Abstract
Of the over 200 identified mammalian microRNAs (miRNAs), only a few have known biological activity. To gain a better understanding of the role that miRNAs play in specific cellular pathways, we utilized antisense molecules to inhibit miRNA activity. We used miRNA inhibitors targeting miR-23, 21, 15a, 16 and 19a to test efficacy of antisense molecules in reducing miRNA activity on reporter genes bearing miRNA-binding sites. The miRNA inhibitors de-repressed reporter gene activity when a miRNA-binding site was cloned into its 3′-untranslated region. We employed a library of miRNA inhibitors to screen for miRNA involved in cell growth and apoptosis. In HeLa cells, we found that inhibition of miR-95, 124, 125, 133, 134, 144, 150, 152, 187, 190, 191, 192, 193, 204, 211, 218, 220, 296 and 299 caused a decrease in cell growth and that inhibition of miR-21 and miR-24 had a profound increase in cell growth. On the other hand, inhibition of miR-7, 19a, 23, 24, 134, 140, 150, 192 and 193 down-regulated cell growth, and miR-107, 132, 155, 181, 191, 194, 203, 215 and 301 increased cell growth in lung carcinoma cells, A549. We also identified miRNA that when inhibited increased the level of apoptosis (miR-1d, 7, 148, 204, 210, 216 and 296) and one miRNA that decreased apoptosis (miR-214) in HeLa cells. From these screens, we conclude that miRNA-mediated regulation has a complexity of cellular outcomes and that miRNAs can be mediators of regulation of cell growth and apoptosis pathways.
INTRODUCTION
Cellular microRNAs (miRNAs) are a class of 17–24 base single-stranded RNA molecules that are expressed in cells from plants to animals (1). MiRNAs are expressed as long precursor RNAs that get processed by a cellular nuclease, Drosha, before being transported by an Exportin-5-dependent mechanism into the cytoplasm (2). Once in the cytoplasm miRNAs are cleaved further by the enzyme DICER (3,4) and the resulting 17–24 nt miRNAs associate with a cellular complex that is at least similar to the RNA-induced silencing complex that participates in RNA interference (5). The complex-bound single-stranded miRNA guides the complex to mRNAs with sequences that are at least partially complementary to the miRNA. The translation of the bound mRNA is inhibited by a mechanism that is not fully understood (6).
MiRNAs are a very prevalent class of cellular RNAs, but because they have only recently been identified, very few miRNAs have known cellular functions. Currently, the best understood miRNA, lin-4, was first identified in Caenorhabditis elegans [reviewed in (7,8)]. Research revealed that lin-4 accumulates during the first and second larval stages and triggers passage to the third larval stage by repressing the translation of at least two genes, lin-14 and lin-28 (9). The activity of lin-4 depends on the partial homology of the miRNA to specific regions of the 3′-untranslated regions (3′-UTRs) of the lin-14 and lin-28 mRNAs (9,10). A second miRNA, let-7 accumulates during C.elegans larval development and triggers passage from late larval to adult cell fates (11,12). A few other miRNAs, such as bantam and miR-14, have at least partial defined roles in cells (13,14). Currently, only a few mammalian miRNAs have been shown to have a defined role in a biological process while associations have implicated others. In one example, the mammalian miRNA, miR-181, was found to be specifically expressed and dynamically regulated in hematopoietic cells, and its expression in hematopoietic stem/progenitor cells increased the fraction of B-cells in both tissue culture and adult mice (15).
Four reports have correlated aberrant miRNA expression with cancer, cancer-associated genomic regions and fragile sites in chromosomes. First, loss at 13q14 constitutes the most frequent chromosomal abnormality in chronic lymphocytic leukemia (CLL), suggesting the involvement of one or more tumor suppressor genes at this locus. Although several groups had performed detailed genetic analyses, including extensive loss of heterozygosity, mutation and expression studies, no consistent involvement of any of the genes with open reading frames located in the deleted region was demonstrated. Interestingly, the genes for miR-15 and miR-16 are located at this locus and appear to be deleted in the majority of B-CLL cases (16). Second, studies of miRNA expression in colonic adenocarcinoma and normal mucosa were used to identify potential links between miRNA expression/maturation and cancer (17). Out of 28 miRNAs identified in human colorectal mucosa, two (miR-143 and miR-145) proved to be significantly down-regulated in 12 adenocarcinoma samples compared with matched, normal tissues. Third, the human BIC RNA is elevated in children with Lymphoma. Metzler and co-workers (18) indicate that the BIC gene encodes miR-155. Using PCR, they demonstrate that the expression of the precursor of miR-155 is found in children with Burkitt Lymphoma, but not patients with pediatric leukemia. Fourth, in a recent study, the chromosomal locations of 186 miRNA genes were mapped and compared with the location of non-random genetic alterations (19). Over 52% of the miRNA genes analyzed are in cancer-associated genomic regions or in fragile sites. This study also found that several miRNAs located in deleted regions are expressed at low levels in cancer samples.
