Abstract
Structural studies suggest that most point mutations in the BCR-ABL kinase domain cause resistance to the ABL kinase inhibitor imatinib by impairing the flexibility of the kinase domain, restricting its ability to adopt the inactive conformation required for optimal imatinib binding, rather than by directly interfering with drug contact residues. BMS-354825, currently in clinical development for imatinib-resistant chronic myelogenous leukemia, is a dual SRC/ABL kinase inhibitor that binds ABL in both the active and inactive conformation. To examine the potential role of conformational binding properties in drug resistance, we mapped the mutations in BCR-ABL capable of conferring resistance to BMS-354825. Through saturation mutagenesis, we identified 10 such BCR-ABL mutations, 8 of which occurred at drug contact residues. Some mutants were unique to BMS-354825, whereas others also conferred imatinib resistance. Remarkably, the identity of the amino acid substitution at either of two contact residues differentially affects sensitivity to imatinib or BMS-354825. The combination of imatinib plus BMS-354825 greatly reduced the recovery of drug-resistant clones. Our findings provide further rationale for considering kinase conformation in the design of kinase inhibitors against cancer targets.
Keywords: BMS-354825, chronic myeloid leukemia, imatinib, mutagenesis
Imatinib (STI-571 or Gleevec) and BMS-354825 are two clinically useful ABL kinase inhibitors that serve as a paradigm for the study of emergence of resistance in targeted cancer therapy. Imatinib is an ABL-specific inhibitor that binds with high affinity to the inactive conformation of the BCR-ABL tyrosine kinase and has been shown to be effective in the treatment of chronic myelogenous leukemia (CML) with little toxicity compared to other cancer therapies (1–3). However, the success of imatinib is hampered by acquired resistance, which occurs over months to years as a result of selection for subclones bearing mutations in the kinase domain, by amplification of the BCR-ABL genomic locus, or, potentially, through reduced BCR-ABL dependence (4–6). Twenty-five amino acid substitutions at 21 positions have been reported to confer imatinib resistance in CML patients undergoing treatment (7). Seven of these 25 mutations map to contact residues and sterically preclude the drug from binding to ABL; however, most do not. These are postulated to cause a conformational change in the conserved phosphate binding (P) loop or the activation loop that favors the active conformation, diminishing imatinib binding (1, 8–10).
BMS-354825, a novel synthetic chemotype, is an ATP-competitive, dual-specific SRC and ABL kinase inhibitor that can bind BCR-ABL in both the active and inactive conformations (11, 12). Mutations in BCR-ABL that favor the adoption of an active, imatinib-resistant conformation are effectively targeted by BMS-354825, as shown in cell lines expressing 14 of 15 imatinib-resistant mutants (12). From a clinical standpoint, BMS-354825 is particularly attractive because it has been shown to induce hematologic and cytogenetic responses in imatinib-resistant CML patients treated in a phase I clinical trial with minimal toxicity (13).
In view of the fact that BMS-354825 can bind both the active and inactive conformations of BCR-ABL, we reasoned that fewer kinase domain mutations are likely to lead to resistance to BMS-354825 compared with imatinib. To address this question, we conducted a saturation mutagenesis screen of BCR-ABL and found that the spectrum of mutations that allow for BMS-354825 resistance is reduced compared with that of imatinib (8, 14). All but two of the mutations causing resistance map to known BMS-354825 contact residues as shown by crystallographic studies (11). Furthermore, we report that screens with a combination of imatinib and BMS-354825 reduce both the total number and the spectrum of recovered mutants. Biochemical and biological characterization of the mutants revealed, surprisingly, that the identity of the particular amino acid substitution at key contact residues selectively controls the sensitivity to each of the kinase inhibitors.
