Abstract
Non-O1, non-O139 Vibrio cholerae can cause gastroenteritis and extraintestinal infections, but, unlike O1 and O139 strains of V. cholerae, little is known about the virulence gene content of non-O1, non-O139 strains and their phylogenetic relationship to other pathogenic V. cholerae. Comparative genomic microarray analysis of four pathogenic non-O1, non-O139 strains indicates that these strains are quite divergent from O1 and O139 strains. Genomic sequence analysis of a non-O1, non-O139 strain (AM-19226) that appeared particularly pathogenic in experimental animals suggests that this strain carries a type III secretion system (TTSS) that is related to the TTSS2 gene cluster found in a pandemic clone of Vibrio parahaemolyticus. The genes for this V. cholerae TTSS system appear to be present in many clinical and environmental non-O1, non-O139 strains, including at least one clone that is globally distributed. We hypothesize that the TTSS present in some pathogenic strains of non-O1, non-O139 V. cholerae may be involved in the virulence and environmental fitness of these strains.
Keywords: genome, virulence, cholera pathogenesis
Pathogenic Vibrio cholerae strains are the etiologic agents of cholera, a severe diarrheal disease of potential high lethality (1). Both pathogenic and nonpathogenic V. cholerae reside in the aquatic ecosystem, but nonpathogenic strains typically dominate this niche (2, 3). No bacterial factors have been correlated with the prevalence of pathogenic strains in the aquatic environment other than resistance to bacteriophages (4).
V. cholerae strains are grouped into two defined biotypes (El Tor and classical) and >200 serogroups (5–7). The El Tor and classical biotypes are differentiated based on biochemical properties and phage sensitivity, whereas serogroup differentiation is based on O-antigen structure. Pathogenic clinical isolates generally express one of two O antigens (O1 or O139). Most nonpathogenic, environmental isolates express other O antigens and are referred to as “non-O1, non-O139.” However, some non-O1, non-O139 V. cholerae are clearly pathogenic and have caused outbreaks or sporadic cases of gastroenteritis and extraintestinal infections in humans (8–12).
Pathogenic O1 and O139 isolates typically encode two critical virulence factors: cholera toxin (CT) and toxin-coregulated pilus (TCP) (1). CT is primarily responsible for the diarrheal purge, whereas TCP is an essential intestinal colonization factor (13). Nontoxigenic O1 strains expressing TCP can efficiently acquire the CT genes (ctxAB) through lysogenic conversion with CTXϕ, a filamentous phage that encodes CT and uses TCP as its receptor (14). However, it is unclear how strains efficiently acquire the chromosomal segment that encodes TCP (15).
Seven cholera pandemics have occurred since 1861, with the first six caused by classical biotype, O1 serogroup strains (16). The seventh pandemic began in 1961 and is caused by a specific clone of El Tor O1 V. cholerae (1). In 1991, the El Tor O1 seventh pandemic strain was introduced from Asia into South America and caused a major cholera epidemic (17). In contrast to the classical O1 strains that caused cholera in South America more than a century ago but eventually vanished from the region, the El Tor O1 seventh pandemic strain has remained an endemic cause of cholera for well over a decade in virtually all of Latin America. In 1992, the O139 serogroup emerged in South Asia as the first non-O1 strain capable of causing epidemic cholera (18), and O139 strains have since established endemicity along with O1 El Tor strains in South Asia. The O139 strain clearly emerged from a CTX+TCP+ seventh pandemic strain through replacement of O1 antigen genes with those encoding the O139 antigen (19). Other than TCP and CTX, we know little about the bacterial factors that contribute to the success of the seventh pandemic and related O139 V. cholerae strains.
The pathogenic mechanisms of non-O1, non-O139 strains is far less clear (3). Some of these strains carry the CTX and TCP genes, whereas others do not (2). Both pathogenic and nonpathogenic non-O1, non-O139 strains encode a toxin referred to as RTX, whereas others encode a heat-stable enterotoxin (nag-ST) (20, 21). Although one CTX+TCP+ clone of the O141 serogroup is apparently globally distributed (9), most pathogenic non-O1, non-O139 strains are CTX-TCP-, do not cause endemic or pandemic disease as clones, and are highly diverse (22, 23). Thus, the genetic relationship between pathogenic non-O1, non-O139 strains and O1 and O139 strains remains unclear.
The gene content of different O1 and O139 strains was recently examined by using DNA microarrays based on the genomic sequence of V. cholerae strain El Tor N16961 (24, 25). These studies revealed a surprisingly high degree of genomic conservation among TCP-positive O1 classical, O1 El Tor, and O139 strains that were previously thought to be quite diverse based on other methodologies (6, 17, 26). The microarrays also identified two chromosomal islands that are uniquely present in O139 and El Tor O1 seventh pandemic strains, suggesting that these regions may encode factors involved in the endemic and pandemic potential of these clones (25).
We performed a microarray-based analysis of four pathogenic, CTX-TCP-, non-O1, non-O139 V. cholerae strains and found that these strains are quite divergent from each other and from typical O1 or O139 strains. We also sequenced most of the genome of a non-O1, non-O139 strain (AM-19226). Annotation suggests that AM-19226 carries a type III secretion system (TTSS) (27) that is related to a Vibrio parahaemolyticus TTSS gene cluster (28). Further analysis indicated that this TTSS is present in many clinical and environmental non-O1, non-O139 strains. We propose that the TTSS in some pathogenic non-O1, non-O139 V. cholerae may be involved in the virulence and environmental fitness of these strains.