As stated above, miRNA bind to mRNA targets and inhibit translation by a currently unknown mechanism. While several publications predict target genes for Drosophila and human miRNA (19,20), only a few have been confirmed using reporter genes. In the most comprehensive study to date, Lewis et al. (19) predicted mRNA targets for human miRNA and used a reporter construct with the 3′-UTRs of 16 target genes to confirm target sites for 8 miRNAs. The system of transfecting miRNA or miRNA expression vectors and reporter constructs with predicted miRNA-binding sites offers the most straightforward method for confirming target sites; however, it does not demonstrate biological significance. In light of the need to identify miRNA targets and their corresponding function, miRNA inhibitors and miRNA expression systems will also be valuable (15,21–23). MiRNA inhibitors have also recently been proven to be valuable in understanding miRNA function (22–24). Not only were the miRNA inhibitors shown to be active in human cells, but the let-7 antisense molecules were injected into C.elegans and found to induce a let-7 loss-of-function phenotype (24), validating the use of these molecules for miRNA inhibition.
We used a library of miRNA inhibitors in functional screening assays to identify miRNAs that affect cell proliferation and apoptosis. Before screening, we validated the efficacy of the miRNA inhibitors using a luciferase reporter bearing miRNA target sequences cloned into its 3′-UTR. In each case, the miRNA inhibitors inhibited the repression of the endogenous miRNA on the reporter gene. Screening our library of miRNA inhibitors targeting over 90 human miRNAs resulted in several interesting observations. First, the hits identified for cell growth were not the same between different cell lines. Second, both increases and decreases in cell number and apoptosis were identified. Our results indicate that miRNA-mediated gene regulatory pathways are complex and have roles in important biological processes.
MATERIALS AND METHODS
Reporter vectors and DNA constructs
To generate reporter vectors bearing miRNA-binding sites, we generated direct match miRNA target sites and cloned these inserts into the multiple cloning sites in the luciferase reporter vector described in Figure 1A. The sense and antisense strands of the oligonucleotides were annealed by adding 2 μg of each oligonucleotide to 46 μl of annealing solution (100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4 and 2 mM Magnesium acetate) and incubated at 90°C for 5 min and then at 37°C for 1 h. The annealed oligonucleotides were digested with HindIII and SpeI and used to ligate into HindIII and SpeI of pmir-Report luciferase (Ambion, Inc.). The oligonucleotides used in these studies were mir 21, 5′-AATGCACTAGTTCAACATCAGTCTGATAAGCTAGCTCAGCAAGCTTAATGC and 5′-GCATTAAGCTTGCTGAGCTAGCTTATCAGACTGATGTTGAACTAGTGCATT; mir 19a, 5′-AATGCACTAGTTCAGTTTTGCATAGATTTGCACAGCTCAGCAAGCTTAATGC and 5′-GCATTAAGCTTGCTGAGCTGTGCAAATCTATGCAAAACTGAACTAGTGCATT; mir 23, 5′-AATGCACTAGTGGAAATCCCTGGCAATGTGATGCTCAGCAAGCTTAATGC and 5′-GCATTAAGCTTGCTGAGCATCACATTGCCAGGGATTTCCACTAGT GCATT; mir15, 5′-AATGCACTAGTCACAAACCATTATGTGCTGCTAGCTCAGCAAGCTTAATGC and 5′-GCATTAAGCTTGCTGAGCTAGCAGCACATAATGGTTTGTGACTAGTGCATT; and mir 16, 5′-AATGCACTAGTCGCCAATATTTACGTGCTGCTAGCTCAGCAAGCTTAATGC and 5′-GCATTAAGCTTGCTGAGCTAGCAGCACGTAAATATTGGCGACTAGTGCATT. A BlpI site (underlined) was added into each insert to test for positive clones.
