was equal to or greater than that for the detection of human prions by bioassay in Tg(MHu2M)5378/Prnp0/0 mice (Fig. 1; see also Fig. 5 and Table 4, which are published as supporting information on the PNAS web site). Within the linear range, there was a good correlation between PrPSc concentration measured by CDI and prion titer measured by bioassay.
Fig. 1.
Correlation between CDI and bioassay in Tg mice. Sandwich CDI protocol for the detection of PrPSc was compared to titration bioassays in Tg(MHu2M)5378/Prnp0/0 mice for three sCJD brains (a) and one fCJD brain (b). Samples were precipitated with PTA and digested with 2.5 μg/ml PK for 1 h at 37°C. The MAR1 mAb was used (11) for capture, and Eu-labeled 3F4 mAb was used for detection. The (D - N) values measured in cpm are directly proportional to the concentration of PrPSc (8, 10). Data points and bars represent the average ± SD obtained from three to four independent measurements. The cutoff (D - N) value of 1,789 cpm for this sandwich CDI protocol was calculated by [mean + 3(SD)] and determined from 100 brain samples obtained from patients who died from nonneurologic disease (n = 6), Alzheimer's disease (n = 7), and other neurologic diseases (n = 5).
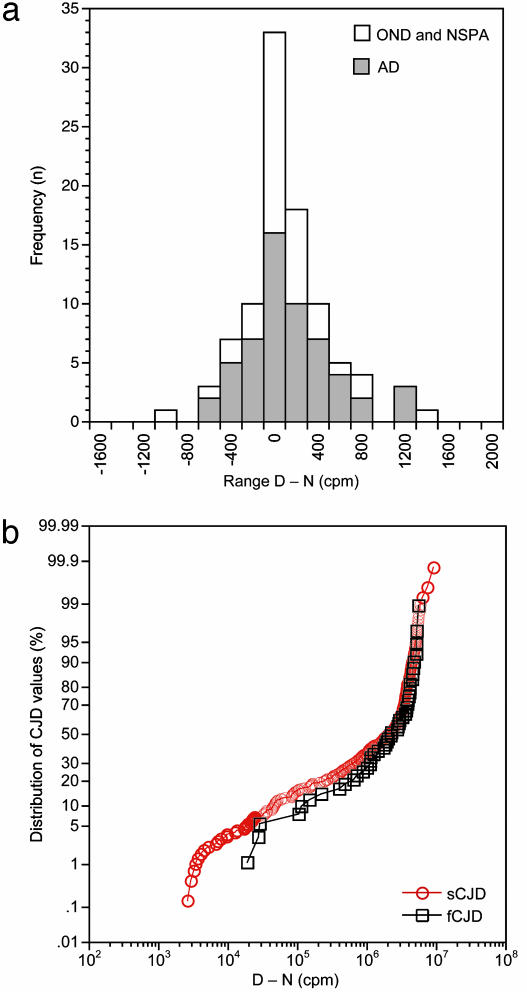
Diagnostic Sensitivity of the CDI for Detection of Human Prions. To evaluate the diagnostic sensitivity of the CDI for the detection of different CJD prions, we performed multiple blind tests on brain tissue from 24 sCJD cases, 3 fCJD cases, 1 iCJD case, and 18 controls (Table 3). Data from the controls exhibited a Gaussian distribution, with the median (D - N) value oscillating around zero, as expected for samples containing only residual PrPC after phosphotungstate (PTA) precipitation (Fig. 2 and Table 5, which is published as supporting information on the PNAS web site). In contrast, median (D - N) values for sCJD and fCJD cases are ≈106, which is six orders of magnitude higher than the median of the control group (Fig. 2). The dynamic range of the CJD data approaches 104, and all values are above threshold. After performing 493 tests, the CDI identified all CJD cases with 100% accuracy, and no false positives occurred in the control group (Table 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Statistical distribution of the CDI data for the detection of PrPSc. Multiple samples from control (n = 18) (a) and CJD brains (n = 27) (b) were tested. The control group included cases of Alzheimer's disease (AD), patients who died from other neurological diseases (OND), and cases with no specific brain pathology at autopsy (NSPA). Each brain sample was tested two to four times. Results are expressed as (D - N) in cpm.
Codon 129 and the Anatomical Distribution of PrPSc in sCJD Brains. By using the CDI, we determined the levels of PrPSc in 24 brain regions for some patients and as few as 10 for others because frozen samples for all regions were not available (Fig. 6, which is published as supporting information on the PNAS web site). Generally, the highest concentrations of HuPrPSc were detected in the primary visual cortex, thalamus, and cerebellum. All other areas of the cortex and subcortical gray matter displayed substantial accumulation of PrPSc. From these quantitative studies, we conclude that, although the codon 129 genotype may influence the levels of PrPSc deposition, the differences among the three codon 129 genotypes (MM, MV, and VV) are less prominent than expected from qualitative lesion profiles and IHC.
