Abstract
Like alternative splicing, leaky ribosomal scanning (LRS), which occurs at suboptimal translational initiation codons, increases the physiological flexibility of the genome by allowing alternative translation. Comprehensive analysis of 22 208 human mRNAs indicates that, although the most important positions relative to the first nucleotide of the initiation codon, −3 and +4, are usually such that support initiation (A−3 = 42%, G−3 = 36% and G+4 = 47%), only 37.4% of the genes adhere to the purine (R)−3/G+4 rule at both positions simultaneously, suggesting that LRS may occur in some of the remaining (62.6%) genes. Moreover, 12.5% of the genes lack both R−3 and G+4, potentially leading to sLRS. Compared with 11 genes known to undergo LRS, 10 genes with experimental evidence for high fidelity A+1T+2G+3 initiation codons adhered much more strongly to the R−3/G+4 rule. Among the intron-less histone genes, only the H3 genes adhere to the R−3/G+4 rule, while the H1, H2A, H2B and H4 genes usually lack either R−3 or G+4. To address in vivo the significance of the previously described LRS of H4 mRNAs, which results in alternative translation of the osteogenic growth peptide, transgenic mice were engineered that ubiquitously and constitutively express a mutant H4 mRNA with an A+1→T+1 mutation. These transgenic mice, in particular the females, have a high bone mass phenotype, attributable to increased bone formation. These data suggest that many genes may fulfill cryptic functions by LRS.
INTRODUCTION
The fewer than expected protein-coding sequences in mammalian genomes (1,2) underscore the importance of gene multi-functionality. The most commonly described mechanism of obtaining several proteins from a single gene is alternative splicing, whereby various mRNAs are derived from the same nascent transcript. Deriving diverse proteins from a single mature transcript via alternative translation is emerging as an additional mechanism by which cells utilize a single gene to fulfill multiple functions (3). One type of alternative translation occurs in a cap-independent manner through internal ribosomal entry sites (4). This mechanism was initially described in viral genomes and later in a growing number of cellular mRNAs (4). A second type of alternative translation is mediated by leaky ribosomal scanning (LRS) and is cap-dependent (3). In these cases, the optimal sequence (5′-CCRCCAUGG-3′) surrounding the first AUG initiator is compromised, leading to LRS and translational initiation at both the imperfect (but first, hence most accessible) AUG and additional, downstream, AUG(s). Infrequent AUG initiators that are boldly different from the consensus are inefficient and allow strong leaky ribosomal scanning. Such is the case when both of the most important consensus nucleotides, R−3 and G+4, are concomitantly absent (5,6).
sLRS caused by poor AUG initiators, frequently found in mRNAs encoding regulatory proteins (7), can be expected to result in the accumulation of alternative translation products to levels comparable with those of the protein translated from the first AUG. Weak LRS, which presumably goes unnoticed in many mammalian mRNAs, would result in significant accumulation of an alternative translation product when the mRNA in question is relatively abundant and the alternative translation product relatively stable. Such is the case for histone H4 mRNA. The good, but imperfect H4 first AUG initiation codon allows significant LRS that supports the alternative translation of the osteogenic growth peptide (OGP) from a second, downstream AUG initiator, which is surrounded by an optimal sequence (17). Thus, histone H4 genes, among the most ancient in eukaryotes, lack introns but encode at least two functionally diverse peptides by a mechanism of LRS: (i) the 103 amino acid histone H4 protein, which participates in DNA packaging and in transcriptional regulation through post-translational modifications of its N-terminal tail (9), and (ii) OGP, a circulating peptide identical to the C-terminus of histone H4, which participates in the regulation of bone formation and hematopoesis (8,10).
In the present paper, we provide a genome-wide estimate of the frequency of genes potentially undergoing LRS due to lack of R−3 and/or G+4. To demonstrate the significance of LRS in vivo, we assessed the outcome of histone H4 alternative translation by generating transgenic mice that express a mutant histone H4 mRNA, H4tTG1, which can produce OGP but not histone H4 protein. Transgenic mice, in particular females, have elevated trabecular bone formation, resulting in increased bone mass and improved structural integrity of the trabecular network. These results portray the diverse functionality of histone H4 alternative translation products and highlight the potential biological significance of LRS, a mechanism that likely gives rise to hundreds of yet unstudied peptides.
MATERIALS AND METHODS
Computational methods
Human mRNA sequences were downloaded from the NCBI ftp website (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/), which contains 27 330 mRNA sequences (refseq). Of these, 22 208 files that contained 5′ untranslated region (5-UTR) sequences were used for analysis. Human and mouse histone mRNA sequences (refseq) were downloaded from the GenBank database for each species using histone[TITL] as keyword, and are listed in Supplementary Table S1. To demonstrate sequence conservation around initiation codons, Plot Sequence Logo diagrams were generated as described previously (11–13).
Functional categorization of genes with predicted sLRS was accomplished using the Onto-Tools database suite (14,15) (http://vortex.cs.wayne.edu/Projects.html). First, a master list of 2290 sLRS genes that each correspond to a single unigene cluster was generated using the Onto-Translate application in the Onto-Tools database suite (14,15) and Microsoft Excel to remove repeats. Second, a reference list of 19 494 human mRNA sequences with unique unigene cluster assignments was generated using the same strategy. The Onto-Compare data mining tool in Onto-Tools was then employed to identify function categories defined by the Gene Ontology (GO) Consortium that are over-represented in the sLRS gene group. Over-representation was examined for statistical significance using the binomial distribution setting with the FDR correction factor.
Transgene construction
For over-expression of the H4 alternative translation product, we used the entire H4 coding sequence, excluding the 3′-UTR, of the X13554 rat H4 gene (16), in which we mutated the ATG initiation codon to tTG. This choice was based on the recurrent observation that the H4–CAT fusion gene pSVrH4Δ87–103CAT(T/A) expressed at higher levels compared with the CAT gene in the absence of H4 sequences (17), suggesting that histone H4 mRNA elements may contribute to message stability. The ATG-to-tTG mutation was introduced by PCR-amplification of the X13554 rat H4 gene with the forward primer 5′-TCTTCTgGaTCCATTACTGCTCTACTAGGAAGtTGTCTGG-3′, where the italicized t represents the mutation in the initiation codon and the other letters in lowercase represent substitutions that generate a BamHI restriction site (underlined). The reverse primer was 5′-CAAGGAGAAttCCTAGACAAACG-3′, where letters in lowercase represent substitutions that generate an EcoRI restriction site (underlined). The PCR product was digested with BamHI and EcoRI and inserted by blunt end ligation into the XhoI site of pCAGGS (18), between the chicken β-actin promoter plus intron and the rabbit β-globin 3′-UTR (Figure 4A). This strategy restored the XhoI site at the H4/β-globin junction. The resultant construct, named H4tTG1, also contains the cytomegalovirus enhancer from pCAGGS (Figure 4A). The transgene was separated from vector sequences by digestion with SalI and HindIII and isolated using a gel purification kit (Qiagen, Inc.).