Materials and Methods
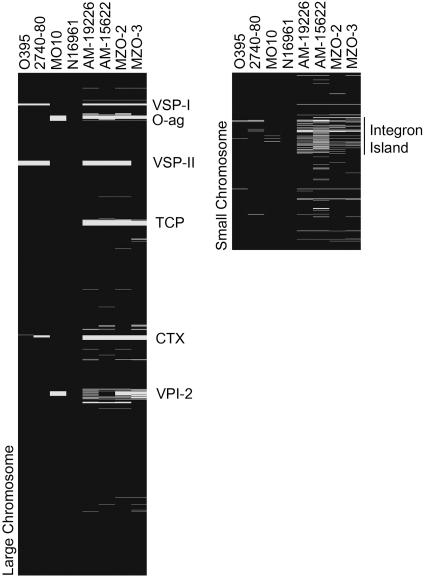
Microarray Analysis. Genomic DNA (gDNA) was extracted using the Easy DNA kit (Invitrogen). The DNA microarrays and their use in comparative genomic studies have been described (25). Briefly, 3 μg of gDNA from the reference strain (N16961) and a test strain were used to generate Cy3- and Cy5-labeled samples that were cohybridized to a single array. Fluorescence intensity ratios for individual genes were used as indicators of gene presence. Data for Fig. 1 are compiled from at least three slides for each strain.
Fig. 1.
Results of microarray-based comparative genomic analyses of four non-O1, non-O139 strains with the O1 El Tor sequenced strain used for array constriction, N16961. Representation of absent (white) and present (black) genes was based on fluorescence intensity ratios compiled from multiple slides from each strain, and was included alongside previously published data for O1 classical, environmental, and O139 strains for comparison.
Infant Mouse Colonization Assay. Colonization of infant mice by non-O1, non-O139 strains in competition with reference strain Bah-2 (CTX-TCP+) was assayed as described (29, 30). Competitive indices were calculated by dividing the output ratios by the input ratios.
Rabbit Diarrheal Assay. Diarrheal response of rabbits to V. cholerae was assayed by the removable intestinal tie-adult rabbit diarrhea (RITARD) model, using adult New Zealand White rabbits weighing 1.5–2.7 kg (30). After inoculation, rabbits were observed at 6-h intervals for 7 days for death, diarrheal severity, and shedding of challenge organisms. All rabbits that died were subjected to postmortem examination to assess the amount of fluid in the intestine.
Sequencing the AM-19226 Genome. An AM-19226 genomic library was constructed according to standard methods (see Supporting Text, which is published as supporting information on the PNAS web site). Sequencing was performed with pSMART primers (Lucigen) in 10-μl reactions in 384-well plates using Applied Biosystems bigdye v 3.1. Products were purified using the cleanseq system (Agencourt) on biomek fx (Beckman Coulter) and applied to an ABI 3730 sequencer. Chromatograms were transferred to the UNIX platform, and quality control/quality assessment scripts were run. Data were analyzed and assembled by using phred, phrap, and consed (31–33), resulting in the generation of 284 contigs.
Sequence Annotation. Sequence data were submitted to the Annotation Engine at The Institute for Genomic Research (Rockville, MD). The glimmer system (34, 35) was used to identify putative ORFs, and predicted translation products were searched against an internal nonredundant amino acid database by using the blast-extend-repraze algorithm (36, 37). All putative proteins were also searched against two sets of hidden Markov models, PFAM (38), and TIGRFAMs (39) by using the program hmmpfam (40). artemis v 6 from the Sanger Institute was used to visualize and further annotate sequence data (41).
Southern Analysis. Five micrograms of gDNA from each strain was digested with EcoRI (New England Biolabs), separated on agarose gels, and transferred to Hybond-N by standard methods. Hybridization was carried out by using the ECL kit (Amersham Pharmacia). Probes were obtained by PCR amplification using V. cholerae AM-19226 as the template; primer pairs are shown in Supporting Text. Blots were probed with a combination of vcsN2 and vcsC2 probes, stripped, and probed again with a combination of vcsV2 and vspD probes.
Results
Selection of Non-O1, Non-O139 Strains for Analysis. Non-O1, non-O139 V. cholerae are easily isolated from the environment, and they cause sporadic disease in humans that is identical to that caused by O1 or O139 strains (2, 3). AM-19226, AM-15622, MZO-2, and MZO-3 were isolated from patients with diarrhea in Bangladesh in 2001. Hybridization analysis using probes for known virulence genes indicated that strains carried sequences encoding RTX, but not genes for CTX, TCP, or nag-ST. We chose to analyze these strains further with respect to their genetic composition and their virulence in experimental animal models for gastroenteritis.
Microarray Analysis Suggests Non-O1, Non-O139 Strains Are Genetically Diverse. Fig. 1 presents the results of microarray analysis of the non-O1, non-O139 clinical isolates AM-19226, AM-15622, MZO-2, and MZO-3, shown next to previously analyzed O1 classical, O1 El Tor and O139 strains (25). The data indicate that each non-O1, non-O139 strain is missing ≈250 genes (≈6% of the genome) compared with the sequenced O1 strain N16961. A significant percentage of the missing genes belong to eight previously identified gene clusters; six of these clusters are located on the large chromosome, and two clusters are found on the small chromosome. Earlier dot-blot and PCR analysis of these four strains indicated that they lacked the TCP and CTX gene clusters, and array analysis confirmed these findings. Because these strains are non-O1, the absence of O-antigen genes was expected (as also seen for the O139 strain MO10). Also, all four strains are missing the Vibrio seventh pandemic (VSP)-I island, previously identified as unique to seventh pandemic strains (25). A second island called VSP-II was also previously identified as unique to seventh pandemic strains (25). It was recently reported that VSP-II is larger than originally determined, encompassing genes VC0490–VC0516 (42). Our analysis suggested that VSP-II is missing in AM-19226, AM-15622, and MZO-2, but is present in MZO-3. These conclusions were confirmed by PCR analysis (data not shown).
