Abstract
The incorporation of C5-amino-modified 2′-deoxyuridine analogues into DNA have found application in nucleic acid labelling, the stabilization of nucleic acid structures, functionalization of nucleic acid aptamers and catalysts, and the investigation of sequence-specific DNA bending. In this study, we describe the physicochemical properties of four different C5-amino-modified 2′-deoxyuridines in which the amino group is tethered to the base via a 3-carbon alkyl, Z- or E-alkenyl or alkynyl linker. Conformational parameters of the nucleosides and their pKa values were deduced using 1H NMR. All of them display the expected anti-conformation of the nucleoside with 2′-endo sugar puckers for the deoxyribose ring. A preference for the cisoid conformation for the Z-alkenyl analogue is found, while the E-alkenyl analogue exists exclusively as its transoid conformation. The pKa values range from 10.0 for the analogue with an aliphatic propyl linker to 8.5 for the propargylamino analogue. The analogues have been used for the synthesis of triple-helix forming oligonucleotides (TFOs) in which they replace thymidine in the natural sequence. Oligonucleotides containing the propargylamino analogue display the highest stability especially at low pH, while those containing analogues with propyl and especially Z-alkenyl linkers are destabilized to a great extent. TFOs containing the analogue with the E-alkenyl linker have stability similar to the unmodified structures. The chemical synthesis of TFOs containing the analogue, 5-(3-hydroxyprop-1-ynyl)-2′-deoxyuridine that possesses a neutral but polar side chain show a remarkable stability, which is higher than that of all TFOs containing the alkylamino or alkenylamino analogues and only slightly lower than that of TFOs containing the propargylamino analogue. Both the hydroxyl and propargylamino substitutions impart enhanced triple-helix stability relative to the analogous sequences containing C5-propynyl-2′-deoxyuridine. Furthermore, a similar dependence of stability on pH is found between TFOs containing the hydroxypropynyl modifications and those containing the propargylamino side chains. This suggests that the major factor responsible for stabilizing such triple helices is due to the presence of the alkyne with an attached electronegative group.
INTRODUCTION
Pyrimidine nucleosides functionalized at C5 with aminoalkyl side chains have been used extensively for attaching labels and reporter groups to both nucleotides and oligonucleotides (1). There has also been great interest in exploiting the properties of such amino-modified DNA, which at the appropriate pH displays cationic side chains. The chemical syntheses and properties of such ‘zwitterionic DNA’ containing C5-aminohexyl-modified cytosines (1,2) and C5-aminohexyl (2) and C5-aminopropyl-modified uracil (3a) (2,3) bases were first reported by Switzer et al. They observed reduced mobilities for zwitterionic oligonucleotides during PAGE, suggesting that the tethered amino groups are largely protonated below pH 8. At very low-salt concentrations, the relative stabilities of duplexes containing C5-amino modifications of U and C were generally higher than those of the natural duplexes (2,3), but at higher ionic strength this situation is reversed. In contrast, C5-propyl and hexyl-modified 2′-deoxyuridines (i.e. simple alkyl chains in the place of cationic side chains) result in a marked destabilization of a DNA duplex (2,3). Most recently, the stabilizing properties of C5-propargylamino-2′-deoxyuridine (4a) when placed within the third strand of DNA triple helices (4,5) have been described. Such stabilization is largely independent of salt concentration but does show a pH-dependence. Further stabilization of triplexes can be achieved by attachment of a 2-aminoethoxy side chain at the 2′-position of this analogue (5). DNA duplexes containing C5-propargylamino-2′-deoxyuridine also show enhanced stabilities relative to the unmodified duplexes (6–8), a trend also found for the corresponding analogue C5-propargylamino-2′-deoxycytidine (7).
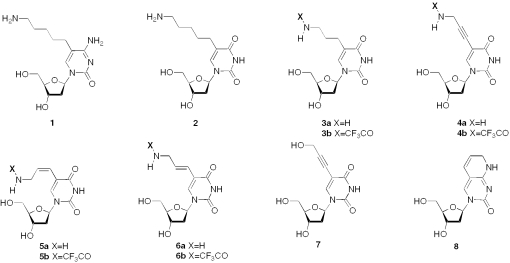
C5-Amino-modified pyrimidine nucleosides (Figure 1) have also been used to investigate the processes of DNA bending that may be induced following multiple incorporation of several such analogues (8–12). Interestingly, DNA containing C5-propargylamino-2′-deoxyuridine shows no propensity to undergo sequence-specific bending in contrast to that containing the corresponding C5-propylamino analogue (8).
Figure 1.
Structures of C5-amino-modified pyrimidine nucleosides (1–8).
The 5′-triphosphates of several C5-amino-modified pyrimidine nucleosides have also been investigated for use in SELEX (13–25) for which the cationic side chains add additional functionality to nucleic acids designed to function as aptamers or catalysts. In a comparative study of the substrate properties of C5-amino-modified dUTPs, in which the amino group is tethered to the nucleobase via 3-carbon linkers of differing flexibility (alkyne, alkene and alkane), we found that both the rigidity and conformation (Z- versus E-alkenyl) of the linker is very important (13). Thus, analogues containing alkynyl and E-alkenyl linkers were considered as the best substrates, while C5-propylamino dUTP was not incorporated by a variety of polymerases.
Clearly, the properties of nucleotides and DNA containing C5-amino-modified-2′-deoxyuridines depend on the type of linker between the amino group of the side chain and the nucleobase. In this regard, the nucleoside conformation and ionization states of the amino group are relevant to studies concerned with nucleotide and nucleic acid labelling, SELEX and the structural properties of such modified nucleic acids. However, there has so far been no detailed comparative study investigating the effect of a variety of linkers differing in both flexibility and conformation on the properties of C5-amino-modified 2′-deoxyuridines and oligonucleotides containing them. In this study, we investigate the physicochemical properties of four different C-5 amino-modified 2′-deoxyuridines in which the amino group is tethered to the nucleobase by one of four different 3-carbon linkers, such as propyl, E-propenyl, Z-propenyl and propargyl, and compare the stabilities of DNA structures that contain these analogues in the place of thymidine in the third strand of triple helices. Furthermore, we investigate the basis of the stabilizing role of the propargylamino side chain in triple helices by comparison with analogous DNA structures containing the novel analogue C5-(3-hydroxypropyn-1-yl)-2′-deoxyuridine.