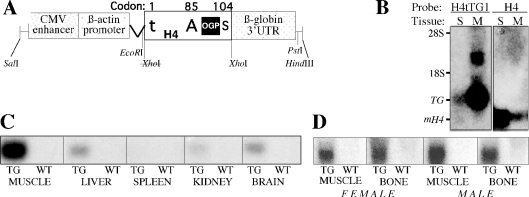
Figure 4.
(A) Schematic illustration of the H4tTG1 transgene. The transgene is composed of the human CMV enhancer, the chicken β-actin promoter and intron, a rat histone H4 coding sequence and the rabbit β-globin 3′-UTR. Within the H4 box, t represents the mutation in the H4 translational initiator (from ATG to tTG at codon #1), A represents the alternative ATG initiation codon (#85), giving rise to preOGP, S represents the stop codon (#104) and the black square depicts the OGP-coding sequence (H490–103). (B) H4tTG1 transgene versus endogenous H4 expression. An aliquot of 20 μg RNA from the spleen (S) and muscle (M) of a 17-week-old transgenic mouse were subjected to northern analysis using as probe either the rabbit β-globin 3′-UTR or a murine histone H4 sequence. 28S and 18S, location of the ribosomal RNA bands; TG, transgenic mRNA; and mH4, endogenous murine H4 mRNA. (C and D) Ubiquitous H4tTG1 transgene expression. Transgene expression was measured by northern analysis of 20 μg RNA from the indicated tissues of a transgenic (TG) and a wild-type (WT) mouse using the rabbit β-globin 3′-UTR as probe. In (D), the RNA was obtained from either a female or a male as indicated.
Animals
Transgenic mice were generated in the transgenic core facility of the University of Southern California. Transgenesis and breeding were performed with a first generation cross of C57Black/6 and DBA2 mice from The Jackson Laboratory. Transgenic mice were identified using tail DNA based on the presence of the rabbit β-globin 3′-UTR, either by Southern blot analysis or by PCR. Distribution of the H4tTG1 transgene followed a Mendelian inheritance pattern. Wild-type and heterozygous transgenic females and males were examined at 8, 17 and 34 weeks of age, 5–12 mice per gender/age/genotype group. To study bone formation, newly formed bone was vitally labeled by the fluorochrome calcein (Sigma) injected intraperitoneally (15 mg/kg) 4 days and 1 day prior to sacrifice. At sacrifice, the femora were dissected, fixed in phosphate-buffered formalin for 48 h and further kept in 70% ethanol. Soft tissues for northern analysis were snap-frozen in liquid nitrogen.
Northern blot analysis
Transgene expression was analyzed in selected tissues by northern blot analysis. The tissues were homogenized in Trizol® solution (Invitrogen Corp., Carlsbad, CA) followed by chloroform extraction and precipitation in isopropanol. The probe was a 0.5 kb XhoI–PstI fragment of pCAGGS, containing the rabbit β-globin 3′-UTR. For endogenous H4 gene expression, the probe was a 0.3 kb cDNA fragment containing the entire murine ‘clone 12’ H4 coding sequence (accession no. X13235) (19).
Micro-computed tomographic (μCT) analysis
Whole femora were examined by a μCT system (μCT 40; Scanco Medical AG, Basserdorf, Switzerland) equipped with a 5 μm focal spot microfocus X-ray tube as a source. A 2D CCD, coupled to a thin scintillator as a detector permitted parallel acquisition of stacks including 20 tomographic images. The long axis of the femur was set parallel to the plane of the X-ray beam axis. The X-ray tube was operated at 50 KVp and 160 μA. The integration time was set to 100 ms. The scans were performed at a resolution of 20 μm in all three spatial dimensions (medium resolution mode). The 2D CT images were reconstructed in 1024 × 1024 pixel matrices from 1000 projections using a standard convolution–backprojection procedure with a Shepp and Logan filter. Images were stored in 3D arrays with an isotropic voxel size of 20 μm. A constrained 3D Gaussian filter (width: σ = 0.8, support: one voxel width) was used to partly suppress the noise in the volumes. The samples were binarized using a global thresholding procedure (20). The threshold was set to 22.4 and 16.0% of the maximal gray scale value for cortical bone and trabecular bone, respectively. Morphometric parameters were determined using a direct 3D approach (21) in (i) secondary spongiosa in the distal metaphysis; and (ii) cortical bone in a 1 mm thick segment in the mid-diaphysis. The overall femoral length was also measured.
Histomorphometry
After μCT image acquisition, the specimens were dehydrated in progressive concentrations of ethanol, cleared in xylene and embedded undecalcified in polymethylmethacrylate (Technovit 9100; Heraeus Kulzer, Wehrheim, Germany). Undeplasticized, longitudinal 5 μm sections were left unstained for fluorescent microscopy and dynamic histomorphometric measurements based on the calcein double labeling. To identify osteoclasts, consecutive sections were deplasticized and stained for tartrate-resistant acid phosphatase [TRAP; (22)] using an acid phosphatase kit (Sigma, St Louis, MO) and counterstained with Mayer's hematoxylin. Histomorphometric analysis was carried out on digital photomicrographic images using Image-Pro Express Version 4.0.1 image analysis software (Media Cybernetics, Silver Spring, MD). The measurements, terminology and units used for the histomorphometric analysis were according to the convention of standardized nomenclature (23).
Statistical analysis
All analyses were carried out using SigmaStat software (SPSS Science, Chicago, IL). The significance of differences in quantitative μCT and histomorphometric studies was determined based on the analysis of variance (ANOVA). When significant differences were indicated by ANOVA, group means were compared using the Tukey test.