All four non-O1, non-O139 strains appeared to lack a significant number of the VPI-2 island genes (VC1758–VC1810) (43) when compared with seventh pandemic O1 El Tor and O139 strains. Some VPI-2 genes may still be present in these strains, but the precise extent of gene deletions in this region requires future analysis. Our results show that the non-O1, non-O139 strains may display a level of diversity within this region not seen in previously analyzed O1 or O139 strains. However, they are consistent with published observations that VPI-2 is most often present in toxigenic strains (43).
The integron island, located on the small chromosome of N16961, appears to be highly variable in the non-O1, non-O139 strains. Whereas O1 and O139 strains share >95% of integron island genes with N16961, the four non-O1, non-O139 strains share only 50–75% of integron island genes with N16961. Also, genes VCA0728–VCA0730, which lie outside the integron island, are absent in the non-O1, non-O139 strains. Previous work identified these genes as absent in O1 classical strains, but as present in O1 El Tor and O139 strains (25).
TCP- Non-O1, Non-O139 Strains Are Proficient for Colonization in Mice. TCP production is essential to the ability of diverse O1 strains of V. cholerae to colonize infant mice (1, 30). We therefore tested the four TCP-, non-O1, non-O139 strains for colonization in infant mice (Table 1), using the CTX-TCP+ O1 strain (Bah-2) as the reference strain (44). Whereas N16961 competed effectively with Bah-2 (competitive index = 1.21), the TCP- control strain TCP-2 showed an ≈10-fold reduction in colonization. In contrast, each of the TCP-, non-O1, non-O139 strains competed effectively with the O1 TCP+ strain Bah-2 in this model. However, the formal possibility existed that the successful colonization of these strains was due in part to the presence of an O1 TCP+ strain during the mixed infection. To determine whether these strains could colonize in a single infection, individual strains were inoculated into infant mice. In these studies, AM-19226 colonized >75% of the mice inoculated, and its colonization level was similar to that of N16961 (data not shown). In contrast, AM-15622, MZO-2, and MZO-3 colonized mice only sporadically and seldom achieved levels of colonization comparable to AM-19226 (data not shown).
Table 1. Infant mouse colonization data for different V. cholerae strains.
| Strain | Serogroup | Competitive index* |
|---|---|---|
| AM-19226 | O39 | 1.42 ± 0.34 |
| AM-15622 | O8 | 0.69 ± 0.11 |
| MZO-2 | O14 | 0.91 ± 0.36 |
| MZO-3 | O37 | 0.8 ± 0.32 |
| N16961 | O1 | 1.21 ± 0.52 |
| TCP-2 | O1 | 0.12 ± 0.09 |
Values represent the mean of six assays in different mice ± SD
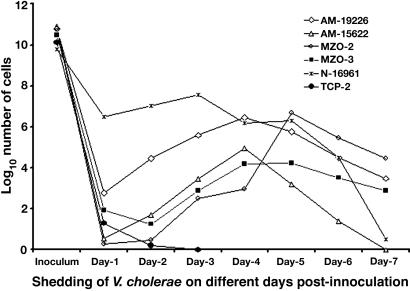
Ability to Induce Diarrhea in the Rabbit Model. The infant mouse assay is a simple test for the colonization ability of V. cholerae but is less reliable in scoring the pathogenicity of strains. We therefore used the rabbit RITARD model to test whether these strains could induce diarrhea and colonize the intestine, as evidenced by long-term shedding (30) (Table 2 and Fig. 2). N16961 was used as a positive control and caused diarrhea in five of the six animals (≈83%). Bacterial shedding occurred throughout the 7-day observation period, with a peak shedding of ≈107 cells per g of feces. TCP-2, a CT-TCP- strain that does not colonize human subjects (13) caused no diarrhea and was shed at low levels for only 2 days after inoculation. AM-19226 induced diarrhea in all nine animals that were challenged (100%) and caused fatal diarrhea in three of the nine animals. Animals challenged with AM-19226 shed bacteria for at least 7 days after inoculation, following a trend similar to that seen for N16961, although fewer cells (≈106 cells per g of feces) were recovered at the peak of shedding. The other non-O1, non-O139 strains colonized and displayed virulence in this model but generally at lower levels than AM-19226 (Fig. 2 and Table 2).
Table 2. Rabbit diarrheal response to different V. cholerae strains in the RITARD model.
| Strain* | No. of animals challenged | No. of animals with fatal diarrhea | No. of animals with nonfatal diarrhea† |
|---|---|---|---|
| AM-19226 | 9 | 3 | 6 |
| MZO-2 | 6 | 1 | 3 |
| MZO-3 | 5 | 0 | 2 |
| AM-15226 | 6 | 0 | 4 |
| N16961 | 6 | 0 | 5 |
| TCP-2 | 6 | 0 | 0 |
All strains are CT- TCP- except N16961, which is CT+ TCP+
Rabbits inoculated with non-O1 strains and TCP+ O1 strain N16961 continued to shed the strains in stools for at least 7 days after inoculation
Fig. 2.
Shedding of V. cholerae strains in stools of adult rabbits challenged with the nontoxigenic TCP negative V. cholerae non-O1, non-O139 strains in the RITARD model. N-16961 is a TCP-positive toxigenic V. cholerae O1 strain used as a positive control. TCP-2 is a TCP- and CT-deleted V. cholerae O1 strain used as a negative control. The prolonged shedding of the challenge organisms suggested that the strains colonized the intestines of the rabbits. Values represent the mean of at least five assays in different rabbits.