Figure 1.
(A) MiRNA reporter vectors used to analyze miRNA activity. The restriction sites for cloning the miRNA-binding sites are located in the 3′-UTR of the luciferase gene in the pmir-Report luciferase vector. Once cloned the miRNA-binding site vector is co-transfected with the pmir-Report β-gal vector as a control for transfection efficiency and a miRNA inhibitor molecule that is specific to the binding site or is mutated as compared with the binding site. (B) Enhanced expression of miRNA-regulated reporter by miRNA inhibitors. HeLa cells were plated at 50 000 cells/well in 24-well plates. Cells were transfected using lipofectamine 2000 in duplicate with pmir-REPORT β-gal, a luciferase reporter construct that contained one target site for either miR-23, miR-21, miR-15a, miR-16 or miR-19a and either inhibitors for these miRNA or a negative control (NC). HeLa cells were also transfected with a control luciferase reporter vector lacking a miRNA-binding site without an inhibitor to demonstrate the level of activity that can be achieved for an unmodified pmir-REPORT luciferase. Twenty-four hours post-transfection cells were assayed for luciferase and β-gal expression, and β-gal is used to normalize for differences in transfection efficiency.
Sequence of miRNA inhibitors
The sequences of the inhibitors used in our studies are listed in the Supplementary Material and are the exact antisense copy of the mature miRNA sequence that can be found in the miRNA Registry (25), where all the nucleotides in the inhibitors contain 2′-OMe modifications at every base and a 3′ C3 containing amino linker.
Transfections
All transfections were carried out in triplicate. To transfect, we diluted 5 pmol inhibitor into 10 μl Opti-Mem into each well. We next diluted 0.3 μl NeoFx (Ambion, Inc.) into 10 μl Opti-MEM for each sample, incubated for 10 min at room temperature and added 10 μl of diluted transfection mixture to wells that already contain the inhibitors and incubate for another 10 min at room temperature. We next added 100 μl of diluted cell suspension mixture containing 8000 cells on top of the complex. After 24 h, the medium was changed and the samples were assayed after 72 h.
To transfect the miRNA inhibitors with reporter vectors, we pre-plated 50–60 K HeLa cells 24 h prior to transfection in 24-well tissue culture plates. The next day ∼200 ng of each vector (control and experimental vector) and 10–30 pmol inhibitor were diluted into 50 μl Opti-MEM into round-bottom polystyrene tubes. Next, 3 μl of lipofectamine 2000 was diluted into 50 μl Opti-MEM and incubated at room temperature for 5 min. The diluted DNA/inhibitor was next added into the transfection agent complex and incubated at room temperature for 20 min. The growth medium was changed and 100 μl of the complex was added to the cells. Twenty-four hours following transfection reporter activity was measured.
Analysis of cell growth
To determine the number of HeLa cells remaining following transfection with the miRNA inhibitors, we fixed cells with 4% paraformaldehyde for 5 min 72 h post-transfection, permeablized with 0.1% Triton X-100 for 5 min and stained with propidium iodide. The cells were next counted using the Acumen Explorer (TTP LabTech). The relative number of cells was normalized against a negative control inhibitor, gapas, used in these experiments. To determine the number of A549 cells remaining following transfection with the miRNA inhibitors, we stained the cells using ViaCount Flex Reagent and analyzed for cell number using the Guava PCA-96 (Personal Cell Analysis). The relative cell numbers obtained from the Guava instrument were also graphed and normalized against a negative control miRNA inhibitor gapas as described above. The negative control inhibitor used in these experiments is the same sequence as a section of the GAPDH mRNA and functionality inhibits GAPDH siRNA activity (data not shown).
Caspase activity assay
The level of apoptosis in the transfected cells was determined by measuring the induction of caspase-3 activity as follows. First, the cells were washed once with phosphate-buffered saline, lysed by adding 40 μl of cold lysis buffer (50 mM HEPES, pH 7.2, 40 mM NaCl, 0.5% NP-40 and 0.5 mM EDTA) to the wells and incubated for 20 min at 4°C. Half the sample was used for analysis of caspase activity and the other half was used for analysis of esterase activity that is described below. To the half that was used for analysis of caspase-3, 160 μl of buffer (50 mM HEPES, pH 7.4, 0.1% CHAPS, 0.1 mM EDTA and 10% sucrose) +5 mM DTT containing 20 μM DEVDafc (fluorogenic substrate N-acetyl-asp-glu-val-asp-afc) substrate was added and fluorescence was measured at 400 nm/500 nm excitation/emission.