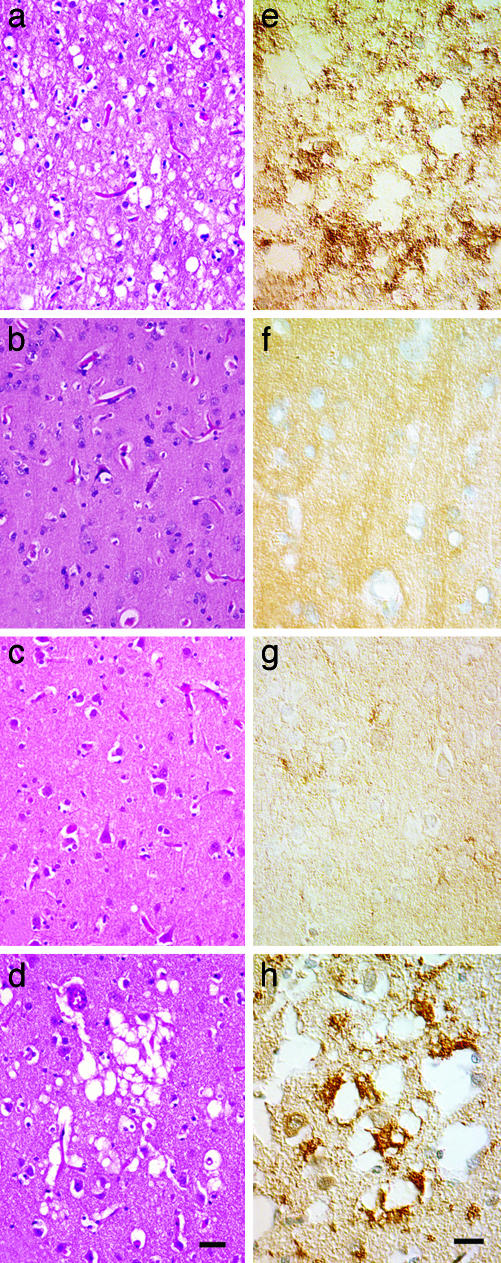
Biopsies of Human Brains. A 52-year-old female experienced memory problems, difficulty with concentration, anxiety, confabulation, and visual hallucinations. A left parietal lobe biopsy was performed ≈3 months after symptoms began to rule out CJD. The biopsy contained a sample of cortex extending from the pial surface to the underlying white matter. No characteristic vacuolation or PrPSc deposits were identified in sections of the biopsy (Fig. 3 b and f).
Fig. 3.
Routine histology and PrPSc IHC on brain sections of two CJD patients. (a and e) Patient with CJD (MV2 subtype) in which vacuolation and PrPSc deposits were identified in all brain regions sampled, and kuru-type plaques were identified in the cerebellar cortex. (b and f and c and g) Patient with CJD (MM1 subtype) in which neither vacuolation nor PrPSc deposits were found in a brain biopsy (b and f) or at routine autopsy (c and g). (d and h) By using high-intensity MRI signals as a guide, a second set of brain sections were prepared from this patient; vacuolation of the neuropil as well as coarse PrPSc deposits were found. (a–d) H&E stain. (Scale bar, 50 μm.) (e–h) IHC for PrPSc. (Scale bar, 30 μm.)
The patient died 3.5 months after the biopsy. Routine sampling of multiple brain regions again failed to reveal sufficient degrees of gray matter vacuolation (Fig. 3c) to make the diagnosis of human prion disease. IHC for PrPSc show rare punctate deposits in the neocortical regions sampled (Fig. 3g). At the time, one of us (S.J.D.) believed such infrequent deposits might be an artifact, and, therefore, the inconclusive IHC and routine histology prevented a definitive diagnosis of CJD. By using the MRI scan performed a week before death as a guide, new samples were obtained from cortical regions with high signal intensities. In these regions, clusters of vacuoles measuring 40–60 μm in diameter were found in cortical layers 2 and 3 (Fig. 3d) and coarse PrPSc deposits were associated with the clusters of vacuoles (Fig. 3h). Small amounts of PrPSc deposits were also found away from the vacuoles. The clusters of vacuolation and deposits of PrPSc tended to be small and highly focal, occupying <1% of the cortical cross-sectional area.
Two years later, when unfixed frozen samples from the right hemisphere corresponding to neuropathologically positive and negative contralateral brain regions were analyzed by the CDI, all 13 of the regions examined were strongly positive for PrPSc. The data from histology with H&E staining, IHC, and the CDI are summarized in Table 7, which is published as supporting information on the PNAS web site. (D - N) values varied from almost 300,000 cpm in the medulla to >3,000,000 cpm in the thalamus, globus pallidus, frontal cortex, occipital cortex, as well as the parietal cortex, the region where the initial biopsy was taken on the contralateral side.
Diagnostic Sensitivity of the CDI and IHC. When brain sections from 10 CJD cases [8 sCJD and 2 fCJD(E200K)] were analyzed by H&E staining, IHC with α-PrP mAbs, and the CDI (Tables 1 and 2), we discovered that, like the biopsied patient reported above, the CDI was vastly superior to both histologic examination for vacuolation of the neuropil and IHC for PrP immunostaining. The microscopic studies were performed on formalin-fixed, paraffin-embedded tissue sections with knowledge that the patients were clinically diagnosed with CJD, but without knowledge of the CDI results.