Materials and Methods
DNA Constructs, BCR-ABL Mutagenesis, and Drug Resistance Screen. WT p210 BCR-ABL cDNA cloned into the EcoRI site of the pMSCV puro retroviral vector (Clontech) was used as a template for mutagenesis. We used a modified strategy for random mutagenesis described by others (8, 14). Briefly, 1–2 μg of WT MSCV p210 was used to transform the DNA-repair-deficient Escherichia coli strain XL-1 Red (Stratagene) and plated on 20–40 ampicillin-agar bacterial plates. After incubation for 36 h, colonies were collected by scraping, and plasmid DNA was purified by using a plasmid MAXI kit (Qiagen). Subsequently, 15 μg of mutagenized p210 plasmid stock and 15 μg of Ecopack packaging plasmid (gift of Richard Van Etten, Harvard University, Cambridge, MA) were cotransfected by the calciumphosphate method into 293T cells grown in DMEM (Cellgro) containing 10% FCS (Omega Scientific). Twenty-four hours after transfection, the medium was changed to Iscove's (Cellgro) supplemented with 10% FCS. Viral supernatants were collected at 48 h, centrifuged to remove cellular debris, and used to infect Ba/F3 cells at a 1:10 dilution of viral supernatant to fresh media. For infection, 1–2 × 106 Ba/F3 cells, 3 ml of the diluted viral stock supplemented with recombinant mouse IL-3 (R & D Systems), and 4 μg/ml polybrene were plated in a 12-well tissue culture dish and centrifuged at 1,000 RCF in a Beckman Coulter GS-6R centrifuge with a microplate carrier for 90 min at 34°C. Centrifuged cells were subsequently transferred to a 37°C incubator for 14–16 h. Infected Ba/F3 cells were washed twice with PBS to remove IL-3 and plated in 3 ml of RPMI medium 1640 (Cellgro) at 5 × 105 cells per well of a six-well dish supplemented with 20% FCS and 1.2% Bacto-agar with drug. After 10 days, individual colonies were plucked from agar and expanded in the presence of appropriate drug (10 μM imatinib and/or 25–100 nM BMS-354825).
Sequencing and Alignment. Expanded colonies were harvested 3–14 days after isolation from agar, and whole genomic DNA was isolated by using the DNeasy kit (Qiagen). BCR-ABL kinase domain was amplified by high-fidelity PCR from whole genomic DNA by using Optimase polymerase (Transgenomic) on an MBS 0.2G (ThermoHybaid) automated cycler. The primers 5′ABL KD (5′-GCGCAACAAGCCCACTGTCTATGG-3′) and 3′ABL KD [(5′-GACGCCTTGTTTCCCCAGCTCCTTTTCCACTTCG-3′) were used for kinase domain amplification and subsequent bidirectional sequencing. Alignments were performed by pairwise blast (NCBI) to the WT BCR-ABL sequence.
Generation of Mutants. Mutants isolated in the screen were engineered into pMSCV puro p210 by using the QuikChange mutagenesis kit (Stratagene). Alternatively, a portion of the kinase domain was amplified by using high-fidelity PCR from the genomic DNA of mutants recovered and sequenced in the screen and cloned into the KpnI–BsrGI site of p210 in a modified pBluescript II KS+ (Stratagene) missing the KpnI site then shuttled into pMSCV puro. In all cases, individual point mutants were confirmed by sequence analysis. In this paper, we use the type Ia human sequence numbering scheme differing by -19 aa from the type Ib residue numbers (15).
Cell-Viability Assay. Stable Ba/F3 lines were generated by using retroviral spinfection outlined above with the appropriate mutated plasmid. At 18 h postinfection, IL-3 was washed twice from the cells with PBS and stables were selected in RPMI medium 1640/10% FCS in the absence of IL-3. Exponentially growing Ba/F3 cells (2 × 104) were plated in each well of a 24-well dish with 1 ml of RPMI medium 1640/10% FCS containing the appropriate drug as indicated in triplicate. Cells were allowed to expand for 3 days and were counted by using a Vi-cell XR automated cell viability analyzer (Beckman Coulter). The mean number of viable cells at varying concentrations of drug was normalized to the mean number of viable cells in the no-drug sample for each mutant. Error bars represent the standard deviations of the mean.
Immunoblotting. Exponentially growing Ba/F3 cells (1 × 106) stably expressing each mutant along with a WT control were plated in RPMI medium 1640/10% FCS supplemented with kinase inhibitor at the indicated concentration. After a 4-h incubation, the cells were lysed in 0.1 ml of 1% Triton-X buffer (12.5 mM EDTA, pH 8.0/25 mM Hepes, pH 7.5/150 mM NaCl/1% Triton X-100/10% glycerol) supplemented with Protease Inhibitor Mixture Set III and Phosphatase Inhibitor Mixture Set II (Calbiochem). Fifteen microliters of each lysate was boiled in 4× sample buffer, run on a 7.5% polyacrylamide gel, and transferred to an Immobilon membrane (Millipore) for Western blot analysis. We used anti-ABL AB-3 (Oncogene Research Products), anti-phosphotyrosine 4G10 (Upstate Biotechnology), and anti-β-actin antibody AC-15 (Sigma-Aldrich) for detection of protein.