To normalize the caspase results, half of the samples were also analyzed for esterase activity. First, the fluorescein diacetate (FDA) substrate (0.4 mg/ml FDA in acetonitrile) was diluted 1:19 into dilution buffer (40 mM Tris–HCl, pH 7.5, 20 mM NaCl, 0.5% NP-40, 0.02 mg/ml final concentration). Samples were incubated for 10 min on ice, and 160 μl of diluted FDA substrate was added to each well. Fluorescence was measured for 30 min using excitation of 488 nm and emission of 529 nm.
RESULTS
Confirmation of miRNA inhibitor activity
To confirm the activity of miRNA inhibitors, we constructed reporter vectors that contained miRNA-binding sites for miR-23, miR-21, miR-15a, miR-16 and miR-19a. These binding sites were made by hybridizing oligonucleotides bearing the miRNA-binding sites and cloning them into the HindIII and SpeI sites of the pmir-REPORT luciferase vector (Figure 1A). This places the miRNA-binding site directly into the 3′-UTR of the luciferase gene. We tested the activity of the miRNA inhibitors by transfecting the luciferase reporter vector bearing the miRNA-binding site with the miRNA inhibitor and the pmir-Report β-gal vector (Figure 1A) as a control for transfection efficiency. In addition, we transfected the luciferase vectors bearing the miRNA-binding sites and β-gal vectors with a negative control inhibitor that does not target any cellular miRNA (gapas). Following transfection into HeLa cells, both luciferase and β-gal activity were measured and the relative level of luciferase was normalized against the β-gal readings to control for transfection variation. In each case, the miRNA inhibitors were effective at inhibiting the ability of the endogenous miRNA to inhibit the expression of the reporter gene containing the miRNA-binding site (Figure 1B). This indicates that the miRNA inhibitors are effective at inhibiting miRNA function. The extent of the induction of luciferase activity is different for several inhibitors. We have found that many of these miRNAs are expressed at varying levels in HeLa cells and this could be what is accounting for the differential effects between inhibitors and reporter vectors (Supplementary Figure 2).
Identification of miRNA involved in cell growth
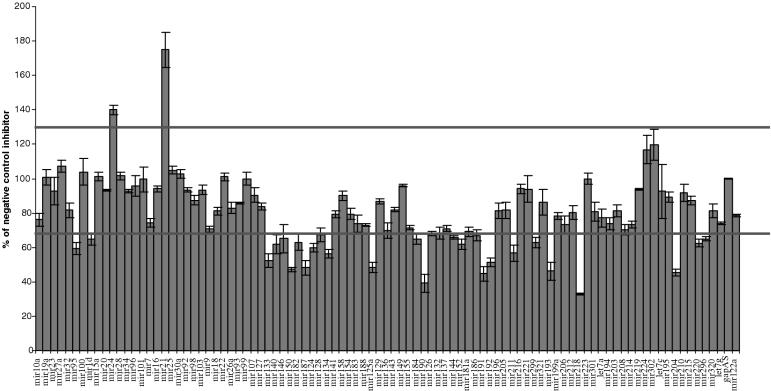
After validating the miRNA inhibitors, we produced a library of over 90 miRNA inhibitors and screened for miRNAs that were important for growth in the cervical cancer-derived cell line, HeLa. In each well of a 96-well plate, an miRNA inhibitor targeting a different miRNA was transfected as described in Materials and Methods. The cells were analyzed 72 h post-transfection for cell number and the numbers obtained were normalized to the negative control gapas inhibitor. We identified a hit as anything that was 30% greater or less than gapas. Each transfection was performed in triplicate, and the standard deviations are represented on the graph shown in Figure 2. Inhibitors that had error bars that were inside the cutoff were not included as hits. Using these criteria, we identified 19 miRNA that inhibited cell growth following inhibition in HeLa cells (miR-95, 124, 125, 133, 134, 144, 150, 152, 187, 190, 191, 192, 193, 204, 211, 218, 220, 296 and 299) and 2 miRNA that had profound increases in cell growth (miR-21 and miR-24).