Table 1. Comparison of the diagnostic sensitivity of the vacuolation profile, IHC, and sandwich CDI in the detection of PrPSc in different anatomical areas of sCJD brains.
| Vacuoles
|
IHC
|
CDI
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive*
|
Score†
|
Positive*
|
Score
|
Positive*
|
D–N, cpm × 10-5
|
|||||||
| Anatomical Area | n+ | % | Mean | S.D. | n+ | % | Mean | S.D. | n+ | % | Mean | S.D. |
| Frontal cortex | 6 | 75 | 32.5 | 34.8 | 8 | 100 | 1.3 | 0.5 | 8 | 100 | 30.1 | 5.0 |
| Entorhinal area | 8 | 100 | 54.5 | 40.3 | 6 | 75 | 1.4 | 0.9 | 8 | 100 | 24.7 | 4.7 |
| Parietal cortex | 7 | 87 | 36.3 | 27.7 | 8 | 100 | 1.6 | 0.7 | 8 | 100 | 30.1 | 5.1 |
| Temporal cortex | 8 | 100 | 32.3 | 29.1 | 8 | 100 | 1.5 | 0.8 | 8 | 100 | 21.6 | 5.4 |
| Hippocampus | 2 | 25 | 4.8 | 10.6 | 3 | 37 | 0.9 | 1.5 | 8 | 100 | 23.7 | 6.8 |
| Amygdala | 4 | 50 | 8.8 | 11.3 | 5 | 63 | 0.9 | 0.8 | 8 | 100 | 19.7 | 5.3 |
| Occipital cortex | 7 | 87 | 27.5 | 27.3 | 4 | 50 | 1.0 | 1.2 | 8 | 100 | 32.1 | 6.4 |
| Sulcus cingulate | 7 | 87 | 40.0 | 33.5 | 7 | 87 | 1.4 | 0.9 | 8 | 100 | 24.1 | 5.8 |
| Gyrus dentate | 1 | 13 | 1.3 | 3.5 | 1 | 13 | 0.3 | 0.4 | 8 | 100 | 5.3 | 3.2 |
| Insula | 6 | 75 | 34.4 | 40.8 | 8 | 100 | 1.3 | 0.5 | 8 | 100 | 25.1 | 4.7 |
| Thalamus | 7 | 87 | 22.6 | 22.3 | 5 | 63 | 0.9 | 0.8 | 8 | 100 | 23.1 | 6.8 |
| Nucleus caudate | 8 | 100 | 38.8 | 23.6 | 5 | 63 | 0.9 | 1.0 | 8 | 100 | 23.8 | 5.1 |
| Globus pallidus | 3 | 37 | 7.3 | 13.9 | 4 | 50 | 0.8 | 1.0 | 8 | 100 | 21.9 | 6.9 |
| Putamen | 7 | 87 | 36.0 | 29.5 | 6 | 75 | 0.9 | 0.6 | 8 | 100 | 20.1 | 5.8 |
| Midbrain | 2 | 25 | 3.2 | 5.9 | 1 | 13 | 0.3 | 0.7 | 8 | 100 | 14.6 | 4.7 |
| Pons | 0 | 0 | 0.0 | 0.0 | 1 | 13 | 0.1 | 0.4 | 8 | 100 | 7.9 | 2.9 |
| Medulla | 1 | 13 | 0.6 | 1.8 | 1 | 13 | 0.1 | 0.4 | 8 | 100 | 7.1 | 2.1 |
| Cerebellum | 4 | 50 | 8.8 | 15.3 | 5 | 63 | 1.3 | 1.2 | 8 | 100 | 17.7 | 4.5 |
Adjacent sections from eight sCJD patients were analyzed in parallel by all three methods.
n+, number of samples with positive diagnosis of eight samples examined. Percentages are based on eight samples
Vacuolation scores are the percentage area of gray matter occupied by vacuoles, determined by the visual estimation of a single rater (S.J.D.)
Table 2. Comparison of the diagnostic sensitivity of the vacuolation profile, IHC, and sandwich CDI for the detection of PrPSc in different anatomical areas of fCJD brains.