MATERIALS AND METHODS
Chemicals and reagents
All reagents were obtained from commercial suppliers and used without purification. Dichloromethane, pyridine and acetonitrile were dried under reflux from calcium hydride, distilled and stored under argon, over 3 Å molecular sieves. MeOH was dried under reflux from magnesium methanolate, distilled and stored under argon, over molecular sieves. Triethylamine was dried under reflux from potassium hydroxide, distilled and stored over potassium hydroxide pellets under argon. Anhydrous N,N-dimethylformamide was purchased from Aldrich. NMR spectra were recorded on a Bruker AC250 spectrometer and chemical shifts are reported in δ values relative to tetramethylsilane (1H and 13C) or 85% phosphoric acid (31P) as an external standard. 1H NMR, 13C NMR and 31P NMR were recorded at 250, 60 and 101 MHz, respectively. Described syntheses were used for phosphoramidites 10b (4) and 12b (26,27).
Full characterization for all the compounds described below can be found in the Supplementary Material.
General method for the synthesis of nucleosides 3a, 4a and 6a from 3b (13), 4b (13,28) and 6b (17,29)
The protected nucleoside (100 mg) was stirred in concentrated aqueous ammonia (5 ml) overnight at room temperature and evaporated as oil (quantitative yield).
5-(Z-3-Aminoprop-1-enyl)-2′-deoxyuridine (5a)
Compound 5b (30) (0.1 g, 0.265 mmol) was stirred in concentrated aqueous ammonia (5 ml) for 45 min at room temperature and the solution was evaporated. The residue was again dissolved in water (1 ml) and acidified to pH 4 by the dropwise addition of trifluoroacetic acid. Evaporation gave yellow crystals (quantitative yield).
3-β-d-(2′-Deoxyribofuranosyl)-7,8-dihydropyrido[2,3-d] pyrimidine (8)
Prepared in the manner similar to 5a but stirred overnight in ammonia solution followed by evaporation.
5′-O-(4,4′-Dimethoxytrityl)-5-(3-trifluoroacetamidopropyl)-2′-deoxyuridine (9a)
To a solution of 5-(3-trifluoroacetamidopropyl)-2′-deoxyuridine (13) (0.260 g, 0.682 mmol) in anhydrous pyridine (5 ml), anhydrous Et3N (0.12 ml, 0.887 mmol) and dimethylaminopyridine (0.004 g, 0.034 mmol) were added. Dimethoxytrityl chloride (0.277 g, 0.819 mmol) was then added and the reaction was stirred for 4 h at room temperature. MeOH (1 ml) was added and the solution was evaporated, diluted in CH2Cl2 (100 ml) and the organic layer was washed with saturated aqueous NaHCO3 solution (150 ml). The organic layer was dried (MgSO4) and the solution was evaporated. Purification by silica column chromatography [CH2Cl2 (1% Et3N) followed by 5% MeOH/CH2Cl2 (1% Et3N)] resulted in a pale yellow foam (0.370 g, 0.542 mmol, 79%).
5′-O-(4,4′-Dimethoxytrityl)-5-(3-trifluoroacetamidopropyl)-2′-deoxyuridine-3′-(2-cyanoethyl-N,N-diisopropyl) phosphoramidite (9b)
To a solution of 5′-O-(4,4′-dimethoxytrityl)-5-(3-trifluoroacetamidopropyl)-2′-deoxyuridine (9a) (0.350 g, 0.512 mmol) in anhydrous CH2Cl2 (2.3 ml), anhydrous diisopropylethylamine (0.36 ml, 2.05 mmol) was added followed by the dropwise addition of 2-cyanoethyl-N,N-diisopropylchlorophosphoamidite (0.23 ml, 1.02 mmol). After 1 h at room temperature, MeOH (0.1 ml) was added and the mixture was stirred for 1 min. EtOAc (100 ml) was added and the organic layer was washed with 5% aqueous NaHCO3 solution (50 ml) followed by saturated aqueous sodium chloride solution (50 ml). The organic layer was then dried (MgSO4) and evaporated as foam. Purification by silica column chromatography using high-performance liquid chromatography (HPLC) grade solvents and a positive pressure of argon (50% EtOAc, 40% CH2Cl2 and 10% Et3N) resulted in the cream-coloured foam (0.350 g, 0.396 mmol, 87%).
5′-O-(4,4′-Dimethoxytrityl)-5-(Z-3-trifluoroacetamidoprop-1-enyl)-2′-deoxyuridine (11a)
Prepared in an analogous way similar to 9a. Yield 77%.
5′-O-(4,4′-Dimethoxytrityl)-5-(Z-3-trifluoroacetamidopropenyl)-2′-deoxyuridine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (11b)
Prepared in an analogous way similar to 9b. Yield 77%.
5′-O-(4,4′-Dimethoxytrityl)-5-(3-tert-butyldimethylsilyloxyprop-1-ynyl)-2′-deoxyuridine (13a)
Copper(I)iodide (0.035 g, 0.183 mmol) was added to a stirred solution of 5′-O-(4,4′-dimethoxytrityl)-5-iodo-2′-deoxyuridine (7) (0.300 g, 0.458 mmol) in anhydrous DMF (10 ml), in the absence of light, and the mixture was stirred for 10 min. Propargyl alcohol (0.160 ml, 2.75 mmol), anhydrous Et3N (0.13 ml, 0.916 mmol) and tetrakis(triphenylphosphine)palladium(0) (0.106 g, 0.092 mmol) were then added sequentially and the reaction was left for overnight stirring. The solvent was removed by evaporation and the reaction was diluted with excess MeOH. The precipitate formed was removed by filtration through hyflo filter aid and the filtrate was evaporated. Treatment with excess MeOH and the subsequent filtration was repeated until no further precipitate was formed. Purification by silica column chromatography (gradient of 0–5% MeOH in CH2Cl2/0.5% pyridine) gave a white foam of 5′-O-(4,4′-dimethoxytrityl)-5-(3-hydroxyprop-1-ynyl)-2′-deoxyuridine (0.440 g, 0.755 mmol, 50%). To the stirred solution of 5′-O-(4,4′-dimethoxytrityl)-5-(3-hydroxyprop-1-ynyl)-2′-deoxyuridine (0.350 g, 0.600 mmol) in anhydrous DMF (10 ml), tert-butyldimethylsilylchloride (0.095 g, 0.630 mmol) and imidazole (0.205 g, 3.00 mmol) were added. After 3.5 h, the reaction was diluted with CH2Cl2 (100 ml) and washed with 5% aqueous sodium carbonate solution (100 ml) and saturated aqueous sodium chloride solution (100 ml). The organic layer was dried (MgSO4), evaporated and purified by silica column chromatography [2% MeOH/CH2Cl2 (0.5% pyridine)] to give 13a as a white foam (0.274 g, 0.393 mmol, 65%).