Structural Modeling. We recognized the similarity in structure between the BMS-354825 and PD173955 compounds and used the program o to model the position of BMS-354825 in the Abl kinase domain based solely on the Abl kinase:PD173955 cocrystal structure (16).
Results
Resistance to BMS-354825 Is Primarily Caused by BCR-ABL Mutations at Contact Residues. We conducted a saturation mutagenesis screen for BMS-354825-resistant BCR-ABL subclones, using a method described previously by Azam et al. (8), in an analogous study of imatinib resistance. This approach utilizes a DNA-repair-deficient E. coli strain to produce random mutagenesis of a BCR-ABL retroviral plasmid, infection of Ba/F3 cells, and selection for Ba/F3 clones expressing drug-resistant BCR-ABL isoforms in agar (8, 14). The mutant BCR-ABL isolates exhibit drug resistance and are capable of transforming Ba/F3 cells, indicating that the mutation does not also abolish kinase activity. We used an optimized BMS-354825 dose of 25–50 nM to select for drug-resistant clones and minimize background. These concentrations are analogous to the imatinib doses used in previously published studies, corresponding to a dose 15- to 30-fold over the IC50 for growth inhibition of a WT BCR-ABL-expressing Ba/F3 line in liquid culture (8). In a series of screens with five independently derived, mutagenized libraries, we isolated 201 individual clones in the presence of 50 nM BMS-354825. Sequence analysis demonstrated 10 different mutations leading to amino acid substitutions at six different residues (Table 1). Four of these six sites (Leu-248, Val-299, Thr-315, and Phe-317) are BMS-354825 contact residues, as shown by crystallographic analysis, and account for 97.5% of isolates (11). The exceptions were four clones of E255K and one clone of Q252H, mutants shown previously to have a BMS-354825 IC50 for growth <10-fold higher than WT BCR-ABL (12). This result is in contrast with the profile of imatinib-resistant BCR-ABL mutants isolated in a published study, in which only 4 of 20 affected amino acids were contact residues at a comparable imatinib dose, 30-fold over the IC50 (8).
Table 1. Point mutants recovered in 50 nM BMS-354825.
| Mutation | Number of clones (%) | Contact residue |
|---|---|---|
| L248R | 1 (0.5) | X |
| Q252H | 1 (0.5) | |
| E255K | 4 (2.0) | |
| V299L | 1 (0.5) | X |
| T315I | 39 (19) | X |
| T315A | 60 (30) | X |
| F317V | 83 (41) | X |
| F317L | 10 (5.0) | X |
| F317I | 1 (0.5) | X |
| F317S | 1 (0.5) | X |
| Total: | 201 (100) |
The total number of recovered clones at 50 nM BMS-354825 with percentage of total indicated parenthetically. Mutations at drug-kinase contact residues are indicated.
We conducted additional experiments to be sure that the conditions of our screen did not preclude the isolation of a more diverse range of mutants. To address the possibility that the limited repertoire of BMS-354825-resistant mutants might be explained by the use of an excessively stringent drug concentration, we reduced the BMS-354825 dose and did not appreciably increase the range of resistant clones recovered (Table 3, which is published as supporting information on the PNAS web site). Second, we conducted a control screen in the presence of 10 μM imatinib and recovered 80 clones including L248R, G250E, Y253H/C, E255K, E279K, and T315I. These are the imatinib-resistant mutants most commonly isolated in an analogous screen (8). Therefore, our mutagenized BCR-ABL retroviral library was of sufficient diversity to recover a similar range of mutants as previously reported for imatinib (8). Finally, we believe that the screen was performed to saturation for the following reasons: All of the mutants described appeared in more than one independent mutagenesis library; certain mutants were the result of different nucleotide changes at the same position (i.e., F317V/L: TTC to GTC/CTC); and a low percentage of mutants demonstrated additional point mutations, including silent base changes, indicating that the same mutant was derived from a different original clone within the same library (Table 3).