The in vivo results showed that all four non-O1 non-O139 strains were capable of promoting a secretory response in rabbits. Most strikingly, AM-19226 appeared as potent as seventh pandemic O1 strain N16961 in its ability to induce diarrhea in the RITARD model, and it was also able to colonize infant mice despite being TCP-negative. This strain was therefore chosen for genome sequence analysis to potentially uncover new virulence factors responsible for the pathogenesis observed in the animal models and in human disease.
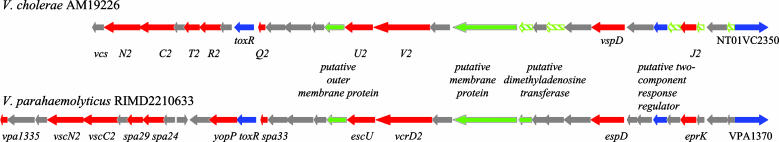
Genomic Sequence Analysis of AM-19226 Reveals Putative ORFs with Homology to V. parahaemolyticus TTSS2. Random shotgun sequencing of AM-19226 generated 284 sequence contigs. Whereas the exact AM-19226 genome size is unknown, sequence coverage is estimated at ≈6.3 fold, based on analogy to the 4.0-Mb sized genome of the sequenced V. cholerae strain N16961. One contig (247) of 31,577 bp has a GC content of 40.13%, which is significantly lower that the average GC content of the entire AM-19226 sequence (47.58%). This contig was of particular interest (Fig. 3) because it contained ORFs with significant similarity to V. parahaemolyticus genes recently shown to encode the functional components of a TTSS (Table 3) (28). We have called these non-O1, non-O139 V. cholerae genes Vibrio cholerae secretion (vcs) loci.
Fig. 3.
Genetic organization of TTSSs from V. cholerae AM19226 and V. parahaemolyticus RIMD2210633. Red arrows designate type III-related genes, blue arrows designate genes encoding putative regulatory and putative effector proteins, and green striped arrows indicate putative ORFs that are not annotated by glimmer. Green arrows designate genes unrelated to TTSS structural components. Gray arrows indicate genes encoding hypothetical proteins.
Table 3.
Results of blast-extend-repraze searches using predicted proteins encoded by V. cholerae AM-19226 ORFs
| AM-19226 ORF | Homolog | Homology span (size) | Identities, % | Positives, % | Description | |
|---|---|---|---|---|---|---|
| VcsN2 (NT01VC2327) | VscN2 (Vp) | VPA1338 | 1–420 (420) | 96 | 98 | ATPase YscN |
| VcsC2 (NT01VC2328) | VscC2 (Vp) | VPA1339 | 1–487 (487) | 93 | 97 | TTSS protein EscC |
| VcsT2 (NT01VC2330) | Spa29 (Vp) | VPA1341 | 1–227 (227) | 90 | 97 | Mxi-Spa secretion protein Spa29 |
| VcsR2 (NT01VC2331) | Spa24 (Vp) | VPA1342 | 20–243 (243) | 86 | 90 | TTSS protein Spa24 |
| VcsQ2 (NT01VC2334) | Spa33 (Vp) | VPA1349 | 1–70 (71) | 94 | 94 | TTSS protein Spa33 |
| VcsU2 (NT01VC2339) | EscU (Vp) | VPA1354 | 13–337 (345) | 87 | 91 | TTSS protein EscU |
| VcsV2 (NT01VC2340) | VcrD2 (Vp) | VPA1355 | 1–627 (627) | 83 | 88 | TTSS protein EscV |
| VspD (NT01VC2345) | EspD (Vp) | VPA1362 | 1–349 (349) | 81 | 85 | Secreted protein EspD |
| VcsJ2 (NT01VC2348) | EprK (Vp) | VPA1367 | 1–177 (177) | 80 | 86 | TTSS lipoprotein precursor EprK |
| ToxR (NT01VC2333) | ToxR (Vp) | VPA1348 | 1–183 (183) | 83 | 90 | ToxR transcriptional activator |
| ToxR (Vp) | VP820 | 8–127 (292) | 33 | 54 | ||
| ToxR (Vp) | VPA1332 | 29–101 (253) | 39 | 58 | ||
| ToxR (V. cholerae) | 20–145 (294) | 34 | 57 | |||
| Tdh | TdhA (Vp) | VPA1314 | 8–188 (189) | 48 | 71 | TDH |
| TdhS (Vp) | VPA1378 | 8–187 (189) | 48 | 70 | ||
| NT01VC2350 | hypothetical (Vp) | VPA1370 | 1–469 (484) | 53 | 67 | WH2 motif domain protein |
The vcs genes are predicted to encode the structural components of the TTSS membrane-associated pore-forming complex (vcsCJRTQVU) and an ATPase that provides energy to drive secretion by means of the apparatus (VcsN). Among the nine conserved proteins in TTSS complexes (CJNQRSTUV) (45), only the gene encoding a VscS homolog was absent from the AM-19226 genome sequence database. VPA1335 from V. parahaemolyticus is weakly similar to RhcS from Bradyrhizobium japonicum (26% identities and 55% positives), and a homolog of these two genes (vcsS) was found outside contig 247. V. parahaemolyticus contains two type III secretions systems, TTSS1 and TTSS2 (28, 46). The V. parahaemolyticus TTSS2 and the TTSS gene cluster on contig 247 from AM-19226 differ slightly, as exemplified by the absence of yopP downstream of toxR. However, sequence annotation of the AM-19226 genome did not identify any ORFs predicted to encode homologs of YscF and YscI, two of the proteins in the needle complex that delivers secreted effector proteins into target eukaryotic cells (47). In other systems, the genes encoding YscI and its homologs are always upstream of the highly conserved YscJ (48). In AM-19226, we found a small ORF upstream of ycsJ that shares characteristics with proteins of the YscI family (49). Finally, although strong homology to V. parahaemolyticus TTSS2 was found, no genes with homology to V. parahaemolyticus TTSS1 were identified in AM-19226. Southern analysis by using probes against V. parahaemolyticus TTSS1 genes VP1671 and VP1690 also failed to identify genes homologous to TTSS1 (data not shown).