| Vacuoles
|
IHC
|
CDI
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive*
|
Score†
|
Positive*
|
Score
|
Positive*
|
D–N, cpm × 10-5
|
|||||||
| Anatomical Area | n+ | % | Mean | S.D. | n+ | % | Mean | S.D. | n+ | % | Mean | S.D. |
| Frontal cortex | 2 | 100 | 20.0 | 14.1 | 2 | 100 | 1.0 | 0.7 | 2 | 100 | 37.0 | 8.7 |
| Entorhinal area | 1 | 50 | 2.5 | 3.5 | 2 | 100 | 1.5 | 0.7 | 2 | 100 | 16.0 | 4.5 |
| Parietal cortex | 2 | 100 | 10.0 | 0.0 | 2 | 100 | 1.5 | 0.7 | 2 | 100 | 35.0 | 17.0 |
| Temporal cortex | 2 | 100 | 10.0 | 2.8 | 2 | 100 | 2.0 | 0.0 | 2 | 100 | 16.0 | 6.3 |
| Hippocampus | 0 | 0 | 0.0 | 0.0 | 2 | 100 | 1.5 | 0.7 | 2 | 100 | 41.0 | 11.0 |
| Amygdala | 0 | 0 | 0.0 | 0.0 | 1 | 50 | 1.0 | 1.4 | 2 | 100 | 25.0 | 23.0 |
| Occipital cortex | 2 | 100 | 13.0 | 9.9 | 2 | 100 | 2.5 | 0.7 | 2 | 100 | 27.0 | 14.0 |
| Sulcus cingulate | 2 | 100 | 25.0 | 7.1 | 2 | 100 | 1.0 | 0.0 | 2 | 100 | 28.0 | 14.0 |
| Gyrus dentate | 0 | 0 | 0.0 | 0.0 | 0 | 0 | 0.0 | 0.0 | 2 | 100 | 5.0 | 3.8 |
| Insula | 0 | 0 | 0.0 | 0.0 | 1 | 50 | 0.5 | 0.7 | 2 | 100 | 35.0 | 7.2 |
| Thalamus | 2 | 100 | 25.0 | 7.1 | 2 | 100 | 2.5 | 0.7 | 2 | 100 | 23.0 | 16.0 |
| Nucleus caudate | 2 | 100 | 60.0 | 28.3 | 1 | 50 | 1.0 | 0.0 | 2 | 100 | 48.0 | 7.8 |
| Globus pallidus | 2 | 100 | 7.5 | 10.6 | 2 | 100 | 1.0 | 0.7 | 2 | 100 | 19.0 | 0.4 |
| Putamen | 2 | 100 | 35.0 | 7.1 | 1 | 50 | 0.5 | 0.7 | 2 | 100 | 37.0 | 1.6 |
| Midbrain | 1 | 50 | 3.5 | 5.0 | 1 | 50 | 0.5 | 0.7 | 2 | 100 | 37.0 | 15.0 |
| Pons | 0 | 0 | 0.0 | 0.0 | 1 | 50 | 0.5 | 0.7 | 2 | 100 | 3.2 | 1.7 |
| Medulla | 1 | 50 | 4.0 | 7.5 | 1 | 50 | 0.5 | 0.7 | 2 | 100 | 8.8 | 2.4 |
| Cerebellum | 1 | 50 | 5.0 | 7.1 | 1 | 50 | 1.5 | 2.2 | 2 | 100 | 25.0 | 21.0 |
Adjacent sections from two fCJD(E200K) patients were analyzed in parallel by all three methods.
n+, number of samples with positive diagnosis of two samples examined. Percentages are based on two samples
Vacuolation scores are the percentage area of gray matter occupied by vacuoles, determined by the visual estimation of a single rater (S.J.D.)
From 18 brain regions of the 8 sCJD cases, we compared the results of histology, IHC, and CDI. Only the CDI gave consistently positive (100%) PrPSc signals for all 18 brain regions in all 8 patients. By routine histology, only 3 of 18 regions were found positive in the 8 sCJD cases (Table 1); this represents a diagnostic sensitivity of 17%. The entorhinal cortex, temporal lobe cortex, and the caudate nucleus were 100% positive from the brains of all eight patients analyzed by histology. The remaining 15 regions varied from 0% to 87% positive for the 8 sCJD brains examined.
The results with IHC were similar to those obtained by histological examination, which was unexpected because IHC is generally thought to be more sensitive than histology. By IHC, only 4 of 18 regions were found positive in all 8 sCJD patients (Table 1); these results represent a diagnostic sensitivity of 22%. The frontal cortex, parietal cortex, temporal lobe cortex, and the insula were 100% positive from the brains of all 8 patients. The remaining 14 regions varied from 13% to 87% positive for the 8 sCJD brains examined. Comparing histology, IHC, and the CDI, only the temporal lobe region gave consistently positive results (100%) for the 8 sCJD patients (Table 1).
Next, we compared histology, IHC, and CDI analysis on the brains of two patients who died of fCJD(E200K). Of the 18 regions examined by histology or IHC, 9 were found positive in the 2 patients (Table 2); these results represent a diagnostic sensitivity of 50%. In contrast, the CDI was positive in all 18 regions for both patients.
Asymmetric Lesions and PrPSc Deposits in the Brain. To address the possibility of sampling bias, we examined the brain of a 79-year-old female in whom rapidly progressive motor and language decline were associated with myoclonic jerks and an electroencephalogram with periodic spikes characteristic of CJD. The patient died 14 days after a diffusion-weighted MRI scan showed high intensity signals in the left cerebral cortex but few or no such signals in the right.
Histologic analysis showed moderately severe vacuolation scores in multiple cortical regions of the left cerebral hemisphere with little or none in analogous regions on the right (Table 8, which is published as supporting information on the PNAS web site). Unbiased stereological counts of neurons in different cerebral cortical layers showed marked loss from all layers of the left frontal cortex (Brodmann areas 44 and 45), but no loss was found on the right (data not shown). Morphometric quantification of IHC for glial fibrillary acidic protein showed marked astrocytic gliosis in the left cerebral cortex and only focal, mild gliosis on the right. We found more PrPSc deposits in the left cortex than in the right but these were not quantified (Table 8). PrPSc was found in all locations of analogous right and left cortical samples by the CDI; levels of PrPSc in the right cortex were 5–50% lower than those from the left. Much lower levels of PrPSc in the right cortex might have been expected based the minimal microscopic changes, reflecting again the incongruity between microscopic and CDI analyses.