RESULTS
Nucleotides flanking ATG initiation codons: whole genome overview
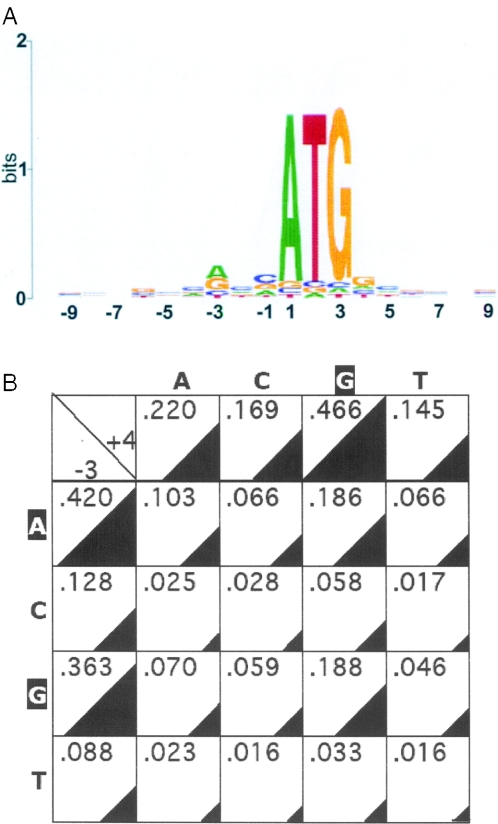
We screened 22 208 human mRNA files available on the NCBI webpage that contain 5′-UTR sequences for nucleotides that occupy positions from −10 to +10 relative to the first nucleotide of the annotated translation initiation codon. As shown in Figure 1, several of these positions (in addition to A+1, T+2 and G+3) exhibit various degrees of conservation. The most conserved position is −3, with A and G present in 42 and 36% of the mRNAs, respectively. Position +4 exhibited a strong predilection for G (47%). Of the 16 possible −3/+4 nucleotide pairs, A−3/G+4 and G−3/G+4 are found most frequently, each approaching 19%, while the least frequent pairs are those without G+4 and without A−3 or G−3 (Figure 1B). Each of these six rare pairs (C−3/A+4, C−3/C+4, C−3/T+4, T−3/A+4, T−3/C+4 and T−3/T+4) is found in 1.6–2.8% of the entire mRNA population, totaling 12.5% (Figure 1B). These trends are consistent with the known contribution of nucleotides at positions −3 and +4 to ribosomal recognition of initiation sites, because a purine at position −3 and a G at position +4 best support translational initiation (5,6). However, the results also highlight a large number of start codons with suboptimal and even unfavorable sequence context for translational initiation.
Figure 1.
Analysis of nucleotides flanking the initiation codons of human genes. (A) Preferred occupancy of positions around the initiation codons of 22 208 human mRNAs was evaluated and diagrammatically illustrated on a Plot Sequence Logo as described previously (11–13). Nucleotides found more frequently than others are indicated, with the size of the corresponding letter reflecting their frequency. Positions with no illustration indicate close-to-equal distribution of all 4 nt. (B) Occupancy of positions −3 and +4 in 22 208 human mRNAs. The left column and the top row indicate the fraction of genes with the indicated nucleotides at positions −3 and +4, respectively. The internal cells represent the 16 possible −3/+4 combinations. Nucleotide frequencies are depicted numerically and illustrated by triangles of corresponding sizes.
Nucleotides flanking ATG initiation codons of genes known to undergo LRS
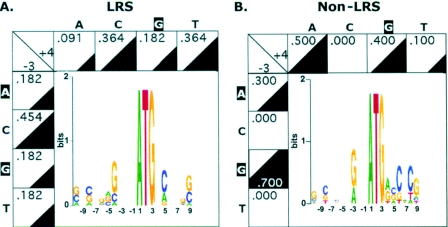
mRNAs with suboptimal initiation codons undergo LRS, which can result in translational initiation at alternative, downstream AUGs [reviewed in (3)]. Although the small number of genes with experimental evidence for LRS does not allow a thorough investigation of nucleotide frequency at positions flanking the first initiation codon of such genes, Figure 2A shows for a small number of such genes that they do not seem to follow the overall R−3 and G+4 genome-wide trends. In fact, only 4 of the 11 genes in the LRS group (Table 1) have a purine at −3 and only 2 have a G at +4. None of the 11 genes has the R−3/G+4 combination. In contrast, each of the 10 genes randomly picked from the published literature, for which evidence exists for lack of LRS, has a purine at position −3, and 4 of them have a G at position +4 (Table 2 and Figure 2B).
Figure 2.
Analysis of nucleotides flanking the initiation codons of human genes known to undergo or not undergo LRS. Preferred nucleotides at the −10/+10 positions is depicted as in Figure 1A. Occupancy of the −3 and +4 positions is depicted as in Figure 1B. (A) LRS group (see Table 1). (B). Non-LRS group (see Table 2).
Table 1.
mRNAs known to undergo LRSa
| Gene name | Accession no. | Sequence flanking first ATG (−10/+10) | References |
|---|---|---|---|
| Glucocorticoid receptor | NM_000176 | ATATTCACTGATGGACTCCA | (39) |
| NFAT | NM_172390 | CGCCGCGCGGATGCCAAGCA | (40) |
| GATA-1 | NM_002049 | CAGAGGCTCCATGGAGTTCC | (41) |
| MxB | M30818 | GAGACAGCACATGTCTAAGG | (42) |
| Ubiquitin-activating enzyme E1 | BC013041 | GGCACAGGGGATGTCCAGCT | (43) |
| Microtubule-associated protein | BC007318 | GAGTGCGCCAATGCCTGGGC | (44) |
| Von Hippel–Lindau gene | NM_000551 | CGCGGAGGGAATGCCCCGGA | (45) |
| p70 S6 kinase | M60724 | GTCCGGGCCCATGAGGCGAC | (46) |
| Folylpolyglutamate synthase | BC064393 | CGCCGGGACTATGTCGCGGG | (47) |
| T-cell receptor gamma chain | AF151103 | AAACAACTTGATGCAGATGT | (48) |
| Embryonic ectoderm development (EED) | NM_003797 | GCGGAGGAATATGTCCGAGA | (49) |
aSelected based on (3) and manual literature search of PubMed.