Other ORFs within the AM-19226 TTSS contig also shared significant homology with V. parahaemolyticus genes (Table 3). These ORFs included two putative regulatory proteins with similarity to the ToxR family of transcriptional regulators, and a gene encoding a WH2 domain-containing protein (NT01VC2350). A protein with ≈70% similarity to the thermostable direct hemolysin (TDH) encoded by V. parahaemolyticus strains of the pandemic O3:K6 serotype (46, 50) was identified outside contig 247.
Distribution of TTSS-Related Genes in V. cholerae Strains. To determine the prevalence of these genes in other non-O1, non-O139 strains, Southern analysis was performed by using TTSS probes corresponding to AM-19226 ORFs predicted to encode proteins with high homology to the V. parahaemolyticus VscN2, VscC2, VcrD2, and EspD proteins (28, 46). O1 strain N16961 was included as a negative control. Positive hybridization was observed for several strains for all four TTSS probes, and the restriction pattern of hybridizing bands was the same in the majority of positive strains (see Table 4 and Fig. 4 and Table 5, which are published as supporting information on the PNAS web site). Surprisingly, all CTX+TCP+ clinical O141 isolates tested were positive for the TTSS. Five strains isolated from the Bangladesh environment were also positive for the four TTSS probes, whereas 20 other strains isolated from clinical and environmental sources in Bangladesh were negative, including MZO-2, MZO-3, and AM-15622. Except for O141, there was no strong correlation between the presence of the TTSS genes and the particular serogroup displayed by these strains. We conclude that the TTSS genes identified in AM-19226 are broadly distributed among various non-O1, non-O139 strains, including some clinical and environmental isolates.
Table 4. Results of Southern analysis.
| Strain | Isolation/souce | Serogroup | TTSS |
|---|---|---|---|
| 624–40 | Bangl. 2002; E | N/A* | — |
| 656–59 | Bangl. 2002; E | O10 | — |
| 523–955 | Bangl. 2002; E | O14 | — |
| 831–65 | Bangl. 2002; E | O14 | + |
| 838–77 | Bangl. 2002; E | O14 | + |
| MZO-2 | Bangl. 2001; C | O14 | — |
| V46 | USA 1978; C | O141 | + |
| V47 | USA 1984; C | O141 | + |
| V48 | USA 1985; C | O141 | + |
| V49 | USA 1985; C | O141 | + |
| V50 | USA 1986; C | O141 | + |
| V51 | USA 1987; C | O141 | + |
| V55 | USA 1978; C | O141 | + |
| 877–163 | Bangl. 2002; E | O16 | + |
| 504–931 | Bangl. 2002; E | O18 | — |
| 622–37 | Bangl. 2002; E | O23 | — |
| 612–29 | Bangl. 2002; E | O37 | — |
| MZO-3 | Bangl. 2001; C | O37 | — |
| AM-19226 | Bangl. 2001; C | O39 | + |
| 652–58 | Bangl. 2002; E | O5 | + |
| 871–147 | Bangl. 2002; E | O6 | — |
| 862–133 | Bangl. 2002; E | O79 | — |
| 641–49 | Bangl. 2002; E | O8 | + |
| 842–86 | Bangl. 2002; E | O8 | + |
| AM-15622 | Bangl. 2001; C | O8 | — |
| VP47† | India 1996; C | O3:K6 | +Weak |
Bangl., Bangladesh; C, clinical; E, environmental.
Serogroup not known, but this strain is non-O1, non-O139
V. parahaemolyticus
Discussion
Numerous phylogenetic studies have shown that pathogenic seventh pandemic El Tor O1 and O139 V. cholerae are very closely related and constitute a globally distributed clone (6, 27, 51). A few pathogenic strains of non-O1, non-O139 serogroups such as O141, O10, and O12 have been isolated in outbreaks of gastroenteritis, and similarly show a clonal structure (9–11). In contrast, environmental non-O1, non-O139 strains have typically been reported to be quite diverse. These conclusions were reached by using a variety of techniques, including ribotyping (22), amplified fragment length polymorphism (3), pulsed-field gel electrophoresis (3), and comparative nucleotide sequence analysis (6, 52). These analytical methods are useful in assessing relatedness of strains but are limited in their ability to define gene content differences that might have biological and phylogenetic significance.
In this report, we used microarray analysis to assess the relatedness of four non-O1, non-O139 clinical isolates to O1 classical, seventh pandemic El Tor O1 and O139 V. cholerae. Genome sequence analysis was then used to define gene content differences that might be associated with unique traits of non-O1, non-O139 strain AM-19226 (i.e., its higher pathogenicity). The four non-O1, non-O139 strains (AM-19226, AM-15622, MZO-2, and MZO-3) are CT-TCP- but displayed some degree of virulence in humans and in experimental animal models for gastroenteritis. They were all isolated from patients presenting with severe watery diarrhea. In addition, they were isolated from a region that supports endemic O1 strains, thus increasing our opportunity to discover genes involved in environmental or host adaptation. Finally, the strains came from a region that has been under continuous environmental and clinical surveillance for V. cholerae for nearly three decades.