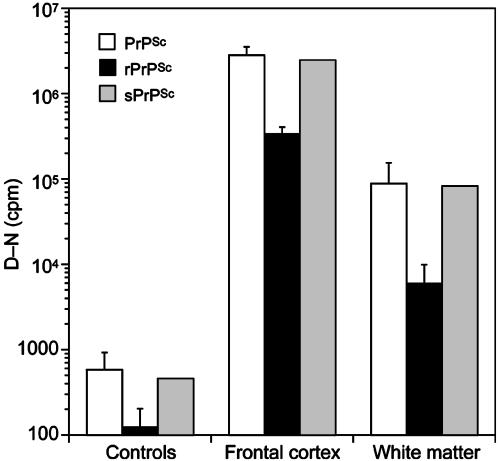
Levels of sPrPSc and rPrPSc. To determine the relationship between sPrPSc and rPrPSc in the brains of sCJD patients, samples were either PTA-precipitated to measure the concentration of total PrPSc or PK-digested then PTA-precipitated to obtain the concentration of rPrPSc, as described in refs. 8 and 9. These treated samples were then subjected to analysis by the CDI. Surprisingly, >80% of total PrPSc was susceptible to proteolytic degradation (Fig. 4). Despite a 20-fold lower concentration of PrPSc in white matter, the ratio between sPrPSc and rPrPSc remained constant. In conclusion, sPrPSc constitutes a major fraction of total PrPSc in the frontal cortex and white matter of the sCJD-infected brains.
Fig. 4.
Most PrPSc in the frontal cortex and white matter of sCJD brains is protease-sensitive [gray bars, calculated from measurements of total PrPSc (white bars) and rPrPSc (black bars)]. Before measurement by the CDI, undigested samples were PTA-precipitated to measure total PrPSc or digested with 50 μg/ml PK at 37°C for 1 h, followed by PTA precipitation to determine rPrPSc (8, 9). The graph shows the means ± SEM obtained from duplicate measurements of samples from the frontal cortex (n = 19) and white matter (n = 12) of sCJD-infected brains.
It is noteworthy that IHC of formalin-fixed, paraffin-embedded tissue sections occasionally showed PrPSc deposits in white matter; however, histoblot analysis, which is our most sensitive and specific tissue-based method, routinely failed to identify rPrPSc in white matter (data not shown). In contrast, the CDI found PrPSc in white matter in all cases of sCJD and fCJD (Fig. 4 and Tables 1 and 2).
Discussion
The clinical diagnosis of human prion disease is often difficult until the patient shows profound signs of neurologic dysfunction. It is widely accepted that the clinical diagnosis must be provisional until a tissue diagnosis either confirms or rules out the clinical assessment. Before the availability of Abs to PrP, a tissue diagnosis was generally made by histologic evaluation of neuropil vacuolation. IHC with anti-glial-fibrillary-acidic-protein Abs in combination with H&E staining preceded the use of anti-PrP Ab staining.
Recently, the role of IHC in the diagnosis of scrapie in the brains of eight clinically affected goats inoculated with the SSBP1 prion isolate has been challenged (14). Thalamic samples taken from seven of eight goats with scrapie were positive for PrPSc by Western blotting but negative by IHC. The eighth goat was negative by Western blotting and IHC. Consistent with these findings in goats are the data reported here, in which IHC of formalin-fixed, paraffin-embedded human brain samples was substantially less sensitive than the CDI.
The CDI was developed to quantify PrPSc in tissue samples from mammals producing prions. Concerned that limited PK digestion was hydrolyzing some or even most of the PrPSc, we developed a CDI that does not require PK digestion. The CDI revealed that as much as 90% of PrPSc is sPrPSc; thus, it was being destroyed during limited proteolytic digestion used to hydrolyze PrPC. sPrPSc comprises ≈80% of PrPSc in the frontal lobe and in the white matter (Fig. 4).
The CDI detected HuPrPSc with a sensitivity comparable to the bioassay for prion infectivity in Tg(MHu2M) mice (Fig. 1). The high sensitivity achieved by the CDI is due to several factors (8, 10, 11, 15). First, both sPrPSc and rPrPSc conformers are specifically precipitated by PTA (Table 5) (8, 9). PTA has also been used to increase the sensitivity of Western blots enabling the detection of rPrPSc in human muscle and other peripheral tissues (16, 17). Second, a sandwich protocol was used with the high-affinity MAR1 mAb (11) to capture HuPrPSc and Eu-labeled 3F4 mAb to detect HuPrPSc (12). Third, the CDI detects PrPSc by Ab-binding to native and denatured forms of the protein and, therefore, does not depend on proteolytic degradation of PrPC. We chose not to perform Western blots on most of the samples used in this study because such immunoblots require denaturation of the sample, which eliminates measurement of the native signal corresponding to PrPC (Table 5). Moreover, a comparison between the CDI and Western blotting on brain samples from sCJD and variant CJD patients showed that the CDI was 50- to 100-fold more sensitive (15). Additionally, Western blots combined with densitometry are linear over a 10- to 100-fold range of concentrations, whereas the CDI is linear over a >104-fold range. The CDI has been automated, which not only improves accuracy and reproducibility (10) but also allows numerous samples to be analyzed, as reported here. Western blots are difficult to automate and are labor intensive.