5′-O-(4,4′-Dimethoxytrityl)-5-(3-tert-butyldimethylsilyloxyprop-1-ynyl)-2′-deoxyuridine-3′-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite (13b)
Prepared in an analogous way similar to 9b. Polymer-bound benzyl alcohol [0.31 g (2–3 mmol/g), 0.775 mmol] was then added, the reaction was stirred for 10 min then diluted with EtOAc (10 ml) and the mixture was filtered washing the solid filtrate with further EtOAc (25 ml). The filtrate was washed with 2 M aqueous sodium carbonate solution (2 × 50 ml), water (4 × 30 ml) and saturated aqueous sodium chloride solution (50 ml), and the organic layer was dried (MgSO4) and evaporated to give a white foam (0.340 g, 0.378 mmol, 97%).
DNA synthesis
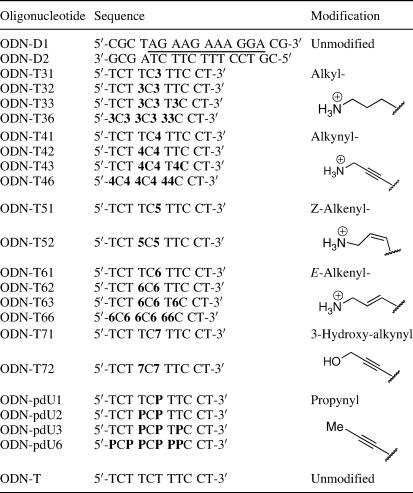
Oligonucleotides were synthesized using standard protocols on an Applied Biosystems 394 automated synthesizer using reagents, columns and phosphoramidites from Applied Biosystems, except for the mild/fast deprotection phosphoramidites and C5-propynyl-2′-deoxyuridine amidite that were purchased from Glen Research. Oligonucleotide syntheses were performed with DMT-ON using 0.1 M solutions of normal phosphoramidites and 0.15 M solutions of modified phosphoramidites. Natural oligonucleotides were deprotected at 65°C overnight while those containing modified dU analogues and N4-acetyl-dC were deprotected at room temperature overnight. All oligonucleotides were purified using reverse-phase HPLC (ODS C18, 300 × 4.6 mm2; Alltech) using a flow rate of 1 ml/min with a gradient of 5–65% CH3CN in 0.1 M triethylammonium acetate, pH 6.5 for more than 30 min. Purified oligonucleotides were then detritylated (20% AcOH at room temperature for 1 h, followed by evaporation) and re-purified by reverse-phase HPLC as above using either gradient A [5–30% CH3CN in 30 min (ODN D1–D3, ODN T41, ODN T61–T63, ODN T71 and ODN T72; see Table 2 for sequences)] or gradient B [t =0 min, 5% CH3CN; t = 20, 14% CH3CN; t = 30, 14% CH3CN; t = 40, 17% CH3CN; and t = 45, 17% CH3CN (all other ODNs)]. Retention times were 20–25 min for gradient A and 30–40 min for gradient B. Solutions of purified oligonucleotides were then evaporated, resuspended in 1 ml of water and dialysed. All the oligonucleotides were characterized using matrix-assisted laser desorption ionization time-of-flight (MALDI–TOF) mass spectra and displayed molecular weights within 4 Da of the expected masses.
Table 2.
Sequences of oligonucleotides (ODNs) used in this study
The suffix D or T refers to ODNs used in duplex formation or triplex formation, respectively. The first number refers to the analogue (see Figure 1) and the second number refers to the number of such modifications in the oligonucleotide. The sequence involved in triplex formation is underlined in ODN-D1. ‘P’ refers to C5-propynyl-2′-deoxyuridine.
UV-melting experiments
UV-melting experiments were carried out on a Cary 500 UV–visible spectrometer with a Cary temperature controller attached. UV-melting profiles were obtained over a pH range of 5.0–9.0 using an appropriate buffer for each pH. For pH 5.0–7.0, pH 7.5–8.0 and pH 8.5–9.0, sodium cacodylate, sodium phosphate and sodium borate buffers were used, respectively. The buffers used consist of 10 mM of the appropriate buffer, 300 mM sodium chloride and 0.1 mM Na2EDTA. Each sample had a total volume of 1 ml and contained 1 nmol of each oligonucleotide strand. Samples were annealed by heating to 90°C and cooling to 15°C at a rate of 2°C/min. The UV-melting curves were measured at 260 nm (in duplicate) while heating the sample from 15 to 90°C at a rate of 1°C/min. Measured melting temperature (Tm) values of duplicate runs were all within 0.5°C of one another. At heating rates of both 0.5 and 1°C/min there were no notable differences in Tm values, suggesting that the system is in equilibrium.