As expected, array analysis confirmed that the non-O1, non-O139 strains did not contain O1 antigen biosynthetic genes, the CTX phage, or the TCP pathogenicity island. The results also suggest that VSP-I and VSP-II, two regions identified as present only in seventh pandemic strains (25), are absent in AM-19226, AM-15622, and MZO-2. MZO-3 appears to carry VSP-II, but not VSP-I. This finding is interesting given the recent report on the presence of VSP-II in Vibrio vulnificus (42). All four strains are also missing a portion of the nanH region (25), recently described as the VPI-2 pathogenicity island (43). Interestingly, this island is found more frequently in toxigenic O1 strains (43, 52) and encodes neuraminidase, an enzyme that can generate additional GM1 receptors for cholera toxin. Each of the four strains appears to lack ≈6% (>250) of the genes that are present in the O1 seventh pandemic strain N16961 (Fig. 1 and Table 6, which is published as supporting information on the PNAS web site). One-third of the putatively absent genes reside on the small chromosome, and approximately half of these genes appear to lie within the integron island, a gene-capture system (53, 54). Collectively, our results suggest that these four non-O1, non-O139 strains have a phylogenetic origin that is quite different from O1 strains. Our earlier microarray analysis found that seventh pandemic O1 El Tor and O139 strains, pre-seventh pandemic O1 El Tor strains, classical biotype strains, and a environmental Gulf Coast O1 strain (2740–80) have strikingly similar gene content (25). By using comparative nucleotide sequence analysis of the mdh gene, O'Shea et al. (52) reached similar conclusions about O1 strains but also noted that most non-O1, non-O139 strains appear to have a different phylogenic origin than O1 strains (52). Indeed, our sequence of the mdh gene of AM-19226 shows five informative polymorphisms (C325T, G606A, T669C, T723C, and C750T) relative to N16961 (data not shown). This pattern most closely matches that of other non-O1, non-O139 strains analyzed by O'Shea et al. (52), such as the O141 strain V46. Interestingly, both AM-19226 and V46 carry the TTSS gene cluster described here (see below).
One limitation of microarray-based comparative genomic studies is the inability to identify genes present in the test strain but absent in the reference strain. For this reason, we sequenced the genome of the non-O1, non-O139 strain AM-19226, which had the most robust response in animal models for colonization and pathogenicity, despite being CTX-TCP-. Our random shotgun sequencing provided ≈6-fold coverage of the AM-19226 genome, assuming that it is comparable in size to the N16961 genome (i.e., 4.0 Mb). As expected, preliminary annotation of the AM-19226 sequence revealed numerous potential virulence genes based on homology to genes in other bacterial species, including those involved in biogenesis of type IV pili and several toxins (data not shown). In particular, contig 247 was found to carry nine genes predicted to encode proteins with significant similarity to the functional components of TTSS (27). Hybridization probes derived from four putative AM-19226 TTSS genes were used to confirm that this system is present in other non-O1, non-O139 strains. Previously, Park et al. (28) concluded that TTSS1 and TTSS2 genes present in V. parahaemolyticus were not present in the V. cholerae strains they analyzed, although TTSS1 gene sequences were found in other Vibrio species.
TTSSs are well known virulence factors and function in several Gram-negative enteric pathogens, including Salmonella, Shigella, Escherichia, Yersinia, Pseudomonas, Vibrio, and Aeromonas species. Recently, the V. parahaemolyticus genome sequence revealed that this species encodes two TTSS gene clusters; TTSS1 is located on the large chromosome, and TTSS2 is located on the small chromosome (28, 46). The TTSS2 genes have been shown to contribute to the enterotoxicity of V. parahaemolyticus, whereas the TTSS1 genes seem to be involved in cytotoxicity (28). The TTSS system of AM-19226 is most similar to the V. parahaemolyticus TTSS2 gene cluster. Although conclusive evidence requires full genetic analysis, these data strongly suggest that the AM-19226 TTSS contributes to this strain's virulence, and possibly also affects virulence in other non-O1, non-O139 strains. However, this conclusion must be tempered by the fact that our sequence analysis did not detect TTSS genes encoding all of the essential components of the needle complex that typically is required for the delivery of TTSS effector proteins into target eukaryotic cells (47). Additional sequence coverage of the AM-19226 genome may reveal these genes. It is also possible that the TTSS found here serves simply to secrete effector proteins rather than deliver them into target cells. In this regard, it should be noted that we identified an ORF outside contig 247 that is predicted to encode a protein with ≈70% similarity to the TDH of V. parahaemolyticus (50). TDH is an important secreted toxin that, like the genes for TTSS2, is most often carried by clinical isolates and pandemic clones of V. parahaemolyticus (50). Also, the V. parahaemolyticus TTSS2 cluster and the AM-19226 TTSS cluster both contain a gene that is highly similar to the toxR gene of V. cholerae. TDH expression is regulated by a toxR-like gene in V. parahaemolyticus (55) and in V. cholerae O1 strains, ToxR controls expression of virulence factors (such as TCP and CT) and other less characterized genes (56). Thus, it will be of interest to determine whether TDH is regulated by ToxR, and secreted by the TTSS in V. cholerae strains that carry these genes. Similarly, the potential exists for these non-O1, non-O139 strains to influence the organization of eukaryotic cell components such as actin through the action of the predicted product of NT01VC2350, which shows homology to the WH2/WASP family of actin-binding proteins (data not shown). Understanding the role of this newly identified V. cholerae TTSS in the pathogenesis of some non-O1, non-O139 strains, and whether the action of putative effector proteins relies on a functioning TTSS, awaits further experimentation.