Table 2.
mRNAs that do not undergo LRSa
| Gene name | Accession no. | Sequence flanking first ATG (−10/+10) | References |
|---|---|---|---|
| HDAC6 | NM_006044 | CTCCTCAACTATGACCTCAA | (50) |
| HDAC5 | AF132608 | CAGAGCCGGCATGAACTCTC | (50) |
| HDAC4 | NM_006037 | ATTGCTAGCAATGAGCTCCC | (50) |
| UDP-glucuronosyl transferase | AJ005162 | TTGCATCAGGATGTCTATGA | (51) |
| Cannabinoid receptor 1 | NM_016083 | GACTGAGGTTATGAAGTCGA | (52) |
| C1-tetrahydrofolate synthase | AY374130 | GTCCCGCGCCATGGGCACGC | (53) |
| Cytokine inducible nuclear protein | X83703 | GTCCCGCGCCATGGGCACGC | (54) |
| Nephrin (NPHS1) | AF035835 | GGGACCTGTGATGGCCCTGG | (55) |
| ALK4 (activin A receptor, type IB) | NM_004302 | GGTGGTTACTATGGCGGAGT | (56) |
| ASK1 | D84476 | GCCCGGAGAGATGAGCACGG | (57) |
aSelected based on published western blots. Approximately 100 genes, for which published western analyses showed a single band, were examined. Of these, the 10 genes depicted in the Table had an in-frame internal AUG codon, which could have resulted in a second protein product, detectable by the antibody used, if LRS had occurred at the first AUG.
To test whether poor first translational initiation codons are preferentially found in genes with a particular function(s), we interrogated the genes with the predicted strongest LRS (sLRS gene group), i.e. those without R−3 and without G+4, for characteristic functions based on the GO Consortium annotations (24). We compared the frequency of each GO designation within this group of genes with the respective frequency within the entire gene pool from which it was derived. We did not find in the sLRS gene group any GO category that was over-represented with a corrected P-value <0.05. Thus, poor first AUG initiation codons do not appear to preferentially associate with any particular gene function.
Nucleotides flanking ATG initiation codons of human and mouse histone genes
We have previously shown that histone H4 mRNAs, which lack the favorite G at position +4, undergo LRS, giving rise to OGP (17). Because histone genes lack introns, LRS is probably the main mechanism by which they might give rise to diverse products. We therefore screened the sequences flanking the AUG initiation codons of all human histone mRNAs available in the GenBank database (Supplementary Table S1). As shown in Figure 3A and B, most of human histone mRNAs do not have the R−3/G+4 combination, and are therefore candidates for LRS. With the exception of histone H3 mRNA, the nucleotide at position +4 is usually a T (for histones H1, H2A and H4) or a C (for histone H2B). These same ‘unpopular’ trends are conserved in mouse histone genes (Figure 3C and D). However, no human histone gene and only four mouse genes (from a total of 77, highlighted in Supplementary Table 1) lack both an R−3 and a G+4 (Figure 3).
Figure 3.
Analysis of nucleotides flanking the initiation codons of histone genes. (A and C) Preferred nucleotides at the −10/+10 positions is depicted as in Figure 1A. (B and D) Occupancy frequency at the −3 and +4 positions, as well as the frequency of each −3/+4 combination, are depicted as in Figure 1B. (A and B), human histone genes (9 H1, 30 H2A, 19 H2B, 14 H3 and 14 H4 genes); (C and D), mouse histone genes (9 H1, 23 H2A, 17 H2B, 15 H3 and 13 H4). Gene list is provided in Supplementary Table S1.
Potential LRS-mediated alternative translation products of human and mouse histone genes
We next examined every human and every mouse histone gene that lacks either R−3 or G+4 for the presence of an alternative open reading frame(s) [ORF(s)], which, following LRS, could give rise to an alternative peptide of 16 or more amino acids. We found such alternative ORFs in every histone gene family in both human and mouse (Supplementary Table 2). Many of the alternative ORFs are in-frame, starting at an internal methionine codon. However, approximately half the alternative ORFs found within the human H1, H2A and H2B families are out of frame, and several of them have similar homologs in the mouse. These are predicted to give rise to a 33 amino acid peptide encoded by nucleotides 206–304 of HIST1H2AB, a 26 amino acid peptide encoded by nucleotides 197–274 of H2AFY and a 21 amino acid peptide encoded by nucleotides 77–139 of each of HIST1H2BB and HIST1H2BM (Supplementary Table 2). Such peptides are prime candidates for future investigation of cryptic peptides encoded by histone genes, which may be highly expressed in proliferating cells and could fulfill functions very different from DNA packaging.
H4tTG1 transgenic mice
The above bioinformatic analysis highlights the potential for synthesis of a plethora of cryptic peptides by a mechanism of LRS. We have previously demonstrated that LRS accounts for the translation of OGP from histone H4 mRNA (17). To address the significance of histone H4 LRS in vivo, transgenic mice were generated that carry the H4tTG1 transgene described in Figure 4A. H4tTG1 encodes OGP but not histone H4, due to a mutation substituting the A+1T+2G+3 H4 initiation codon for t+1T+2G+3. The transgene regulatory sequences were designed to promote ubiquitous (β-actin promoter) high-level [cytomegalovirus (CMV) enhancer] expression. In addition, the H4tTG1 mRNA was designed to be equally stable in proliferating and quiescent cells, by replacing the histone H4 3′-UTR, which is responsible for cell cycle-dependent transcript stability (25), with the rabbit β-globin 3′-UTR. Results presented here are from one of the two independent transgenic lines, which had a similar expression pattern and a similar phenotype. Figure 4B presents northern blot analysis of the H4tTG1 transgene next to endogenous histone H4 in muscle and spleen of a transgenic mouse. The results show that the H4tTG1 transgene is expressed in muscle on a minimal background of endogenous histone H4 expression, reflecting the post-mitotic state of most muscle cells and the independence of H4tTG1 expression on proliferation. Expression of the H4tTG1 transgene was much lower in spleen than in muscle (Figure 4B), with liver, brain and kidney displaying intermediate levels (Figure 4C). Expression in bone was high, similar to that seen in muscle, and, important for the interpretation of data presented below, no significant difference was observed between females and males (Figure 4D).
High bone mass (HBM) phenotype in H4tTG1 transgenic mice
The H4tTG1 transgenic mice displayed no overt phenotype at birth and followed normal growth curves (data not shown). Several transgenic mice maintained up to the age of 18 months were grossly indistinguishable from littermate controls. Because OGP has been implicated in osteoblast differentiation and bone formation (10), we compared femora from transgenic and control mice. μCT analysis performed at 8, 17 and 34 weeks of age showed that the H4tTG1 transgene induced a HBM phenotype, and that this phenotype was much more pronounced in females than in males (Figure 5).