To validate that the mutants recovered played a causal role in BMS-354825 resistance, we generated individual BCR-ABL retroviral constructs containing the point mutation of interest, introduced these into Ba/F3 cells, and measured the IC50 for growth in BMS-354825 (Table 2). T315I conferred the greatest degree of resistance to BMS-354825 with an IC50 >750-fold over WT BCR-ABL. T315A and F317V, two previously uncharacterized mutants, show the next highest IC50s, 40- to 90-fold over WT. Of note, these three mutants were the most frequently recovered in our screen. L248R, E255K, F317L, and the previously uncharacterized mutant V299L show modest changes in IC50, increasing ≈10- to 15-fold from WT BCR-ABL. The appearance of previously uncharacterized mutations, not previously reported in imatinib studies, raised the question of whether combination treatment could further reduce the spectrum of recovered mutants.
Table 2. IC50 for growth of Ba/F3 stable lines grown in the presence of BMS-354825 and imatinib.
| Ba/F3 Clone | BMS-354825 IC50, nM (fold WT IC50) | Imatinib IC50, nM (fold WT IC50) |
|---|---|---|
| p210 WT | 1.34 (1) | 323 (1) |
| L248R | 16 (12) | >10,000 (>30) |
| Y253H | 10 (7.5) | >10,000 (>30) |
| E255K | 13 (9.7) | 8,400 (26) |
| V299L | 18 (13.4) | 540 (1.7) |
| T315I | >1,000 (>750) | >10,000 (>30) |
| T315A | 125 (93) | 760 (2.4) |
| F317L | 18 (13.4) | 810 (2.5) |
| F317V | 53 (40) | 350 (1.1) |
Viable cell counts normalized to the no-drug control were preformed after 3 days of growth in triplicate. Stable Ba/F3 cells were exposed to no drug, BMS-354825 (5, 10 25, 50, 100, 200, and 500 nM), and imatinib (500, 1,000, 5,000, and 10,000 nM). Biological IC50 was determined from semilogarithmic graphing of the dose–response curve for each mutant. The adjacent column represents the fold change in IC50 as compared with WT BCR-ABL.
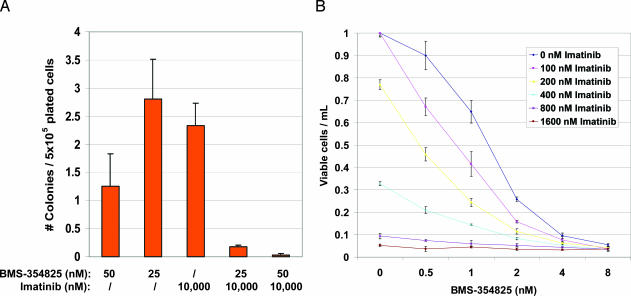
Combination of BMS-354825 and Imatinib Reduces the Number of Resistant Clones and Limits the Range of Drug-Resistant BCR-ABL Mutations. Combination treatment reduced the recovery of resistant clones by 10- to 60-fold when 25 or 50 nM BMS-354825 was combined with a fixed dose of 10 μM imatinib compared with imatinib alone (Fig. 1A). In addition, the range of drug-resistant BCR-ABL mutations was severely reduced. At the 50 nM BMS-354825 concentration, only one clone expressing the T315I mutation was recovered. At 25 nM BMS-354825, we found six clones expressing T315I and five clones expressing E255K.
Fig. 1.
BMS-354825 works in concert with imatinib to reduce the number of resistant clones. (A) Graphical representation of the frequency of drug-resistant Ba/F3 clones obtained from four independent screens. For each experiment, 75–90 × 106 Ba/F3 cells infected with mutagenized p210 BCR-ABL were divided into five treatment groups (50 nM BMS-354825, 25 nM BMS-354825, 10,000 nM imatinib, and combinations of each) and plated at a density of 5 × 105 cells per well (30–36 wells per treatment group). The number of clones obtained per well was averaged for each experimental condition, and an error bar represents the standard error between the four screens. (B) WT p210 Ba/F3 line grown in increasing concentrations of BMS-354825 in the presence of 0, 100, 200, 400, 800, and 1600 nM imatinib, as indicated, for 3 days. Normalized viable cell counts of the average of the triplicates are plotted with respect to the no-drug control. An error bar represents the standard deviation of the mean for each dose.