Figure 2.
Identification of miRNAs that alter cell proliferation in HeLa cells. In 96-well plates, 8000 HeLa cells were reverse transfected with miRNA inhibitors (5 pmol) in triplicates using Ambion siPORT Neo-FX. Seventy-two hours post-transfection, cells were fixed with 4% paraformaldehyde, permeablized with 0.1% TritonX-100 and stained with propidium iodide to look at total cell number. The plates were scanned using the TTP LabTech Acumen Explorer.
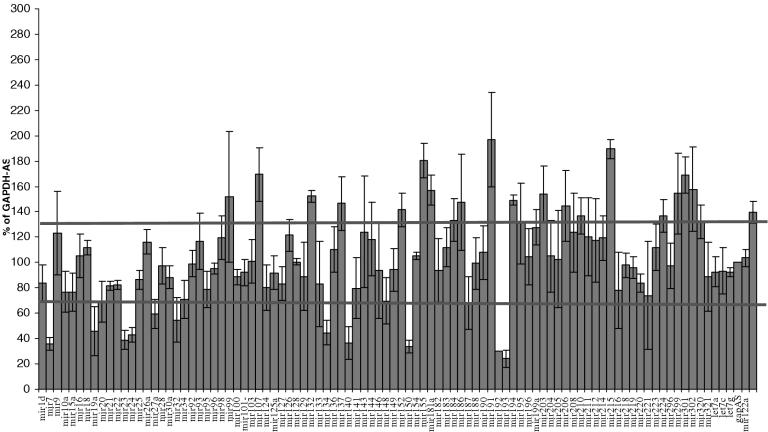
In order to analyze whether the same miRNAs are involved in cell growth in different cell lines, we transfected the library of miRNA inhibitors into the lung carcinoma cell line, A549. Following transfection and analysis of cell number, we used the same criteria for a hit that was used above (30% above and below the gapas control transfection). As shown in Figure 3, 9 miRNAs inhibitors that down-regulated cell growth include miR-7, 19a, 23, 24, 134, 140, 150, 192 and 193 miRNAs, and 9 miRNA inhibitors that were found to increase cell growth include miR-107, 132, 155, 181a, 191, 194, 203, 215 and 301. Among these 18 miRNAs, miR-24, 134, 150, 191, 192 and 193 were also identified in the HeLa screen, miR-24, inhibitors down-regulated growth in A549 but increased cell growth in HeLa, while the other miRNAs decreased cell growth in both A549 and HeLa cells. These data demonstrate the complexity of the regulatory role miRNA play in cell growth regulation between cells and is discussed in further detail below.
Figure 3.
Screen for miRNA involved in cell viability in A549 cells. In 96-well plates, 8000 HeLa cells were reverse transfected with miRNA inhibitors (5 pmol) in triplicates using Ambion siPORT Neo-FX. Seventy-two hours post-transfection, cells were trypsinized and transferred to a non-tissue culture plate, so that cells would not adhere. Cells were stained using ViaCount Flex Reagent and analyzed for total cell number using the Guava PCA-96 (Personal Cell Analysis).
Identification of miRNA involved in apoptosis
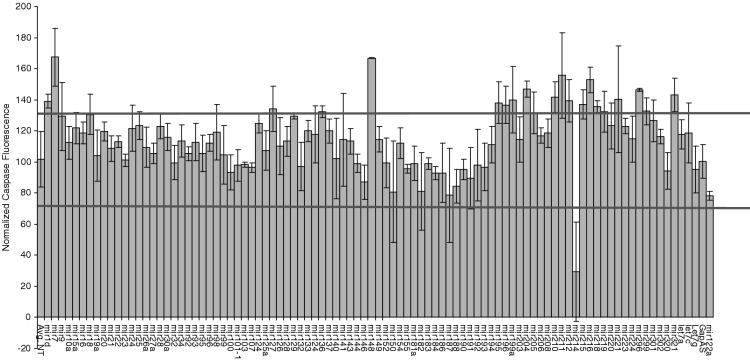
We next screened for miRNAs that were involved in apoptosis in HeLa cells to identify correlations between hits that were found to reduce/increase cell number with miRNAs that reduced/increased relative amounts of apoptosis. To perform this experiment, we transfected HeLa cells with the library of miRNA inhibitors and analyzed the effects the inhibitors had on the activation of caspase-3 activity. Activation is one marker often used for analyzing the induction of apoptosis. The amount of caspase activation was compared relative with that observed with the negative control inhibitor molecule (gapas). Any miRNA that changed caspase-3 activity greater than or less than 30% compared with gapas was considered a hit. In Figure 4, we found 7 miRNA that increased the level of apoptosis (miR-1d, 7, 148, 204, 210, 216 and 296) and one miRNA that decreased apoptosis (miR-214). This data suggest that specific miRNAs are involved in the cell death response.