V. cholerae are found in contact with eukaryotic cells not only within the human host but also in the marine environment where the bacteria can associate with insects, plankton, copepods, and shrimp (57–59). Considering the prevalence and diversity of non-O1, non-O139 strains in the environment, acquisition of a TTSS may represent one of several strategies that V. cholerae employs to compete for survival within its various niches. It is therefore of interest that O141 strains carry the TTSS genes because this serogroup is unusual in having a global distribution and a clonal phylogeny (9, 60). Thus, it is possible that the TTSS2 system in V. parahaemolyticus pandemic clones (46, 50) and the V. cholerae TTSS may be critical for environmental fitness and therefore for pandemic spread.
Supplementary Material
Acknowledgments
We thank Tanja Davidsen and Dana Boyd for assistance with annotation; and Su Chiang for manuscript preparation and editing. This work was supported by National Institutes of Health Grants AI-18045 and GM-068851 and Ellison Medical Foundation Grant ID-T-0007-01, under a subagreement between Harvard Medical School and the International Centre for Diarrhoeal Disease Research, Bangladesh. The International Centre for Diarrhoeal Disease Research, Bangladesh is supported by countries and agencies that share its concern for the health problems of developing countries. The Department of Energy provided funding for the annotation services offered by The Institute for Genomic Research.
Author contributions: M.D., S.M.F., R.K., and J.J.M. designed research; M.D., D. Serruto, V.C.T., D. Sturtevant, P.D., S.M.F., M.H.R., K.T.M., and G.G. performed research; S.M.F., J.F.H., and R.K. contributed new reagents/analytic tools; M.D., D. Serruto, V.C.T., D. Sturtevant, P.D., S.M.F., M.H.R., J.F.H., K.T.M., G.G., R.K., and J.J.M. analyzed data; and M.D., V.C.T., and J.J.M. wrote the paper.
Abbreviations: CT, cholera toxin; TCP, toxin-coregulated pilus; TTSS, type III secretion system; VSP, Vibrio seventh pandemic; TDH, thermostable direct hemolysin; gDNA, genomic DNA; RITARD, removable intestinal tie-adult rabbit diarrhea.
References
- 1.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Dziejman, M., Rahman, M. H., Sack, D. A., Nair, G. B. & Mekalanos, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh, D. V., Matte, M. H., Matte, G. R., Jiang, S., Sabeena, F., Shukla, B. N., Sanyal, S. C., Huq, A. & Colwell, R. R. (2001) Appl. Environ. Microbiol. 67, 910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faruque, S. M., Nasser, I. B., Islam, M. J., Faruque, A. S. G., Ghosh, A. N., Nair, G. B., Sack, D. A., Makalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA, 102, 1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada, T., Arakawa, E., Itoh, K., Okitsu, T., Matsushima, A., Asai, Y., Yamai, S., Nakazato, T., Nair, G. B., Albert, M. J. & Takeda, Y. (1994) Curr. Microbiol. 28, 175-178. [Google Scholar]
- 6.Karaolis, D. K., Lan, R. & Reeves, P. R. (1995) J. Bacteriol. 177, 3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque, S. M., Sack, D. A., Sack, R. B., Colwell, R. R., Takeda, Y. & Nair, G. B. (2003) Proc. Natl. Acad. Sci. USA 100, 1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris, J. G. (1990) Epidemiol. Rev. 12, 179-191. [DOI] [PubMed] [Google Scholar]
- 9.Dalsgaard, A., Serichantalergs, O., Forslund, A., Lin, W., Mekalanos, J., Mintz, E., Shimada, T. & Wells, J. G. (2001) J. Clin. Microbiol. 39, 4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudra, S., Mahajan, R., Mathur, M., Kathuria, K. & Talwar, V. (1996) Indian J. Med. Res. 103, 71-73. [PubMed] [Google Scholar]
- 11.Bagchi, K., Echeverria, P., Arthur, J. D., Sethabutr, O., Serichantalergs, O. & Hoge, C. W. (1993) J. Clin. Microbiol. 31, 1315-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou, T. Y., Liu, J. W. & Leu, H. S. (2003) J. Microbiol. Immunol. Infect. 36, 117-122. [PubMed] [Google Scholar]
- 13.Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K. & Levine, M. M. (1988) J. Exp. Med. 168, 1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldor, M. K. & Mekalanos, J. J. (1996) Science 272, 1910-1914. [DOI] [PubMed] [Google Scholar]
- 15.Faruque, S. M., Zhu, J., Asadulghani, Kamruzzaman, M. & Mekalanos, J. J. (2003) Infect. Immun. 71, 2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barua, D. & Greenough, W. B. (1992) Cholera (Plenum, New York).