NMR studies of nucleosides
Nuclear Overhauser Enhancement (NOE) experiments
All NOE spectra were obtained at 400 MHz and were performed at 298 K with D2O as the solvent, except the experiment to determine double bond orientation within 5-(E-3-aminoprop-1-enyl)-2′-deoxyuridine, which was performed in (CD3)2SO owing to spectral overlap of the alkenyl peaks in D2O. 5-(Z-3-Trifluoroactamidoprop-1-enyl)-2′-deoxyuridine was measured in CD3OD because of solubility problems. d6-Acetone (10%) was introduced into each sample measured in D2O as an internal lock. The nucleoside solutions at ∼0.05 M were degassed with argon for 20 min before use. 5-(3-Aminoprop-1-ynyl)-2′-deoxyuridine, 5-(E-3-aminoprop-1-enyl)-2′-deoxyuridine in D2O, 5-(Z-3-trifluoroactamidoprop-1-enyl)-2′-deoxyuridine and 5-(3-aminopropanyl)-2′-deoxyuridine were all performed under identical processing spectral parameters applying the NOEMULT pulse sequence of the Bruker software package. An irradiation time of 5 s with an irradiation power of 76 dB <50 W gave saturation of ≤95%. 5-(Z-3-Aminoprop-1-enyl)-2′-deoxyuridine was subjected to an irradiation time of 0.2 s and 5-(E-3-aminoprop-1-enyl)-2′-deoxyuridine in (CD3)2SO was subjected to an irradiation time of 10 s. Analysis of the spectral data was performed by sequential exponential multiplication (line broadening 3) and Fourier transform of two FIDs (one for a desired irradiation point and one for the resonance control point) followed by subtraction of the two spectra. All NOE percentages were obtained by peak height measurements from the difference spectra divided by peak height measurements from the control spectra.
pKa determinations
All spectra were obtained at 400 MHz with D2O as the solvent, using its deuterium as an internal lock and a nucleoside concentration of ∼0.05 M. The pH of each solution was adjusted by the addition of 0.1 M sodium deuteroxide or deuterium chloride solution and measured using a pH meter calibrated to pH 4.01 and 7.04 using known standards.
The pKa values were determined following non-linear regression fitting to the following equation using Kaleidagraph software (Synergy Software, Reading, PA).
| 1 |
where m0 is the ppm at a given pH, m1 is pKa, m2 is the ppm of H6 at low pH and m3 is the ppm of H6 at high pH. For fitting of two pKa values to the data, the equation used is as follows:
| 2 |
where m0 is the ppm at given pH, m1 is pKa1, m2 is pKa2, m3 is the ppm of H6 at high pH (double deprotonated species), m4 is the ppm of H6 of monodeprotonated species (double deprotonation) and m5 is the ppm of H6 of monodeprotonated species (double deprotonation).
RESULTS AND DISCUSSION
Synthesis of C5-amino-modified 2′-deoxyuridines
The trifluoroacetylated derivative of 5-(propargylamino)-2′-deoxyuridine (4b) was prepared as described by Hobbs (28) and was used as the precursor for the corresponding Z-alkenyl (5b) and 5-(3-aminopropyl) (3b) analogues as described previously (13,30). The 5-aminoallyl derivative (6a) was prepared according to the modification described by Dey and Sheppard (29) from the original palladium-catalysed coupling procedure described by Sakthivel and Barbas (17).
Treatment of the trifluoroacetyl protected nucleosides 3b, 4b and 6b with aqueous ammonia solution at room temperature overnight afforded the corresponding amino nucleosides 3a, 4a and 6a. In contrast, when the cis-alkenyl derivative 5b was treated with ammonia, a novel bicyclic nucleoside 8 was formed which was characterized using both 1H NMR and mass spectrometry. Analysis of the reaction mixture by reverse-phase HPLC indicated that the amount of the less polar, later-eluting peak corresponding to nucleoside 8 was ∼90% after ammonolysis for 4 h at room temperature. However, the deprotected compound 5a could be obtained as its ammonium salt by reducing the time of the ammonolysis to 45 min, followed by subsequent acidification to pH 4 with trifluoroacetic acid. Compound 5a was found to be stable as its ammonium salt, but underwent complete conversion into the bicyclic nucleoside 8 if the sample was left overnight in aqueous solution above pH 8.
Conformational analysis of C5-amino-modified 2′-deoxyuridines
The glycosidic bond angle (χ) defined as the torsion angle between the O4′–C1′ and N1–C2 bonds of pyrimidine nucleosides defines two major conformations in dynamic equilibrium, either anti or syn in which the base H6 or O2 atoms, respectively, lie above the sugar. The normal conformation in pyrimidine nucleosides is anti. Seela and co-workers (31) have described a simple semi-quantitative method for estimating the population of molecules of a given nucleoside occupying these conformations, which may be derived from proton NOE values between the H-6 proton (for pyrimidines) and the sugar H1′, H2′ and H3′ protons. By using this method, the determined NOE values for the nucleosides 3a–6a (see Table 1 in Supplementary Material) were used to estimate the percentage of the anti conformer for each respective nucleoside (Table 1). The amount of anti conformer estimated for thymidine using this method (31) is 78 ± 10%. As shown in Table 1, all compounds 3a–6a exist predominantly in the anti conformation within the normal expected range. Thus, the side chains do not have any appreciable effect on the glycosylic torsion angles that these analogues display compared with thymidine. All the analogues also display the same conformational preference of the deoxyribose ring for the 2′-endo (S-conformation) that is found for thymidine (32) (see Table 1).
Table 1.