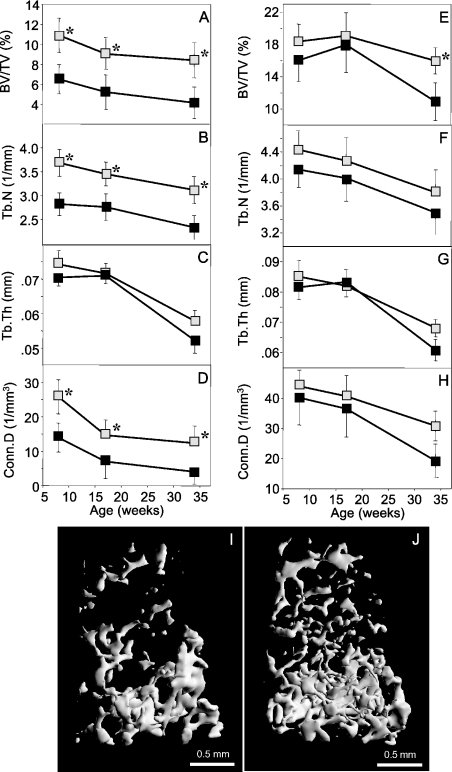
Figure 5.
High trabecular bone mass in female H4tTG1 transgenic mice. μCT analysis was performed in the distal femoral metaphysis of 8-, 17- and 34-week-old female (A–D) and male (E–H) mice. (A and E) Trabecular bone volume density (BV/TV); (B and F) trabecular number (Tb.N); (C and G) trabecular thickness (Tb.Th); (D and H), trabecular connectivity density (Conn.D). (I–J) Representative 3D μCT images of distal femoral metaphyseal trabecular bone from a 17-week-old wild-type (I) and transgenic (J) female mice with median BV/TV values. Data are mean ± SEM. Shaded squares, H4tTG transgenic; solid squares, wild-type mice; *P < 0.05.
In H4tTG1 transgenic females of all three ages, the μCT analysis showed an ∼2-fold higher trabecular bone volume density (BV/TV) as compared with controls (Figure 5A). The higher BV/TV was attributable mainly to increased trabecular number (Tb.N; Figure 5B), while the differences in trabeculcar thickness (Tb.Th) were small and statistically insignificant (Figure 5C). The connectivity density (Conn.D), a measure of the number of trabecular connections that can be broken before the structure collapses (26), was also markedly increased in female transgenic mice (Figure 5D). The computed differences between the H4tTG1 transgenic and wild-type females are evident in 3D reconstituted femoral metaphyseal images that show a substantially denser, internally more connected structure compared with a representative control image (Figure 5I and J). In males, a significant albeit smaller increase in trabecular bone volume density was observed only in the 34-week age group (Figure 5E). Notably, all the structural changes were restricted to trabecular bone; cortical bone parameters, as well as the overall femoral dimensions, had similar values in transgenic and wild-type mice (Supplementary Tables S3 and S4).
Increased bone formation in H4tTG1 transgenic mice
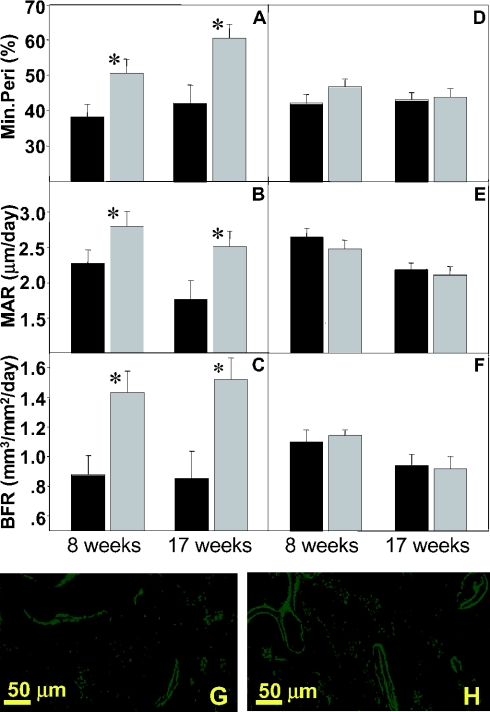
Parameters of bone formation and bone resorption were compared between H4tTG1 transgenic and control mice at the ages of 8 and 17 weeks. Bone formation was evaluated based on calcein double labeling of actively forming bone surfaces (23). Osteoclast number, a surrogate for bone resorption, was determined based on staining for TRAP (22). The H4tTG1 transgenic mice exhibited significantly higher values of bone formation parameters, and consistent with the HBM phenotype, these effects were observed in females (Figure 6A–C) but not in males (Figure 6D–F). Specifically, H4tTG1 females exhibited a 32–42% higher mineralizing perimeter (Min.Peri), i.e. the percentage of trabecular surfaces engaged in bone formation (Figure 6A) and thus a surrogate for the number of osteoblasts. The increase in the Min.Peri is illustrated by the extensive calcein labeling of bone surfaces in a trabecular bone section from a representative H4tTG1 female (Figure 6H) as compared with a non-transgenic control (Figure 6G). In addition, the H4tTG1 females showed a 23–39% increase in the mineral appositional rate (MAR), measured as the distance between the two lines at doubly labeled surfaces (Figure 6B, G and H), which represents the activity of osteoblasts. The overall bone formation rate (BFR), the numerical product of the Min.Per and MAR, was ∼60% higher in the H4tTG1 transgenic females of both age groups as compared with their age-matched controls (Figure 6C). No significant differences were observed in the osteoclast number per trabecular surface area between the H4tTG1 transgenic and control mice (Table 3). Thus, the HBM phenotype is attributable to a net bone anabolic effect of the H4tTG1 transgene product(s).
Figure 6.
Increased trabecular bone formation in female H4tTG1 transgenic mice. Dynamic histomorphometric analysis, based on fluorescing calcein labels, in the distal femoral metaphysical trabecular bone of 8- and 17-week-old female (A–C) and male (D–F) mice. (A and D) mineralizing perimeter (Min.Peri); (B and E) mineral appositional rate (MAR); (C and F) bone formation rate (BFR). (G–H) Fluorescent images from calcein-labeled distal femoral metaphyses of a 17-week-old wild-type (G) and transgenic (H) females with median BFR values. Data are mean ± SEM. Shaded bars, H4tTG1 transgenic mice; solid bars, wild-type mice; *P < 0.05.