We also examined the effect of dual treatment on the growth of Ba/F3 cells expressing WT BCR-ABL. Increasing doses of imatinib in the presence of BMS-354825 shifted the dose–response curve to the left, indicating at least an additive effect of the two drugs on growth inhibition of a WT BCR-ABL Ba/F3 cell line (Fig. 1B). The reduced total number of clones, the reduced range of mutants, and the additive growth inhibition on a WT BCR-ABL cell line all suggest that the two drugs in combination could delay the emergence of resistance.
Identity of Contact-Residue Substitution Determines the Sensitivity to Imatinib or BMS-354825. In cataloging the identity of the BMS-354825-resistant BCR-ABL mutants, we were surprised to find that two of the clones that appeared at high frequency in our screen targeted residues previously implicated in imatinib resistance, yet the substituted amino acid change had never been recovered in imatinib resistance screens. Specifically, T315I and F317L are well described imatinib-resistant mutants, but T315A and F317V have never been reported. Interestingly, Thr-315 and Phe-317 are contact residues for both imatinib and BMS-354825, raising the possibility that the identity of the new amino acid substitution at the contact residue might differentially alter sensitivity to one but not the other ABL kinase inhibitor (1, 10, 11).
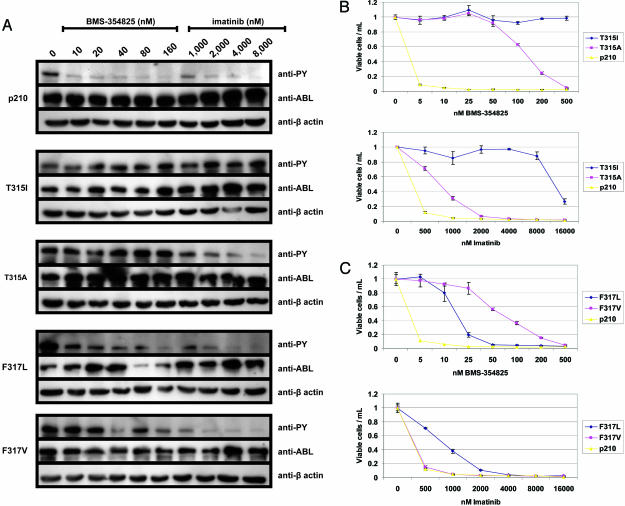
We directly compared the ability of each compound to inhibit BCR-ABL kinase activity and growth of Ba/F3 clones bearing different mutations at Thr-315 and Phe-317. Consistent with the IC50 growth data reported above, T315A dramatically alters BMS-354825 sensitivity, because 90-fold higher concentrations are required to inhibit kinase activity and growth relative to WT BCR-ABL-expressing cells (Fig. 2 A and B). However, T315A is only 2- to 3-fold more resistant to inhibition by imatinib than WT BCR-ABL, both biochemically (Fig. 2 A) and biologically (Fig. 2B Lower). As reported previously, T315I is highly resistant to both compounds (Fig. 2 A and B). Similarly, the identity of the mutation at neighboring Phe-317 differentially affects inhibition by each of the drugs. Imatinib is equipotent, both biologically and biochemically, against F317V and WT BCR-ABL. However, F317V is 40-fold less sensitive to BMS-354825 than WT (Fig. 2 A and C). Conversely, F317L shows moderate resistance to both compounds (Fig. 2 A and C). These data indicate that the identity of the mutated contact residue modulates the ability of a specific compound to inhibit kinase activity and growth.
Fig. 2.
The identity of the mutation at Thr-315 and Phe-317 determines the sensitivity or resistance of BCR-ABL to each of the kinase inhibitors. (A) Western blot analysis of stable Ba/F3 clones expressing the indicated BCR-ABL p210 isoform. In each case, cells were normalized by cell count and lysed in equal volumes of lysis buffer after exposure to the indicated concentration of drug for 3–4 h. Western blots were analyzed with antibody to antiphosphotyrosine (anti-PY), ABL, and β-actin. Comparison of growth inhibition by BMS-354825 and imatinib on mutations at Thr-315 (B) and Phe-317 (C). WT p210 is plotted as a control.