Our studies show that only the CDI detected PrPSc in all regions examined in 24 sCJD and 3 fCJD(E200K) brains (Figs. 2 and 6). Comparative analyses demonstrated that the CDI was vastly superior to histology and IHC. When 18 regions of 8 sCJD and 2 fCJD(E200K) brains were compared, we discovered that histology and IHC were unreliable diagnostic tools except for samples from a few brain regions. In contrast, the CDI was a superb diagnostic procedure because it detected PrPSc in all 18 regions in 8 of 8 sCJD and 2 of 2 fCJD(E200K) cases (Tables 1 and 2).
Histologic changes in prion disease have been shown to follow the accumulation of prions as measured by bioassay of infectivity and by PrPSc accumulation (18–22). Because low levels of PrPSc are not associated with neuropathologic changes, some discrepancy between vacuolation and PrPSc was expected. In contrast to histology, IHC measures PrP immunostaining after autoclaving tissue sections exposed to formic acid. Because IHC measures PrP, we expected the sensitivity of this procedure might be similar to the CDI, but that proved not to be the case. Whether exposure of formic acid-treated tissue sections to elevated temperature destroys not only PrPC but also sPrPSc and only denatures rPrPSc remains to be determined. Such a scenario could account for the lower sensitivity of IHC compared with CDI or bioassay (Tables 1 and 2).
Studies of the white matter in CJD brains were particularly informative with respect to the sensitivity of the CDI, where PrPSc levels were low but readily detectable, 10- to 100-fold above the threshold value (Fig. 4). Because animal studies have shown that PrPSc and infectivity are transported anterogradely from one brain region to another along neuroanatomical pathways (23–25), we expected to find PrPSc in white matter as demonstrated by the CDI but not IHC. Axonal transport of PrPSc is also suggested by diffusion-weighted MRI scans of CJD cases, which show high-intensity signals in analogous neocortical regions of the right and left cerebral hemispheres (26). This symmetry of neuroradiological abnormalities is consistent with spread of PrPSc to the contralateral cortex by means of callosal commissural pathways.
Most immunoassays that detect HuPrPSc do so only after subjecting the sample to limited proteolysis to form PrP 27–30, followed by denaturation. Because the CDI measures the immunoreactivity before and after denaturation to an epitope that is exposed in native PrPC but buried in PrPSc, limited proteolysis to eliminate PrPC is unnecessary. Assays based on limited proteolysis underestimate the level of PrPSc because they digest sPrPSc, which represents 80–90% of PrPSc in CJD and scrapie brains (Fig. 4 and Table 5).
Gerstmann–Sträussler–Scheinker, an inherited human prion disease, is caused by the P102L mutation in the PRNP gene. In mice expressing the Gerstmann–Sträussler–Scheinker mutant PrP transgene, the CDI detected high levels of sPrPSc(P101L) as well as low levels of rPrPSc(P101L) long before neurodegeneration and clinical symptoms occurred (9). sPrPSc(P101L) as well as low concentrations of rPrPSc(P101L) previously escaped detection (27). Whether a similar situation applies in other genetic forms of prion disease, sCJD, or variant CJD remains to be determined. Because most of the PrPSc in the brains of sCJD patients is protease-sensitive (Fig. 4), it is likely that the lower sensitivity of IHC is due to its inability to detect sPrPSc. Presently, we have no information about the kinetics of either sPrPSc or rPrPSc accumulation in human brain. Limited information on the kinetics of PrPSc accumulation in livestock comes from studies of cattle, sheep, and goats inoculated orally, but most of the bioassays were performed in non-Tg mice (28–30) in which prion titers were underestimated by as much as a factor of 104 (10).
The studies reported here are likely to change profoundly the approach to the diagnosis of prion disease in both humans and livestock (31–33). The superior performance of the CDI in diagnosing prion disease compared to routine neuropathologic examination and IHC demands that the CDI be used in future diagnostic evaluations of prion disease. Prion disease can no longer be ruled out by routine histology or IHC. Moreover, the use of IHC to confirm cases of bovine spongiform encephalopathy after detection of bovine PrPSc by the CDI (10) seems an untenable approach in the future. Clearly, the CDI for HuPrPSc is as sensitive or more sensitive than bioassays in Tg(MHu2M) mice (Fig. 1).
Our results suggest that using the CDI to test large numbers of samples for human prions might alter the epidemiology of prion diseases. At present, there is limited data on the frequency of subclinical variant CJD infections in the U.K. population (34). Because appendixes and tonsils were evaluated only by IHC, many cases might have escaped detection (Tables 1 and 2). Equally important may be the use of CDI-like tests to diagnose other neurodegenerative disorders, such as Alzheimer's disease, Parkinson's disease, and the frontotemporal dementias. Whether IHC underestimates the incidence of one or more of these common degenerative diseases is unknown. Moreover, CDI-like tests may help determine the frequency with which these disorders and the prion diseases occurs concomitantly in a single patient (35, 36).