Table 3.
Osteoclast numbera in distal femoral metaphysic of H4tTG1 transgenic mice
| Age (weeks) | Gender | WT | H4tTG1 |
|---|---|---|---|
| 8 | F | 4.4 ± 0.8 | 3.4 ± 0.6 |
| 17 | 2.4 ± 0.4 | 2.4 ± 0.5 | |
| 8 | M | 3.6 ± 0.7 | 2.4 ± 0.4 |
| 17 | 2.0 ± 0.8 | 3.3 ± 0.9 |
aN.Oc/BS (mm−2).
Data are mean ± SEM. Differences between H4tTG1 and WT mice are not statistically significant (P > 0.05).
DISCUSSION
We scanned most human genes for nucleotides occupying the positions that flank their annotated initiation codons. The preferences for the respective purine and guanine residues at positions −3 and +4 are consistent with results reported previously for a smaller dataset (7). The R−3/G+4 combination has been shown to confer high fidelity on an AUG as a translational initiator (5,6). In addition, our analysis indicates a preference, albeit to a lesser degree, for a C at position −1, which also contributes to the fidelity of translational initiation (5). Finally, we observe a secondary preference for A at position +4. The functional significance of this observation remains to be examined.
Only 37.4% of human mRNAs have both R at position −3 and G at position +4, and can thus be expected to be translated starting exclusively at the annotated AUG initiator. A significant fraction of the mRNAs, 12.5%, which have neither an R−3 nor a G+4, can be expected to experience relatively strong LRS, potentially yielding significant concentrations of alternatively translated, biologically significant products. The remaining 50% of human genes, without an R−3 or without a G+4, but with the preferred nucleotide at the other key position, may undergo weaker LRS, although its frequency, magnitude and biological significance remain to be examined. In some of these cases, alternatively translated peptides would accumulate to biologically significant levels in cases of abundant mRNAs. Such may be the case for the ancient histone mRNAs, which accumulate to high levels in proliferating cells, inasmuch as most of them, with the exception of H3, usually lack either R−3 or G+4 (Figure 3). While productive alternative translation has been experimentally demonstrated only for histone H4 mRNA (17), sequence analysis suggests that a similar mechanism likely gives rise to cryptic peptides encoded by additional histone genes (Supplementary Table 2).
The potential biological significance of LRS is demonstrated in the present paper by over-expression of a histone H4 alternative translation product. The increased bone mass observed in the H4tTG1 transgenic mice cannot be ascribed to the full-length H4 gene product, or to histone H4-derived proteolytic products, because the H4tTG1 transgene lacks the first AUG codon that initiates histone H4 translation. Instead, the HBM phenotype is most likely mediated by OGP or a related histone H4 alternative translational product(s). Unfortunately, loss-of-function approaches to study the physiological significance of H4 alternative translation are problematic. First, molecular targeting of OGP, such as by gene ablation or RNA interference, would also target histone H4 expression. Second, gene replacement studies (knock-in) for histones are unrealistic given their multiple copies in mammalian genomes (27).
Mammalian histone H4 genes are tightly regulated by both transcriptional and post-transcriptional mechanisms that couple histone H4 mRNA levels to DNA synthesis. Thus, the translation of OGP, an osteogenic and hematopoietic regulator (8,28–30), from histone H4 mRNA provides a functional link between cell proliferation and osteogenesis/hematopoiesis. This linkage may be important to replenish the osteogenic and blood cell pools during the healing processes that follow osseous and hematopoietic insults, processes inevitably associated with strong proliferative responses and hence robust histone gene expression. Indeed, OGP has been implicated in the osteogenic and hematopoietic reaction to bone marrow ablation as well as massive blood loss (8,31,32). The present study illustrates that the role of histone H4 alternative translation in regulating bone mass can be recapitulated in the absence of injury, raising the possibility that histone H4 alternative translation product(s) can be exploited pharmacologically. For example, OGP or its analogs may simulate cell proliferation and bone formation in osteoporotic patients, in addition to their demonstrated potential to enhance fracture healing (32–34). The selective effect of OGP on trabecular bone is not disadvantageous for the potential treatment of osteoporosis because this osseous compartment is the one most critically affected in osteoporosis. The stimulation of hematopoiesis by OGP and OGP derivatives and its therapeutic potential has been shown elsewhere (28–30), and clinical trials to evaluate the beneficial hematopoietic activity of OGP are in progress. Finally, the present study suggests that the long-term and continuous OGP over-production does not have any gross adverse effects.
Selective effects on the trabecular, not the cortical, bone compartment have been observed for other regulators of bone mass, such as leptin and Wnt ligands (35,36). The increased sensitivity of trabecular bone may result from the higher turnover rate of this osseous compartment, leading to increased requirement for osteoblastic cell renewal. Remarkably, the effect of the H4tTG1 transgene was substantially stronger in females compared with males, although the two genders expressed the transgene at comparable levels. The gender bias may be secondary to fundamental differences between male and female bone mass and bone cell regulation, as suggested by the present and other studies (37,38). The gender bias may indicate that OGP targets a bone anabolic mechanism, which operates in female mice at a suboptimal level; in males, this mechanism may be saturated and thus insensitive to increased OGP concentrations.