Physical properties of C5-modified 2′-deoxyuridines determined using 1H NMR
| Compound | Percentage of anti-conformation | Percentage of S-conformera (2′-endo) | Orientation of alkenyl group | pKa valueb |
|---|---|---|---|---|
| 3a (alkane) | 78 | 59 | — | 10.00 ± 0.04 (8.96) |
| 4a (alkyne) | 71 | 58 | — | 8.53 ± 0.09 (8.05) |
| 5a (Z-alkene) | 89 | 56 | Slight cisoid preference | 9.79 ± 0.15 (9.31) |
| 6a (E-alkene) | 76 | 58 | Cisoid | 9.69 ± 0.04 (9.34) |
| Thymidine | 78c | 62d | — | 10.42 ± 0.04 (9.96) |
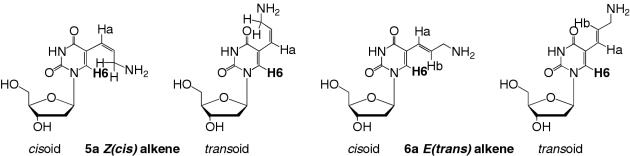
Oligonucleotides containing an amino group tethered by an alkenyl linker are likely to have very different properties depending on the stereochemistry of the double bond and its orientation with respect to the heterocyclic base. Thus, for compounds 5a and 6a both cisoid and transoid orientations are possible (Figure 2) owing to restricted rotation around the C5–C7 single bond resulting from the conjugated π system in these molecules. 1H NOE was used to estimate the populations of the respective conformations. For the E-alkene (6a), 1% NOE was seen between Hb and H6, while no NOE was observed between Ha and H6, demonstrating a strong preference for the cisoid conformation. In contrast, for the Z-alkene (5a), upon irradiation of Ha, NOEs to both H6 and the methylene protons (1.3%) indicate a slight preference for the cisoid conformation. The corresponding NOEs between H6 and Ha (2.6%) and the methylene protons (3.5%) for the trifluoroacetyl protected compound 5b indicate the same preference of the protected Z-alkene for the cisoid conformation. The cyclization of 5a to give nucleoside 8 must occur through the transoid conformation, suggesting a low-energy barrier to the interconversion between this and the cisoid conformation.
Figure 2.
Structures of cisoid and transoid conformations of C5-amino-alkenyl modified 2′-deoxyuridines 5a and 6a.
Determination of the pKa values of C5-amino-modified 2′-deoxyuridines
To our knowledge, the C5-amino-modified 2′-deoxyuridines, 3a–6a, are expected to be cationic at physiological pH but their pKas have not been determined to date. The respective NMR signals associated with the H6 protons of C5-aminoalkyl-2′-deoxyuridines are highly sensitive to the nature of the C5 aminoalkyl side chain (see Materials and Methods). For this reason, we chose to determine the pKa values of these compounds using 1H NMR by measuring the chemical shift of H6 over a range of pH values. The respective plots are shown in Supplementary Figure 1 and the pKa values derived from them using Equation 1 (see Materials and Methods) are shown in Table 1. At high pH (>11), a second transition appears to be present in the titration curve for the Z-alkenyl analogue 5a. This presumably arises owing to the cyclization of 5a to form the bicyclic compound 8.
The reported pKa values for propargylamine, allylamine and propylamine are 8.15 (33), 9.49 (34) and 10.53 (34), respectively, and may be interpreted based on the alkynyl group being the best electron withdrawing group, thereby producing the least basic amino group. Although the same trend is observed for the nucleosides, those bearing the alkyne- and alkene-tethered amino groups are more basic than expected, while the pKa of the aminopropyl-modified analogue 3a is less basic than expected. The determined pKa of thymidine under our experimental conditions was found to be 10.42 (data not shown). The pKas measured in these experiments represent macroscopic pKa values for these nucleosides and cover the double-deprotonation event, i.e. from cationic nucleoside to anionic nucleoside in which the side chain is neutral and the N1 imino proton has dissociated. Our attempts to fit the titration data to two pKa values provides an estimate of the lower value of these two pKas, below which the side chain is protonated and the base is neutral. This value is also displayed in Table 1. The range of pKa values calculated indicates that during UV-melting studies of DNA triple helices containing modified oligonucleotides (see below) the base analogues are largely protonated over the range of pH values investigated (pH 5–7).
Chemical synthesis of oligonucleotides containing C5-modified 2′-deoxyuridines
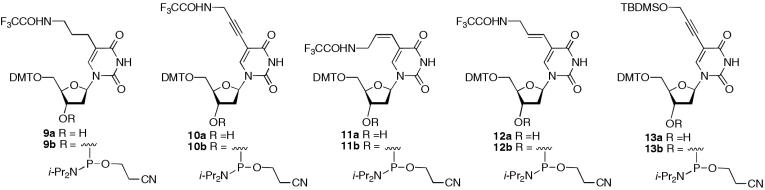
The trifluoroacetyl protected nucleosides 3b–6b were converted into their corresponding 5′-dimethoxytrityl derivatives (9a–12a) and the phosphoramidites 9b–12b (Figure 3) were then prepared by phosphitylation with 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite. The alkyne 10b and E-alkene derivative 12b have been prepared previously by others (4,26,27) and together with compounds 9b and 11b showed the presence of two diastereoisomers using 31P NMR. All of the phosphoramidites display the expected accurate masses following analysis using electrospray mass spectrometry (see Materials and Methods). The phosphoramidite 13b was prepared from 5′-O-dimethoxytrityl-5-iodo-2′-deoxyuridine, by palladium-catalysed coupling with propargyl alcohol. This was followed by protection of the side chain alcohol function as its t-butyldimethylsilyl ether to give 13a, which was subsequently phosphitylated as described above. 31P NMR and accurate mass measurement were used to characterize compound 13b.
Figure 3.
Structures of phosphoramidites of C5-modified 2′-deoxyuridines (9b–13b) used in DNA synthesis.
In order to investigate the effects of C5 modifications on the stability of DNA triple helices, we synthesized the oligonucleotides shown in Table 2. The triple-helix structure chosen for our study has been used previously by others (35) and provides clear melting transitions for the triplex and duplex components. The polypyrimidine third strand of the triplex-forming structures shown in Table 2 contained the bases cytosine, thymine and one or more C5-modified uracil. The phosphoramidites 9b–13b were employed in standard oligonucleotide synthesis, using 0.15 M rather than the standard 0.1 M solutions in acetonitrile and typically resulted in coupling yields ∼95%. To avoid normal deprotection conditions, which were particularly problematic for oligonucleotides containing analogue 5 (see below), N4-acetyl-protected 2′-deoxycytidine phosphoramidite was used and deprotection of DMT-ON oligonucleotides following synthesis was achieved using concentrated aqueous ammonia solution at room temperature for 12 h (4 h for oligonucleotides containing 5a). The crude oligonucleotides containing analogues 3, 4, 6 and 7 were all purified by reverse-phase HPLC and detritylated using 20% aqueous acetic acid, re-purified by HPLC and then dialysed. However, when oligonucleotides containing 5-(Z-3-aminoprop-1-enyl)-2′-deoxyuridine 5 were deprotected, multiple peaks were observed using HPLC owing to the formation of compound 8 within the oligonucleotide. Analysis using MALDI mass spectrometry of the two major peaks observed following the deprotection of ODN-T51 indicated that the minor peak (∼30%) contained the desired analogue 5a, while the later-eluting major peak contains an oligonucleotide that is 18 mass units lower owing to the conversion of 5a into the bicyclic analogue 8. Owing to the complexity of the HPLC traces obtained for oligonucleotides containing analogue 5, it was not possible to purify an oligonucleotide containing more than two such modifications.