Figure 4.
Effects of miRNA inhibitors on caspase activity in HeLa. In 96-well plates, 8000 HeLa cells were reverse transfected with miRNA inhibitors (5 pmol) in triplicates using Ambion siPORT Neo-FX. Seventy-two hours post-transfection, cells were analyzed for caspase and esterase activity as described in the Materials and Methods. Esterase activity was used to normalize for differences in cell number that may exist owing to the miRNA inhibitor causing defects in cell growth.
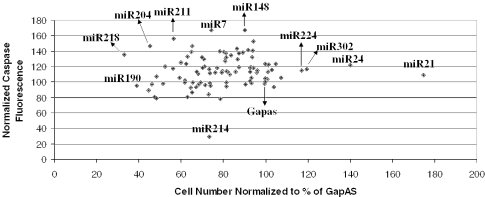
In order to analyze interactions between miRNA that influenced cell death in HeLa cells with those that influenced apoptosis, we graphed the miRNA as a function of cell number compared with caspase-3 activity. This representation of the data quickly points out the hits that cause increases/decreases in cell growth and increases/decreases in caspase activity (Figure 5). In particular, miR-218 was found to inhibit cell growth and induce apoptosis. Thus, for miR-218, growth inhibition may be due to the induction of apoptosis. While for miR-190, cell growth inhibition did not coincide with the induction of apoptosis, suggesting that it may be inhibiting cell growth through cell cycle arrest. These data further point out the complexity of the regulatory circuitry of miRNA on biological processes.
Figure 5.
Effects of miRNA inhibitors on caspase activity compared with cell growth in HeLa cells. We graphed the data that were generated from Figures 2 and 4 together to compare hits that affected cell proliferation with those that affected apoptosis.
DISCUSSION
Reporter genes have been useful to confirm predicted miRNA-binding sites (19,20) making them a useful tool for the initial analysis of potential miRNA-binding sites. However, without evidence to demonstrate that the endogenous miRNA is changing the expression of the endogenous gene, it is difficult to know whether these results are biologically relevant. This will require that the antibody against the predicted products is available and this is not always the case. Since predicted miRNA targets are a good roadmap for possible targets, but will not always be correct, we used miRNA-binding sites that contain direct matches to the endogenous miRNA to ensure that the miRNA will function on the reporter gene (21,24,26). Since it has been shown that a miRNA can act like a siRNA and cleave the miRNA and target it for degradation, we believe this is an effective approach to have taken to verify activity of the miRNA inhibitors (27).
We synthesized over 90 miRNA inhibitors and used these to screen for miRNAs that were involved in cell growth and apoptosis processes that are among the widely studied gene cell pathways and that have direct relevance to cancer and development. In each assay, the relative cutoff for a hit was made at 30% above and below the level observed for the negative control inhibitor gapas. We could have been less stringent and pulled out additional hits; however, making this cutoff stringent allowed for the identification of only major differences in the biological activity of particular miRNA. We also did not include hits that had standard deviations that fell within the cutoff for a hit thus dropping out additional hits. Although we are not counting many miRNAs as hits due to our stringent cutoff does not mean they are not biologically relevant. It is important to mention that the miRNAs that were hits in our screens caused a phenotype by being inhibited. The miRNA may be mediating this effect through increasing protein expression of their targets that then leads to changes in growth or apoptosis. Thus, it is the gain of protein activity rather than the loss of protein activity that is the possible mechanism involved. Knowing the targets of the miRNA will yield important information about the mechanism of regulation of these processes and how they may be able to be fine-tuned with the help of miRNAs.