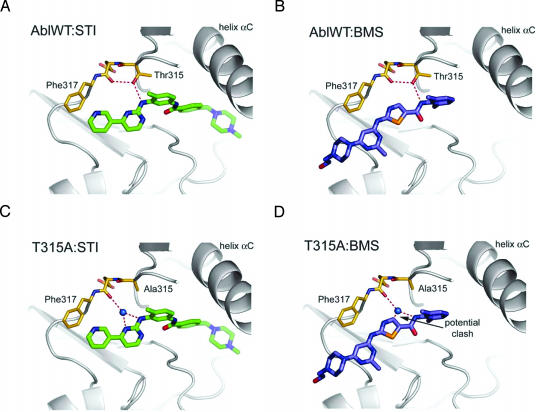
The availability of cocrystal structures of each compound bound to ABL presents an opportunity to postulate how amino acid substitutions at contact residues affect drug sensitivity. Structural models indicate that Thr-315 makes a hydrogen bond with each of the drugs in the ATP-binding pocket (Fig. 3 A and B) (1, 10, 11). As reported with imatinib, we predict that T315I resistance to BMS-354825 is caused by a loss of this hydrogen bond and steric clash between BMS-354825 and the bulky hydrocarbon side chain of isoleucine (6). However, substitution to alanine at this critical residue causes significantly more resistance to BMS-354825 than imatinib. Structural models suggest that relatively more of the surface area of imatinib is buried in the ABL kinase domain compared with BMS-354825. Consequently, the interaction of BMS-354825 and Thr-315 could represent a greater proportion of the total interactions between the drug and kinase. Loss of this one interaction by mutation to T315A, therefore, would cause a greater relative change in BMS-354825 binding affinity compared with imatinib. Additionally, substitution to T315A presumably also eliminates a direct hydrogen bond between each drug and ABL. In the case of imatinib, however, the pocket can accommodate a water molecule that could conceivably bridge hydrogen-bonding interactions between the drug and BCR-ABL (Fig. 3C). T315A modeled with BMS-354825 suggests that a water molecule is not accommodated by a carbon group on the inhibitor, eliminating this critical hydrogen bond and destroying favorable binding energetics (Fig. 3D). These speculative predictions will need to be directly addressed by cocrystallizing the mutants with each of the compounds.
Fig. 3.
Structural modeling of the T315A mutation provides rationale for continued inhibition of BCR-ABL kinase activity by imatinib but not BMS-354825. WT ABL complexed with imatinib (A) and BMS-354825 (B) illustrates key hydrogen-bonding interactions between each inhibitor and the ATP-binding pocket of ABL. In C, mutation to T315A allows a water molecule to bridge the hydrogen-bonding interactions of imatinib and ABL. (D) In the case of BMS-354825 and T315A, the drug displaces the water molecule, loses the critical hydrogen bonds, and prevents binding.
Discussion
This study demonstrates that mutations in BCR-ABL that confer resistance to the dual SRC/ABL kinase inhibitor BMS-354825 map almost exclusively to structural contact points between the kinase domain and the drug. This contrasts with parallel studies of imatinib, where mutations at non-contact residues, primarily in the P-loop and activation domain, account for most resistant clones (8). Because imatinib binds exclusively to the inactive conformation of ABL and induces further structural changes in the P-loop to optimize binding affinity, any mutation that interferes with this flexibility can cause resistance. In contrast, BMS-354825 has recently been shown to bind ABL in the active conformation, and modeling based on these cocrystallographic findings also predicts binding to the inactive conformation (11). Our finding that resistance to this conformation-tolerant compound occurs primarily through mutation at contact residues suggests that such inhibitors may offer advantages over conformation-specific inhibitors. Although conformation-tolerant binding may limit the opportunities for developing drug resistance through kinase domain mutations for acquired resistance, one significant tradeoff is likely to be reduced specificity for the intended kinase target. Whether these off-target activities will represent a significant liability will obviously depend on the specific kinases affected.
Our results have immediate implications for CML therapy. In a phase I clinical study, treatment with BMS-354825 induced remissions in a large fraction of CML patients with resistance to imatinib without significant side effects (13). As clinical studies of this compound in CML expand to include larger numbers of patients, our mutagenesis screen may anticipate mechanisms of clinical resistance. Specifically, we would predict that the three BCR-ABL mutations recovered in >90% of the clones in our screen (T315I, T315A, and F317V) should also account for most cases of clinical resistance. Because T315A and F317V remain sensitive to imatinib, early studies of combination therapy with BMS-354825 and imatinib seem warranted. In addition to delaying the emergence of resistance, combination therapy may also enhance the reduction in tumor burden in newly treated patients because of the increased potency of the combination. However, optimism for this combination must be tempered based on the fact that the T315I mutant is cross-resistant to both compounds, highlighting the need for a third inhibitor with activity against this mutant. Encouragingly, despite the observation that E255K was recovered at low frequency with BMS-354825 alone and in combination with imatinib in our study, this common imatinib-resistant mutant has been effectively targeted in patients with the doses of BMS-354825 achieved in the clinical trial (13).