- 17.Wachsmuth, I. K., Evins, G. M., Fields, P. I., Olsvik, Ø., Popovic, T., Bopp, C. A., Wells, J. G., Carrillo, C. & Blake, P. A. (1993) J. Infect. Dis. 167, 621-626. [DOI] [PubMed] [Google Scholar]
- 18.Ramamurthy, T., Garg, S., Sharma, R., Bhattacharaya, S. K., Nair, G. B., Shmada, T., Takeda, T., Karasawa, T., Kurazano, H., Pal, A. & Takeda, Y. (1993) Lancet 341, 703-704. [DOI] [PubMed] [Google Scholar]
- 19.Li, M., Shimada, T., Morris, J. G., Jr., Sulakvelidze, A. & Sozhamannan, S. (2002) Infect. Immun. 70, 2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalsgaard, A., Serichantalergs, O., Shimada, T., Sethabutr, O. & Echeverria, P. (1995) J. Med. Microbiol. 43, 216-220. [DOI] [PubMed] [Google Scholar]
- 21.Lin, W., Fullner, K. J., Clayton, R., Sexton, J. A., Rogers, M. B., Calia, K. E., Calderwood, S. B., Fraser, C. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalsgaard, A., Forslund, A., Mortensen, H. F. & Shimada, T. (1998) Epidemiol. Infect. 121, 535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma, C., Thungapathra, M., Ghosh, A., Mukhopadhyay, A. K., Basu, A., Mitra, R., Basu, I., Bhattacharya, S. K., Shimada, T., Ramamurthy, T., et al. (1998) J. Clin. Microbiol. 36, 756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., Haft, D. H., Hickey, E. K., Peterson, J. D., Umayam, L., et al. (2000) Nature 406, 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dziejman, M., Balon, E., Boyd, D., Fraser, C. M., Heidelberg, J. F. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faruque, S. M., Saha, M. N., Asadulghani, Sack, D. A., Sack, R. B., Takeda, Y. & Nair, G. B. (2000) J. Infect. Dis. 182, 1161-1168. [DOI] [PubMed] [Google Scholar]
- 27.Lee, C. A. (1997) Trends Microbiol. 5, 148-156. [DOI] [PubMed] [Google Scholar]
- 28.Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T. & Honda, T. (2004) Infect. Immun. 72, 6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angelichio, M. J., Spector, J., Waldor, M. K. & Camilli, A. (1999) Infect. Immun. 67, 3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faruque, S. M., Kamruzzaman, M., Meraj, I. M., Chowdhury, N., Nair, G. B., Sack, R. B., Colwell, R. R. & Sack, D. A. (2003) Infect. Immun. 71, 1020-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 32.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 33.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 34.Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. (1998) Nucleic Acids Res. 26, 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 37.Smith, T. F. & Waterman, M. S. (1981) J. Mol. Biol. 147, 195-197. [DOI] [PubMed] [Google Scholar]
- 38.Bateman, A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. (2000) Nucleic Acids Res. 28, 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haft, D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T. & White, O. (2001) Nucleic Acids Res. 29, 41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy, S. R. (1998) Bioinformatics 14, 755-763. [DOI] [PubMed] [Google Scholar]
- 41.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944-945. [DOI] [PubMed] [Google Scholar]
- 42.O'Shea Y, A., Finnan, S., Reen, F. J., Morrissey, J. P., O'Gara, F. & Boyd, E. F. (2004) Microbiology 150, 4053-4063. [DOI] [PubMed] [Google Scholar]
- 43.Jermyn, W. S. & Boyd, E. F. (2002) Microbiology 148, 3681-3693. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, D. N., Killeen, K. P., Hack, D. C., Kenner, J. R., Coster, T. S., Beattie, D. T., Ezzell, J., Hyman, T., Trofa, A., Sjogren, M. H., et al. (1994) J. Infect. Dis. 170, 1518-1523. [DOI] [PubMed] [Google Scholar]
- 45.Alfano, J. R. & Collmer, A. (1997) J. Bacteriol. 179, 5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., Iijima, Y., Najima, M., Nakano, M., Yamashita, A., et al. (2003) Lancet 361, 743-749. [DOI] [PubMed] [Google Scholar]
- 47.Marlovits, T. C., Kubori, T., Sukhan, A., Thomas, D. R., Galan, J. E. & Unger, V. M. (2004) Science 306, 1040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimbrough, T. G. & Miller, S. I. (2002) Microbes Infect. 4, 75-82. [DOI] [PubMed] [Google Scholar]
- 49.Hueck, C. J. (1998) Microbiol. Mol. Biol. Rev. 62, 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, H. C., Liu, S. H., Wang, T. K., Lee, C. L., Chiou, C. S., Liu, D. P., Nishibuchi, M. & Lee, B. K. (2000) Appl. Environ. Microbiol. 66, 3981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wachsmuth, I. K., Olsvik, O., Evins, G. M. & Popovic, T. (1994) in Vibrio Cholerae and Cholera: Molecular to Global Perspectives, eds. Wachsmuth, I. K., Blake, P. A. & Olsvik, O. (Am. Soc. Microbiol, Washington, DC), pp. 357-370.
- 52.O'Shea, Y. A., Reen, F. J., Quirke, A. M. & Boyd, E. F. (2004) J. Clin. Microbiol. 42, 4657-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazel, D., Dychinco, B., Webb, V. A. & Davies, J. (1998) Science 280, 605-608. [DOI] [PubMed] [Google Scholar]
- 54.Rowe-Magnus, D. A., Guerout, A. M. & Mazel, D. (2002) Mol. Microbiol. 43, 1657-1669. [DOI] [PubMed] [Google Scholar]
- 55.Lin, Z., Kumagai, K., Baba, K., Mekalanos, J. J. & Nishibuchi, M. (1993) J. Bacteriol. 175, 3844-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S. B. & Mekalanos, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Halpern, M., Broza, Y. B., Mittler, S., Arakawa, E. & Broza, M. (2004) Microb. Ecol. 47, 341-349. [DOI] [PubMed] [Google Scholar]
- 58.Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R. & Colwell, R. R. (1983) Appl. Environ. Microbiol. 45, 275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalsgaard, A., Huss, H. H., H.-Kittikun, A. & Larsen, J. L. (1995) Int. J. Food Microbiol. 28, 101-113. [DOI] [PubMed] [Google Scholar]
- 60.Dalsgaard, A., Albert, M. J., Taylor, D. N., Shimada, T., Meza, R., Serichantalergs, O. & Echeverria, P. (1995) J. Clin. Microbiol. 33, 2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.