The histone H4 gene structure, with a suboptimal upstream translational initiation codon and an optimal, in-frame, downstream initiation codon, is shared by most if not all mammalian and plant cells (17). Evolutionary conservation of the upstream initiation codon at a suboptimal environment for translational initiation may result from a need to preserve the T+4C+5N+6 second codon encoding a serine. However, it is not impossible that a need for H4 LRS has played a role as well. Consistent with this notion, some ciliate species display the interesting combination of a second G+4C+5C+6 alanine codon, likely conferring high fidelity of translational initiation on the first AUG, along with the absence of the downstream in-frame AUG codon, which in most other eukaryotes would be responsible for the alternative translation of OGP (17). Regardless of the evolutionary mechanisms, the stimulation of bone formation and other potential roles of histone alternative translation products add to the growing plethora of mechanisms of gene multi-functionality, which enable complex organisms to develop and function with a limited selection of protein-coding genes (1,2).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors are thankful to Dr Jun-ichi Miyazaki (Osaka University Medical School) for the pCAGGS vector, to Dr Robert E. Maxson and to Ms Lan Ying Wu for oocyte microinjection and transfer, and to Lian Liang, Vijaya Rao and Abel Valdovinos for technical assistance. Funding for this project was provided by grants from the American Cancer Society (IRG-58-007-40), the Health Research Association (Los Angeles, CA) and the Zumberge Fund (Los Angeles, CA), and through a collaborative research agreement between Yissum Research Development Company of The Hebrew University of Jerusalem and R & D Pharmaceuticals (USA), Inc. Funding to pay the Open Access publication charges for this article was provided by the Department of Orthopaedic Surgery, USC Keck School of Medicine.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 6.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bab I., Gazit D., Chorev M., Muhlrad A., Shteyer A., Greenberg Z., Namdar M., Kahn A. Histone H4-related osteogenic growth peptide (OGP): a novel circulating stimulator of osteoblastic activity. EMBO J. 1992;11:1867–1873. doi: 10.1002/j.1460-2075.1992.tb05238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 10.Bab I., Chorev M. Osteogenic growth peptide: from concept to drug design. Biopolymers. 2002;66:33–48. doi: 10.1002/bip.10202. [DOI] [PubMed] [Google Scholar]
- 11.Schneider T.D., Stormo G.D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 12.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khatri P., Bhavsar P., Bawa G., Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draghici S., Khatri P., Bhavsar P., Shah A., Krawetz S.A., Tainsky M.A. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe S.A., Anderson J.V., Grimes S.R., Stein G.S., Stein J.S. Comparison of the structural organization and expression of germinal and somatic rat histone H4 genes. Biochim. Biophys. Acta. 1989;1007:140–150. doi: 10.1016/0167-4781(89)90032-8. [DOI] [PubMed] [Google Scholar]
- 17.Bab I., Smith E., Gavish H., Attar-Namdar M., Chorev M., Chen Y.C., Muhlrad A., Birnbaum M.J., Stein G., Frenkel B. Biosynthesis of osteogenic growth peptide via alternative translational initiation at AUG85 of histone H4 mRNA. J. Biol. Chem. 1999;274:14474–14481. doi: 10.1074/jbc.274.20.14474. [DOI] [PubMed] [Google Scholar]
- 18.Kawamoto S., Niwa H., Tashiro F., Sano S., Kondoh G., Takeda J., Tabayashi K., Miyazaki J. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 19.Meier V.S., Bohni R., Schumperli D. Nucleotide sequence of two mouse histone H4 genes. Nucleic Acids Res. 1989;17:795. doi: 10.1093/nar/17.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller R., Ruegsegger P. Micro-tomographic imaging for the nondestructive evaluation of trabecular bone architecture. Stud. Health Technol. Inform. 1997;40:61–79. [PubMed] [Google Scholar]
- 21.Hildebrand T., Laib A., Muller R., Dequeker J., Ruegsegger P. Direct three-dimensional morphometric analysis of human cancellous bone: microstructural data from spine, femur, iliac crest, and calcaneus. J. Bone Miner. Res. 1999;14:1167–1174. doi: 10.1359/jbmr.1999.14.7.1167. [DOI] [PubMed] [Google Scholar]
- 22.Kartsogiannis V., Udagawa N., Ng K.W., Martin T.J., Moseley J.M., Zhou H. Localization of parathyroid hormone-related protein in osteoclasts by in situ hybridization and immunohistochemistry. Bone. 1998;22:189–194. doi: 10.1016/s8756-3282(97)00278-0. [DOI] [PubMed] [Google Scholar]
- 23.Parfitt A.M., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., Recker R.R. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominski Z., Marzluff W.F. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- 26.Odgaard A., Gundersen H.J. Quantification of connectivity in cancellous bone, with special emphasis on 3-D reconstructions. Bone. 1993;14:173–182. doi: 10.1016/8756-3282(93)90245-6. [DOI] [PubMed] [Google Scholar]
- 27.Marzluff W.F., Gongidi P., Woods K.R., Jin J., Maltais L.J. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 28.Gurevitch O., Slavin S., Muhlrad A., Shteyer A., Gazit D., Chorev M., Vidson M., Namdar-Attar M., Berger E., Bleiberg I., et al. Osteogenic growth peptide increases blood and bone marrow cellularity and enhances engraftment of bone marrow transplants in mice. Blood. 1996;88:4719–4724. [PubMed] [Google Scholar]
- 29.Fazzi R., Galimberti S., Testi R., Pacini S., Trasciatti S., Rosini S., Petrini M. Bone and bone marrow interactions: hematological activity of osteoblastic growth peptide (OGP)-derived carboxy-terminal pentapeptide. II. Action on human hematopoietic stem cells. Leuk. Res. 2002;26:839–848. doi: 10.1016/s0145-2126(02)00008-5. [DOI] [PubMed] [Google Scholar]
- 30.Fazzi R., Pacini S., Testi R., Azzara A., Galimberti S., Testi C., Trombi L., Metelli M.R., Petrini M. Carboxy-terminal fragment of osteogenic growth peptide in vitro increases bone marrow cell density in idiopathic myelofibrosis. Br. J. Haematol. 2003;121:76–85. doi: 10.1046/j.1365-2141.2003.04250.x. [DOI] [PubMed] [Google Scholar]
- 31.Lucas T.S., Bab I.A., Lian J.B., Stein G.S., Jazrawi L., Majeska R.J., Attar-Namdar M., Einhorn T.A. Stimulation of systemic bone formation induced by experimental blood loss. Clin. Orthop. 1997:267–275. doi: 10.1097/00003086-199707000-00034. [DOI] [PubMed] [Google Scholar]