Although the studies presented here only address the question of kinase conformation with ABL inhibitors, we believe our results are likely to have broader implications. Recent efforts to sequence the kinome in a broad range of tumors have expanded the list of oncogenic kinase mutations in human cancers (17–21). The likelihood of clinical benefit for patients whose tumors bear such mutations is very high when treated with appropriate inhibitors, but so is the risk of acquired resistance (22). Although the number of such diseases for which the molecular basis for resistance has been resolved is small (CML and gastrointestinal stromal tumor), the mechanism is consistent: selection for tumor subclones bearing secondary kinase domain mutations that block drug action. The recent discovery that mutations in the epidermal growth factor receptor (EGFR) confer sensitivity to EGFR inhibitors in non-small-cell lung cancer patients raises the possibility that acquired resistance to these drugs may occur through an analogous mechanism (20, 23). The two most widely studied EGFR inhibitors, gefitinib and erlotinib, are thought to bind EGFR in the active conformation, whereas lapatinib, a third compound in clinical trials, binds EGFR in an inactive-like conformation (24, 25). Comparative studies of these compounds may reveal distinct resistance mechanisms and could provide rationale for combination therapy, or for a search for additional, more conformation-tolerant EGFR inhibitors.
Supplementary Material
Acknowledgments
We thank B. Nagar and J. Kuriyan for providing structural modeling data, and M. Gorre, J. Nicoll, and members of the C.L.S. laboratory for helpful discussions. M.R.B. was supported by National Institutes of Health National Institute of General Medical Sciences Training Grant GM08042, the Medical Scientist Training Program, and the Aesculapians Fund of the David Geffen School of Medicine. This work was supported by grants from the Leukemia and Lymphoma Society (to N.P.S. and C.L.S.). C.L.S. is an investigator of the Howard Hughes Medical Institute and a Doris Duke Distinguished Clinical Scientist.
Author contributions: M.R.B. and C.L.S. designed research; M.R.B. and B.J.S. performed research; N.P.S., F.Y.L., and C.L.S. contributed new reagents/analytic tools; M.R.B., B.J.S., and C.L.S. analyzed data; and M.R.B. and C.L.S. wrote the paper.
Abbreviation: CML, chronic myelogenous leukemia.
References
- 1.Schindler, T., Bornmann, W., Pellicena, P., Miller, W. T., Clarkson, B. & Kuriyan, J. (2000) Science 289, 1938-1942. [DOI] [PubMed] [Google Scholar]
- 2.Druker, B. J., Talpaz, M., Resta, D. J., Peng, B., Buchdunger, E., Ford, J. M., Lydon, N. B., Kantarjian, H., Capdeville, R., Ohno-Jones, S. & Sawyers, C. L. (2001) N. Engl. J. Med. 344, 1031-1037. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien, S. G., Guilhot, F., Larson, R. A., Gathmann, I., Baccarani, M., Cervantes, F., Cornelissen, J. J., Fischer, T., Hochhaus, A., Hughes, T., et al. (2003) N. Engl. J. Med. 348, 994-1004. [DOI] [PubMed] [Google Scholar]
- 4.Donato, N. J., Wu, J. Y., Stapley, J., Lin, H., Arlinghaus, R., Aggarwal, B. B., Shishodia, S., Albitar, M., Hayes, K., Kantarjian, H., et al. (2004) Cancer Res. 64, 672-677. [DOI] [PubMed] [Google Scholar]