We first screened for miRNAs that influenced cell growth in two different types of cancer-derived cell lines, HeLa and A549 cells. From these experiments, we identified a complexity of activity for miRNAs in these different cell lines. For example, in HeLa, we found that inhibition of miR-95, 124, 125, 133, 134, 144, 150, 152, 187, 190, 191, 192, 193, 204, 211, 218, 220, 296 and 299 caused a decrease in cell growth and that inhibition of miR-21 and miR-24 had a profound increase in cell growth. The miR-21 and 24 gene deletions have been shown to be associated with chromosomal deletion sites in cancers. Since the inhibition of these genes was found to increase cell growth, it suggests that the expression of these two miRNA may be an important growth regulator in cells (19). On the other hand, miR-7, 19a, 23, 24, 134, 140, 150, 192 and 193 down-regulated cell-growth and miR-107, 132, 155, 181, 191, 194, 203, 215 and 301 increased cell growth when inhibited in A549 cells. Common miRNAs that decreased cell growth include miR-134, 192 and 193. There were no common miRNAs that increased cell growth. MiR-24 increased cell growth in HeLa but was found to decrease cell growth in A549, while miR-191 increased cell growth in A549 and decreased in HeLa. These differences observed between the same miRNA in different cell lines suggest that the targets of the miRNA may be different, the targets of the miRNA have different activities, the miRNAs are differentially expressed or due to differential transfection efficiency in the two cells types. We have ruled out different levels of transfection efficiency between the two cells lines and have identified significant differences in expression profiles between A549 and HeLa cells. Given this, one reason for observing the different hits is due to different levels of expression (Supplementary Figure 2). In addition, since no common miRNAs were found to increase cell growth between the two cell lines, another reason for observing different effects could still be due to miRNA target differences. It should also be noted that these are primary hits and require substantial downstream validation to sort out false-positive hits that may have occurred.
We also used screening to identify miRNAs involved in induction or inhibition of steady state levels of apoptosis in HeLa cells. For these studies, we identified miR-1d, 7, 148, 204, 210, 218, 296 and 381 as hits that increased the level of apoptosis and miR-214 that decreased apoptosis. In another case, miR-218 caused a decrease in cell growth in HeLa but its inhibition increased the level of apoptosis, suggesting that inhibition of miR-218 may be inhibiting cell growth by inducing apoptosis. In other sets of hits, apoptosis was found unchanged while cell growth either increased or decreased. The majority of those that did have an effect on cell growth but not apoptosis caused inhibition of growth, while only a few hits in general caused increase in growth.
The availability of libraries of miRNA inhibitors as well as methods for high-throughput delivery will make it possible to identify miRNAs involved in any cellular process for which there is a quantitative phenotypic assay. This will ultimately lead not only to a greater understanding of the regulatory mechanisms used to control cell processes but might also reveal key pathways that might be exploited for disease detection, prevention or treatment.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dave Brown and Manu Labourier for critical reading of the manuscript and Joe Krebs for the help with the Caspase and esterase assays. Funding to pay the Open Access publication charges for this article was provided by Ambion, Inc.
REFERENCES
- 1.Bartel D.P. MiRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates miRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MiRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutvagner G., Zamore P.D. A miRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasquinelli A.E., Ruvkun G. Control of developmental timing by miRNAs and their targets. Annu. Rev. Cell Dev. Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. miRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 9.Lee R.C., Feinbaum R.L., Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C.elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 12.Slack F.J., Basson M., Liu Z., Ambros V., Horvitz H.R., Ruvkun G. The lin-41 RBCC gene acts in the C.elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor, Mol. Cell. 2000;4:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 13.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 14.Xu P., Vernooy S.Y., Guo M., Hay B.A. The Drosophila MiRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MiRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 16.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. Frequent deletions and down-regulation of miRNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michael M.Z., O'Connor S.M., van Holst Pellekaan N.G., Young G.P., James R.J. Reduced accumulation of specific miRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 18.Metzler M., Wilda M., Busch K., Viehmann S., Borkhardt A. High expression of precursor miRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;2:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 19.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian miRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 20.Enright A., John B., Gaul U., Tuschl T., Sander C., Marks D. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 22.Boutla A., Delidakis C., Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G., Landthaler M., Dorsett Y., Tuschl T. Sequence-specific inhibition of miRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutvagner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;4:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones S. The miRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Y., Yi R., Cullen B.R. MiRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.