Thermal stability studies of DNA triple helices containing C5-amino-modified 2′-deoxyuridines
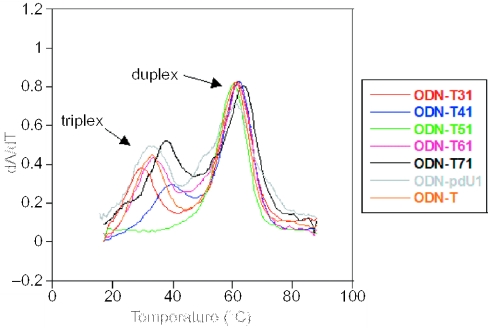
The UV-melting curves for the triplex structures in which the third pyrimidine-rich strand contains one or more C5-amino-modified 2′-deoxyuridine were measured between pH 5.0 and 7.0 and compared with the melting of the unmodified triplex. The triplex structure and sequences of oligonucleotides are shown in Table 2. At low pH values cytosine becomes protonated (pKa ∼4.5) and the unmodified polypyrimidine strand participates in triplex formation resulting in two distinct melting transitions (triplex to duplex and duplex to single-stranded) with increased temperature. Figure 4 shows the first derivatives of the melting curves for the unmodified and modified triplexes measured at pH 5, in which a single thymidine in the third strand has been replaced by one of the analogues 3a–6a, 7 or C5-propynyl-2′-deoxyuridine. In agreement with the previous work (4), the propargylamino-modified 2′-deoxyuridine has a dramatic effect on the stability of the triplex with an increase in Tm > 6°C with just a single substitution. When a single thymidine is replaced by the C5-aminoalkene analogue containing a trans (E) double bond in the linker (6a) the Tm is essentially unchanged (ODN-T61), while an aminopropyl-modified analogue significantly decreases the Tm (ODN-T31). Interestingly, an oligonucleotide containing the aminoalkene analogue possessing a cis (Z) double bond (5a) (ODN-T51) is unable to form a triplex even at pH 5 despite the fact that under such conditions the amino group is essentially fully protonated. All triplex structures containing analogues 3a, 5a or 6a become less stable as the number of amino-modified dU analogues is increased (see Table 3). This may result from intrinsic destabilizing properties of the analogues themselves or from a destabilization owing to repulsion between cationic side chains. In contrast, triple helices containing the propargylamino-modified analogue 4a exhibit a dramatic increase in stability that is proportional to the number of substitutions (compare triplexes containing ODN-T46 and ODN-T41).
Figure 4.
First derivative of melting curves for triplex structures containing a single C5-amino-modified dU analogue in the third strand. Spectra were recorded at 260 nm and at pH 5. For complete experimental details see Materials and Methods and for sequences see Table 2.
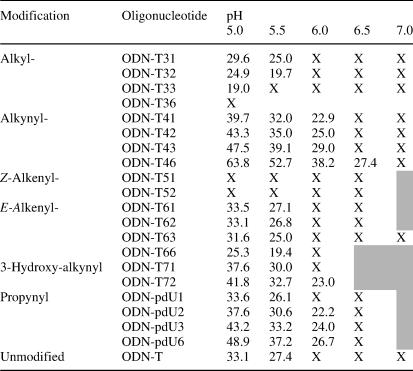
Table 3.
Thermal melting midpoint (Tm) values for triplex melting at various pH values
The oligonucleotide refers to the third strand of the triplex structure indicated in Table 1. ‘X’ indicates that no melting transition was observed. The shaded sections of the table indicate that no data was collected. For full experimental details see Materials and Methods.
The stabilities of all the triple-helical structures decrease with increasing pH and only when the third strand contains amino-modified dU analogues containing the alkyne linker triplexes are formed above pH 5.5 (see Table 3). Over the pH range studied, the relative stabilities of triplexes containing the amino-modified dU analogues are preserved; alkynyl > trans-alkenyl ∼ unmodified > alkyl > cis-alkenyl (Table 3). Below pH 7, all the amino-modified side chains of the dU analogues are largely protonated and it is quite clear that the relative abilities of such analogues to stabilize triple helices depend less on the ionization state of the base, but rather on the type of linker between the protonated amino group and the nucleobase. The ability of C5-propynyl-2′-deoxyuridine (i.e. with no appended cations to the nucleobase) to stabilize triple helices through enhanced base stacking is well-documented (36,37). In DNA duplexes and triplexes, a C5-propynyl-modified cytosine or uracil can enhance stability (7). Replacement with the propargylamino analogue 4a is also known to increase stability further (6,8). When the amino group is protected as an amide, lower duplex stabilities result (8,38). These studies with duplexes clearly demonstrate the effect of an appended amino group on stability. However, the exact role it plays, in particular whether it enhances base stacking of the analogue or whether it forms favourable electrostatic interactions, e.g. with neighbouring phosphates is not clear.