- 5.Donato, N. J., Wu, J. Y., Stapley, J., Gallick, G., Lin, H., Arlinghaus, R. & Talpaz, M. (2003) Blood 101, 690-698. [DOI] [PubMed] [Google Scholar]
- 6.Gorre, M. E., Mohammed, M., Ellwood, K., Hsu, N., Paquette, R., Rao, P. N. & Sawyers, C. L. (2001) Science 293, 876-880. [DOI] [PubMed] [Google Scholar]
- 7.Nardi, V., Azam, M. & Daley, G. Q. (2004) Curr. Opin. Hematol. 11, 35-43. [DOI] [PubMed] [Google Scholar]
- 8.Azam, M., Latek, R. R. & Daley, G. Q. (2003) Cell 112, 831-843. [DOI] [PubMed] [Google Scholar]
- 9.Shah, N. P., Nicoll, J. M., Nagar, B., Gorre, M. E., Paquette, R. L., Kuriyan, J. & Sawyers, C. L. (2002) Cancer Cell 2, 117-125. [DOI] [PubMed] [Google Scholar]
- 10.Nagar, B., Bornmann, W. G., Pellicena, P., Schindler, T., Veach, D. R., Miller, W. T., Clarkson, B. & Kuriyan, J. (2002) Cancer Res. 62, 4236-4243. [PubMed] [Google Scholar]
- 11.Tokarski, J. S., Newitt, J., Lee, F. Y., Lombardo, L., Borzilleri, R., Kish, K., Xie, D., Zhang, Y., Cheng, J. D., Kiefer, S. E., Chang, C. Y. J., Klei, H. E. & (Intr. by Sawyers, C. L. (2004) Blood 104, abstr. 553.
- 12.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D. & Sawyers, C. L. (2004) Science 305, 399-401. [DOI] [PubMed] [Google Scholar]
- 13.Sawyers, C. L., Shah, N. P., Kantarjian, H. M., Donato, N., Nicoll, J., Bai, S. A., Huang, F., Clark, E., DeCillis, A. P. & Talpaz, M. (2004) Blood 104, abstr. 1.
- 14.Azam, M., Raz, T., Nardi, V., Opitz, S. L. & Daley, G. Q. (2003) Biol. Proc. Online 5, 204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fainstein, E., Einat, M., Gokkel, E., Marcelle, C., Croce, C. M., Gale, R. P. & Canaani, E. (1989) Oncogene 4, 1477-1481. [PubMed] [Google Scholar]
- 16.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 17.Samuels, Y., Wang, Z., Bardelli, A., Silliman, N., Ptak, J., Szabo, S., Yan, H., Gazdar, A., Powell, S. M., Riggins, G. J., et al. (2004) Science 304, 554. [DOI] [PubMed] [Google Scholar]
- 18.Bardelli, A., Parsons, D. W., Silliman, N., Ptak, J., Szabo, S., Saha, S., Markowitz, S., Willson, J. K., Parmigiani, G., Kinzler, K. W., et al. (2003) Science 300, 949. [DOI] [PubMed] [Google Scholar]
- 19.Stephens, P., Hunter, C., Bignell, G., Edkins, S., Davies, H., Teague, J., Stevens, C., O'Meara, S., Smith, R., Parker, A., et al. (2004) Nature 431, 525-526. [DOI] [PubMed] [Google Scholar]
- 20.Paez, J. G., Janne, P. A., Lee, J. C., Tracy, S., Greulich, H., Gabriel, S., Herman, P., Kaye, F. J., Lindeman, N., Boggon, T. J., et al. (2004) Science 304, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 21.Davies, H., Bignell, G. R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M. J., Bottomley, W., et al. (2002) Nature 417, 949-954. [DOI] [PubMed] [Google Scholar]
- 22.Sawyers, C. L. (2003) Genes Dev. 17, 2998-3010. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, T. J., Bell, D. W., Sordella, R., Gurubhagavatula, S., Okimoto, R. A., Brannigan, B. W., Harris, P. L., Haserlat, S. M., Supko, J. G., Haluska, F. G., et al. (2004) N. Engl. J. Med. 350, 2129-2139. [DOI] [PubMed] [Google Scholar]
- 24.Stamos, J., Sliwkowski, M. X. & Eigenbrot, C. (2002) J. Biol. Chem. 277, 46265-46272. [DOI] [PubMed] [Google Scholar]
- 25.Wood, E. R., Truesdale, A. T., McDonald, O. B., Yuan, D., Hassell, A., Dickerson, S. H., Ellis, B., Pennisi, C., Horne, E., Lackey, K., et al. (2004) Cancer Res. 64, 6652-6659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.