Acknowledgments
We thank the staff of the Hunters Point Animal Facility for their expert mouse studies and ZLB Behring for making the MAR1 mAb available. This work was supported by National Institutes of Health Contract NS02328 and National Institutes of Health Grants AG02132, AG010770, and AG021601. M.D.G. is supported by the John Douglas French Foundation for Alzheimer's research, the McBean Foundation; National Institute on Aging Grants AG021989, AG019724, and AG023501; the Alzheimer's Disease Research Center of California; and National Institutes of Health Grant M01 RR00079 to the General Clinical Research Center. B.L.M. is supported by National Institute on Aging Grants AG019724 and AG023501.
Author contributions: J.G.S. and S.B.P. designed research; M.D.G., C.D., S.D., M.S., H.S., and S.J.D. performed research; A.S., M.V., and H.B. contributed new reagents/analytic tools; J.G.S., K.G., B.L.M., S.J.D., and S.B.P. analyzed data; and J.G.S., S.J.D., and S.B.P. wrote the paper.
Abbreviations: CDI, conformation-dependent immunoassay; CJD, Creutzfeldt–Jakob disease; sCJD, sporadic CJD; fCJD, familial CJD; iCJD, iatrogenic CJD; IHC, immunohistochemistry; PrP, prion protein; PrPC, normal cellular PrP; PrPSc, disease-causing PrP; HuPrP, human PrP; MHu2M, chimeric mouse–human transgene; sPrPSc, protease-sensitive PrPSc; rPrPSc, protease-resistant PrPSc; PTA, phosphotungstate; PK, proteinase K; H&E, hematoxylin and eosin; Tg, transgenic; (D - N), difference in Ab-binding between native and denatured samples.
References
- 1.Prusiner, S. B. (1989) Annu. Rev. Microbiol. 43, 345-374. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, P., Gibbs, C. J., Jr., Rodgers-Johnson, P., Asher, D. M., Sulima, M. P., Bacote, A., Goldfarb, L. G. & Gajdusek, D. C. (1994) Ann. Neurol. 35, 513-529. [DOI] [PubMed] [Google Scholar]
- 4.Telling, G. C., Scott, M., Mastrianni, J., Gabizon, R., Torchia, M., Cohen, F. E., DeArmond, S. J. & Prusiner, S. B. (1995) Cell 83, 79-90. [DOI] [PubMed] [Google Scholar]
- 5.Telling, G. C., Scott, M., Hsiao, K. K., Foster, D., Yang, S.-L., Torchia, M., Sidle, K. C. L., Collinge, J., DeArmond, S. J. & Prusiner, S. B. (1994) Proc. Natl. Acad. Sci. USA 91, 9936-9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asante, E. A., Linehan, J. M., Desbruslais, M., Joiner, S., Gowland, I., Wood, A. L., Welch, J., Hill, A. F., Lloyd, S. E., Wadsworth, J. D. & Collinge, J. (2002) EMBO J. 21, 6358-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korth, C., Kaneko, K., Groth, D., Heye, N., Telling, G., Mastrianni, J., Parchi, P., Gambetti, P., Will, R., Ironside, J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 4784-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safar, J., Wille, H., Itri, V., Groth, D., Serban, H., Torchia, M., Cohen, F. E. & Prusiner, S. B. (1998) Nat. Med. 4, 1157-1165. [DOI] [PubMed] [Google Scholar]
- 9.Tremblay, P., Ball, H. L., Kaneko, K., Groth, D., Hegde, R. S., Cohen, F. E., DeArmond, S. J., Prusiner, S. B. & Safar, J. G. (2004) J. Virol. 78, 2088-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safar, J. G., Scott, M., Monaghan, J., Deering, C., Didorenko, S., Vergara, J., Ball, H., Legname, G., Leclerc, E., Solforosi, L., et al. (2002) Nat. Biotechnol. 20, 1147-1150. [DOI] [PubMed] [Google Scholar]
- 11.Bellon, A., Seyfert-Brandt, W., Lang, W., Baron, H., Groner, A. & Vey, M. (2003) J. Gen. Virol. 84, 1921-1925. [DOI] [PubMed] [Google Scholar]
- 12.Kascsak, R. J., Rubenstein, R., Merz, P. A., Tonna-DeMasi, M., Fersko, R., Carp, R. I., Wisniewski, H. M. & Diringer, H. (1987) J. Virol. 61, 3688-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muramoto, T., DeArmond, S. J., Scott, M., Telling, G. C., Cohen, F. E. & Prusiner, S. B. (1997) Nat. Med. 3, 750-755. [DOI] [PubMed] [Google Scholar]