In order to assess the contribution towards stability imparted by the combination of the amino group and the propynyl group of 4a, we decided to investigate the ability of oligonucleotides containing the novel dU analogue 7 and those containing C5-propynyl-2′-deoxyuridine to form triple-helical structures. Compound 7 retains the propynyl side chain that is present in analogue 4a, but also bears a tethered electronegative hydroxyl group. Thus, it is analogous to the protonated amino group of 4a but lacking a positive charge. Interestingly, incorporation of analogue 7 into the third strand of the triplex results in an increase in stability that is only slightly lower than that observed for triplexes that contain the propargylamino modification (compare ODN T41 and 71 and ODN T42 and 72). Furthermore, essentially the same pH-dependence of triple-helix stability is observed for structures containing either analogue 4a or 7 (Supplementary Figure 2), which presumably results from the requirement to protonate cytosines within the triple-helix forming oligonucleotide (TFO). The analogous triplex structures containing C5-propynyl-2′-deoxyuridine display enhanced stabilities relative to the unmodified sequence but these are much less than those containing 4a or 7. The results suggest that a major factor in the ability of the propargylamino-modified dU analogue to stabilize triple helices was derived mainly from the nature of the alkynyl group itself with a lesser contribution being derived from the possible favourable electrostatic interactions. Indeed, modelling studies (8) suggest that the amino group of the propargylamino analogue 4a displays a preference for location within the major groove of DNA rather than for salt bridge formation with nearby phosphate groups. Therefore, it seems most probable that other electron-withdrawing groups appended to the alkyne would also result in a similar stability increase.
In the previous studies, the effects of ionic strength on the stabilities of DNA duplexes containing C5-aminoalkyl-modified dU analogues have been undertaken (2,3,6). At lower ionic strengths, unmodified duplexes show a greater relative decrease in stability when compared with the same sequences containing such appended cationic residues. In their study of triple-helix stabilization by the analogue 6a, Brown and Fox (4) obtained data at 100 mM NaCl concentration using a TFO containing the analogue within an oligothymidine sequence. In this study, we used a mixed sequence TFO (i.e. containing cytosine and thymine bases) and did not observe triplex formation at 100 mM NaCl. We were also unable to measure Tm values at 200 mM NaCl for the doubly modified sequences, ODN-T72 and ODN-T42, which contain the hydroxypropynyl and aminopropynyl-modified analogues, respectively. Therefore, a comparison of the ionic strength dependence on the stability of TFOs containing the two analogues was not possible. Therefore, in comparison with the previous studies using the same triplex sequence (35), we measured the triplex melting at 300 mM NaCl. At this ionic strength, it might be argued that the differences in stabilities of TFOs containing these two analogues might be masked to some extent. While this cannot be entirely ruled out, the fact that TFOs containing propynyl dU are much less stable (compare ODN-T42 and ODN-T72 with ODN-pdU2 in Table 3) suggests that the combination of amino or hydroxyl group and the propynyl substituent are particularly favourable.
The reason for the low stabilities of triplexes containing C5-amino-modified dU analogues that have alkyl or Z-alkenyl linker arms is unclear. However, for the propyl analogue 3a interaction of the amino group with the O4-carbonyl group of the nucleobase is possible, which would severely disrupt the ability of these analogues to form Hoogsteen base pairs required for triplex formation. Indeed it is clear from PAGE, modelling and NMR studies (12,39–41) that the side chain of this analogue prefers to interact with the major groove of a DNA duplex rather than the phosphate diester backbone. For the Z-alkenyl analogue, the transoid conformation would be required for such an interaction with the carbonyl group. Although NMR experiments indicate the presence of both the transoid and cisoid conformations, it is quite possible that for the latter conformation the charged amino group lies outside the plane of the base in order to minimize unfavourable steric interactions with the H6 proton of the base. Such a conformation would also likely be highly disruptive to DNA secondary structures. In the previous studies, we have noted that the triphosphates of analogues 3a and 5a are not incorporated by a variety of DNA polymerases (13) while those of analogues 4a and 6a are good substrates. Furthermore, we have been unable to form duplex structures that contain the Z-alkene 5a (Brazier,J.A. and Williams,D.M., unpublished data). We attribute these findings to the positively charged amino group, which for each analogue is placed in a position that is highly destabilizing to base pair formation. In contrast, for the E-alkene analogue 6a (cisoid conformation) and alkyne analogue 4a such destabilizing interactions cannot take place since the amino group is located much further away from the base and is able to reside in the same plane. The results of the current study imply that this latter feature is the most important in terms of stability and that the effects of a charged amino group are to enhance the intrinsic stabilizing properties of C5-propynyl group in nucleic acid structures.
CONCLUSIONS
We have reported on the conformational properties and pKa values of four different C5-amino-modified 2′-deoxyuridines that differ in the nature of the linker between the amino group and the nucleobase. We have prepared oligonucleotides containing these analogues and have shown that the relative stabilities of DNA triplexes containing these analogues are in the following order: alkyne > E-alkene > alkane > Z-alkene. We have also shown that the stabilizing effects of the propargylamino modifications are likely to arise principally from favourable base stacking interactions since we have found a high stability for triplex structures containing 5-(3-hydroxyprop-1-ynyl)-2′-deoxyuridine. The results are relevant to the design of both triplex and duplex structures that contain analogues designed to enhance their stabilities. In addition, the physicochemical properties of the analogues will be relevant to understanding the properties of nucleic acid structures and catalysts designed using the SELEX technique.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Simon Thorpe for obtaining mass spectral data and Drs Sham Haq and Jane Grasby for valuable discussions. Funding to pay the open access publication charges for this article was provided by The Department of Chemistry, University of Sheffield.