- 32.Brager M.A., Patterson M.J., Connolly J.F., Nevo Z. Osteogenic growth peptide normally stimulated by blood loss and marrow ablation has local and systemic effects on fracture healing in rats. J. Orthop. Res. 2000;18:133–139. doi: 10.1002/jor.1100180119. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y.Q., Ashhurst D.E. Osteogenic growth peptide enhances the rate of fracture healing in rabbits. Cell Biol. Int. 1998;22:313–319. doi: 10.1006/cbir.1998.0253. [DOI] [PubMed] [Google Scholar]
- 34.Gabet Y., Muller R., Regev E., Sela J., Shteyer A., Salisbury K., Chorev M., Bab I. Osteogenic growth peptide modulates fracture callus structural and mechanical properties. Bone. 2004;35:65–73. doi: 10.1016/j.bone.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Ducy P., Amling M., Takeda S., Priemel M., Schilling A.F., Beil F.T., Shen J., Vinson C., Rueger J.M., Karsenty G. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100:197–207. doi: 10.1016/s0092-8674(00)81558-5. [DOI] [PubMed] [Google Scholar]
- 36.Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D., et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 37.Frenkel B., Capparelli C., Van Auken M., Baran D., Bryan J., Stein J.L., Stein G.S., Lian J.B. Activity of the osteocalcin promoter in skeletal sites of transgenic mice and during osteoblast differentiation in bone marrow-derived stromal cell cultures: effects of age and sex. Endocrinology. 1997;138:2109–2116. doi: 10.1210/endo.138.5.5105. [DOI] [PubMed] [Google Scholar]
- 38.Kawano H., Sato T., Yamada T., Matsumoto T., Sekine K., Watanabe T., Nakamura T., Fukuda T., Yoshimura K., Yoshizawa T., et al. Suppressive function of androgen receptor in bone resorption. Proc. Natl Acad. Sci. USA. 2003;100:9416–9421. doi: 10.1073/pnas.1533500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yudt M.R., Cidlowski J.A. Molecular identification and characterization of a and b forms of the glucocorticoid receptor. Mol. Endocrinol. 2001;15:1093–1103. doi: 10.1210/mend.15.7.0667. [DOI] [PubMed] [Google Scholar]
- 40.Lyakh L., Ghosh P., Rice N.R. Expression of NFAT-family proteins in normal human T cells. Mol. Cell. Biol. 1997;17:2475–2484. doi: 10.1128/mcb.17.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calligaris R., Bottardi S., Cogoi S., Apezteguia I., Santoro C. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc. Natl Acad. Sci. USA. 1995;92:11598–11602. doi: 10.1073/pnas.92.25.11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mevel-Ninio M., Fouilloux E., Guenal I., Vincent A. The three dominant female-sterile mutations of the Drosophila ovo gene are point mutations that create new translation-initiator AUG codons. Development. 1996;122:4131–4138. doi: 10.1242/dev.122.12.4131. [DOI] [PubMed] [Google Scholar]
- 43.Shang F., Deng G., Obin M., Wu C.C., Gong X., Smith D., Laursen R.A., Andley U.P., Reddan J.R., Taylor A. Ubiquitin-activating enzyme (E1) isoforms in lens epithelial cells: origin of translation, E2 specificity and cellular localization determined with novel site-specific antibodies. Exp. Eye Res. 2001;73:827–836. doi: 10.1006/exer.2001.1091. [DOI] [PubMed] [Google Scholar]
- 44.Su L.K., Qi Y. Characterization of human MAPRE genes and their proteins. Genomics. 2001;71:142–149. doi: 10.1006/geno.2000.6428. [DOI] [PubMed] [Google Scholar]
- 45.Schoenfeld A., Davidowitz E.J., Burk R.D. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl Acad. Sci. USA. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grove J.R., Banerjee P., Balasubramanyam A., Coffer P.J., Price D.J., Avruch J., Woodgett J.R. Cloning and expression of two human p70 S6 kinase polypeptides differing only at their amino termini. Mol. Cell. Biol. 1991;11:5541–5550. doi: 10.1128/mcb.11.11.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi H., Atkinson I., Xiao S., Choi Y.J., Tobimatsu T., Shane B. Folylpoly-gamma-glutamate synthetase: generation of isozymes and the role in one carbon metabolism and antifolate cytotoxicity. Adv. Enzyme Regul. 1999;39:263–273. doi: 10.1016/s0065-2571(98)00023-5. [DOI] [PubMed] [Google Scholar]
- 48.Wolfgang C.D., Essand M., Vincent J.J., Lee B., Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus. Proc. Natl Acad. Sci. USA. 2000;97:9437–9442. doi: 10.1073/pnas.160270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuzmichev A., Jenuwein T., Tempst P., Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 50.Brush M.H., Guardiola A., Connor J.H., Yao T.P., Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J. Biol. Chem. 2004;279:7685–7691. doi: 10.1074/jbc.M310997200. [DOI] [PubMed] [Google Scholar]
- 51.Kurkela M., Garcia-Horsman J.A., Luukkanen L., Morsky S., Taskinen J., Baumann M., Kostiainen R., Hirvonen J., Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J. Biol. Chem. 2003;278:3536–3544. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- 52.Farrens D.L., Dunham T.D., Fay J.F., Dews I.C., Caldwell J., Nauert B. Design, expression, and characterization of a synthetic human cannabinoid receptor and cannabinoid receptor/G-protein fusion protein. J. Pept. Res. 2002;60:336–347. doi: 10.1034/j.1399-3011.2002.21066.x. [DOI] [PubMed] [Google Scholar]
- 53.Prasannan P., Pike S., Peng K., Shane B., Appling D.R. Human mitochondrial C1-tetrahydrofolate synthase: gene structure, tissue distribution of the mRNA, and immunolocalization in Chinese hamster ovary calls. J. Biol. Chem. 2003;278:43178–43187. doi: 10.1074/jbc.M304319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mochida Y., Takeda K., Saitoh M., Nishitoh H., Amagasa T., Ninomiya-Tsuji J., Matsumoto K., Ichijo H. ASK1 inhibits interleukin-1-induced NF-kappa B activity through disruption of TRAF6-TAK1 interaction. J. Biol. Chem. 2000;275:32747–32752. doi: 10.1074/jbc.M003042200. [DOI] [PubMed] [Google Scholar]
- 55.Huber T.B., Kottgen M., Schilling B., Walz G., Benzing T. Interaction with podocin facilitates nephrin signaling. J. Biol. Chem. 2001;276:41543–41546. doi: 10.1074/jbc.C100452200. [DOI] [PubMed] [Google Scholar]
- 56.Bianco C., Adkins H.B., Wechselberger C., Seno M., Normanno N., De Luca A., Sun Y., Khan N., Kenney N., Ebert A., et al. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial cells. Mol. Cell. Biol. 2002;22:2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu W., Burns D.K., Swerlick R.A., Presky D.H. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J. Biol. Chem. 1995;270:10236–10245. doi: 10.1074/jbc.270.17.10236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.