- 14.Foster, J., Goldmann, W., Parnham, D., Chong, A. & Hunter, N. (2001) J. Gen. Virol. 82, 267-273. [DOI] [PubMed] [Google Scholar]
- 15.Minor, P., Newham, J., Jones, N., Bergeron, C., Gregori, L., Asher, D., Van Engelenburg, F., Stroebel, T., Vey, M., Barnard, G. & Head, M. (2004) J. Gen. Virol. 85, 1777-1784. [DOI] [PubMed] [Google Scholar]
- 16.Wadsworth, J. D., Joiner, S., Hill, A. F., Campbell, T. A., Desbruslais, M., Luthert, P. J. & Collinge, J. (2001) Lancet 358, 171-180. [DOI] [PubMed] [Google Scholar]
- 17.Glatzel, M., Abela, E., Maissen, M. & Aguzzi, A. (2003) N. Engl. J. Med. 349, 1812-1820. [DOI] [PubMed] [Google Scholar]
- 18.Baringer, J. R., Bowman, K. A. & Prusiner, S. B. (1981) J. Neuropathol. Exp. Neurol. 40, 329. [DOI] [PubMed] [Google Scholar]
- 19.Bruce, M. E., McBride, P. A. & Farquhar, C. F. (1989) Neurosci. Lett. 102, 1-6. [DOI] [PubMed] [Google Scholar]
- 20.Jendroska, K., Heinzel, F. P., Torchia, M., Stowring, L., Kretzschmar, H. A., Kon, A., Stern, A., Prusiner, S. B. & DeArmond, S. J. (1991) Neurology 41, 1482-1490. [DOI] [PubMed] [Google Scholar]
- 21.Taraboulos, A., Jendroska, K., Serban, D., Yang, S.-L., DeArmond, S. J. & Prusiner, S. B. (1992) Proc. Natl. Acad. Sci. USA 89, 7620-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecker, R., Taraboulos, A., Scott, M., Pan, K.-M., Torchia, M., Jendroska, K., DeArmond, S. J. & Prusiner, S. B. (1992) Genes Dev. 6, 1213-1228. [DOI] [PubMed] [Google Scholar]
- 23.Kimberlin, R. H. & Walker, C. A. (1979) J. Comp. Pathol. 89, 551-562. [DOI] [PubMed] [Google Scholar]
- 24.Brandner, S., Isenmann, S., Raeber, A., Fischer, M., Sailer, A., Kobayashi, Y., Marino, S., Weissmann, C. & Aguzzi, A. (1996) Nature 379, 339-343. [DOI] [PubMed] [Google Scholar]
- 25.Bouzamondo-Bernstein, E., Hopkins, S. D., Spilman, P., Uyehara-Lock, J., Deering, C., Safar, J., Prusiner, S. B., Ralston, H. J., III, & DeArmond, S. J. (2004) J. Neuropathol. Exp. Neurol. 63, 882-899. [DOI] [PubMed] [Google Scholar]
- 26.Collie, D. A., Sellar, R. J., Zeidler, M., Colchester, A. C. F., Knight, R. & Will, R. G. (2001) Clin. Radiol. 56, 726-739. [DOI] [PubMed] [Google Scholar]
- 27.Hsiao, K. K., Groth, D., Scott, M., Yang, S.-L., Serban, H., Rapp, D., Foster, D., Torchia, M., DeArmond, S. J. & Prusiner, S. B. (1994) Proc. Natl. Acad. Sci. USA 91, 9126-9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadlow, W. J., Kennedy, R. C., Race, R. E. & Eklund, C. M. (1980) Vet. Pathol. 17, 187-199. [DOI] [PubMed] [Google Scholar]
- 29.Hadlow, W. J., Kennedy, R. C. & Race, R. E. (1982) J. Infect. Dis. 146, 657-664. [DOI] [PubMed] [Google Scholar]
- 30.Wells, G. A. H. (2002) in Update of the Opinion on TSE Infectivity Distribution in Ruminant Tissues (European Commission, Brussels), pp. 10-37.
- 31.Head, M. W., Bunn, T. J., Bishop, M. T., McLoughlin, V., Lowrie, S., McKimmie, C. S., Williams, M. C., McCardle, L., MacKenzie, J., Knight, R., et al. (2004) Ann. Neurol. 55, 851-859. [DOI] [PubMed] [Google Scholar]
- 32.Parchi, P., Giese, A., Capellari, S., Brown, P., Schulz-Schaeffer, W., Windl, O., Zerr, I., Budka, H., Kopp, N., Piccardo, P., et al. (1999) Ann. Neurol. 46, 224-233. [PubMed] [Google Scholar]
- 33.DeArmond, S. J., Ironside, J. W., Bouzamondo-Bernstein, E., Peretz, D. & Fraser, J. R. (2004) in Prion Biology and Diseases, ed. Prusiner, S. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 777-856.
- 34.Hilton, D. A., Ghani, A. C., Conyers, L., Edwards, P., McCardle, L., Ritchie, D., Penney, M., Hegazy, D. & Ironside, J. W. (2004) J. Pathol. 203, 733-739. [DOI] [PubMed] [Google Scholar]
- 35.Hansen, L. A., Masliah, E., Terry, R. D. & Mirra, S. S. (1989) Acta Neuropathol. 78, 194-201. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto, M. & Masliah, E. (1999) Brain Pathol. 9, 707-720. [DOI] [PMC free article] [PubMed] [Google Scholar]