REFERENCES
- 1.Ruth J. Oligonucleotides with reporter groups attached to the base. In: In Eckstein F., editor. Oligonucleotides and Analogues, 1st edn. NY: Oxford University Press; 1991. pp. 255–282. [Google Scholar]
- 2.Hashimoto H., Nelson M.G., Switzer C. Zwitterionic DNA. J. Am. Chem. Soc. 1993;115:7128–7134. [Google Scholar]
- 3.Hashimoto H., Nelson M.G., Switzer C. Formation of chimeric duplexes between zwitterionic and natural DNA. J. Org. Chem. 1993;58:4194–4195. [Google Scholar]
- 4.Bijapur J., Keppler M.D., Bergqvist S., Brown T., Fox K.R. 5-(1-Propargylamino)-2′-deoxyuridine (u-p): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sollogoub M., Darby R.A.J., Cuenoud B., Brown T., Fox K.R. Stable DNA triple helix formation using oligonucleotides containing 2′-aminoethoxy,5-propargylamino-U. Biochemistry. 2002;41:7224–7231. doi: 10.1021/bi020164n. [DOI] [PubMed] [Google Scholar]
- 6.Heystek L.E., Zhou H.Q., Dande P., Gold B. Control over the localization of positive charge in DNA: the effect on duplex DNA and RNA stability. J. Am. Chem. Soc. 1998;120:12165–12166. [Google Scholar]
- 7.Seela F., Ramzaeva N., Leonard P., Chen Y., Debelak H., Feiling E., Kroschel R., Zulauf M., Wenzel T., Frohlich T., Kostrzewa M. Phosphoramidites and oligonucleotides containing 7-deazapurines and pyrimidines carrying aminopropargyl side chains. Nucleosides Nucleotides Nucleic Acids. 2001;20:1421–1424. doi: 10.1081/NCN-100002568. [DOI] [PubMed] [Google Scholar]
- 8.Hardwidge P.R., Lee D.K., Prakash T.P., Iglesias B., Den R.B., Switzer C., Maher L.J. DNA bending by asymmetrically tethered cations: influence of tether flexibility. Chem. Biol. 2001;8:967–980. doi: 10.1016/s1074-5521(01)00065-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang F., Li Z.J., Scholdberg T.A., Li J.S., Gold B., Stone M.P. Incorporation of the cationic 5-(3-aminopropyl)-2′-deoxyuridine (z3dU) into the Dickerson dodecamer: evidence for site-specific DNA bending. Biochemistry. 2003;42:73. [Google Scholar]
- 10.Strauss J.K., Prakash T.P., Roberts C., Switzer C., Maher L.J. DNA bending by a phantom protein. Chem. Biol. 1996;3:671–678. doi: 10.1016/s1074-5521(96)90135-0. [DOI] [PubMed] [Google Scholar]
- 11.Strauss J.K., Roberts C., Nelson M.G., Switzer C., Maher L.J. DNA bending by hexamethylene-tethered ammonium ions. Proc. Natl Acad. Sci. USA. 1996;93:9515–9520. doi: 10.1073/pnas.93.18.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z.J., Huang L., Dande P., Gold B., Stone M.P. Structure of a tethered cationic 3-aminopropyl chain incorporated into an oligodeoxynucleotide: evidence for 3′-orientation in the major groove accompanied by DNA bending. J. Am. Chem. Soc. 2002;124:8553–8560. doi: 10.1021/ja0201707. [DOI] [PubMed] [Google Scholar]
- 13.Lee S.E., Sidorov A., Gourlain T., Mignet N., Thorpe S.J., Brazier J.A., Dickman M.J., Hornby D.P., Grasby J.A., Williams D.M. Enhancing the catalytic repertoire of nucleic acids: a systematic study of linker length and rigidity. Nucleic Acids Res. 2001;29:1565–1573. doi: 10.1093/nar/29.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masud M.M., Kuwahara M., Ozaki H., Sawai H. Sialyllactose-binding modified DNA aptamer bearing additional functionality by selex. Bioorg. Med. Chem. 2004;12:1111–1120. doi: 10.1016/j.bmc.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Matulic-Adamic J., Daniher A.T., Karpeisky A., Haeberli P., Sweedler D., Beigelman L. Functionalized nucleoside 5′-triphosphates for in vitro selection of new catalytic ribonucleic acids. Bioorg. Med. Chem. Lett. 2000;10:1299–1302. doi: 10.1016/s0960-894x(00)00226-2. [DOI] [PubMed] [Google Scholar]
- 16.Matulic-Adamic J., Daniher A.T., Karpeisky A., Haeberli P., Sweedler D., Beigelman L. Synthesis of modified nucleoside 5′-triphosphates for in vitro selection of catalytic nucleic acids. Nucleosides Nucleotides Nucleic Acids. 2001;20:1113–1115. doi: 10.1081/NCN-100002500. [DOI] [PubMed] [Google Scholar]
- 17.Sakthivel K., Barbas C.F. Expanding the potential of DNA for binding and catalysis: highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem. Int. Ed. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Vaish N.K., Fraley A.W., Szostak J.W., McLaughlin L.W. Expanding the structural and functional diversity of RNA: analog uridine triphosphates as candidates for in vitro selection of nucleic acids. Nucleic Acids Res. 2000;28:3316–3322. doi: 10.1093/nar/28.17.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewey T.M., Zyzniewski M.C., Eaton B.E. RNA world: functional diversity in a nucleoside by carboxyamidation of uridine. Nucleosides Nucleotides. 1996;15:1611–1617. [Google Scholar]
- 20.Latham J.A., Johnson R., Toole J.J. The application of a modified nucleotide in aptamer selection—novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res. 1994;22:2817–2822. doi: 10.1093/nar/22.14.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoetzau T., Langner J., Moyroud E., Roehl I., Vonhoff S., Klussmann S. Aminomodified nucleobases: functionalized nucleoside triphosphates applicable for SELEX. Bioconjug. Chem. 2003;14:919–926. doi: 10.1021/bc0256547. [DOI] [PubMed] [Google Scholar]
- 22.Perrin D.M., Garestier T., Helene C. Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucleosides Nucleotides Nucleic Acids. 1999;18:377–391. doi: 10.1080/15257779908043083. [DOI] [PubMed] [Google Scholar]
- 23.Perrin D.M., Garestier T., Helene C. Bridging the gap between proteins and nucleic acids: a metal-independent RNase A mimic with two protein-like functionalities. J. Am. Chem. Soc. 2001;123:1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]
- 24.Battersby T.R., Ang D.N., Burgstaller P., Jurczyk S.C., Bowser M.T., Buchanan D.D., Kennedy R.T., Benner S.A. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc. 1999;121:9781–9789. doi: 10.1021/ja9816436. [DOI] [PubMed] [Google Scholar]
- 25.Roychowdhury A., Illangkoon H., Hendrickson C.L., Benner S.A. 2′-Deoxycytidines carrying amino and thiol functionality: synthesis and incorporation by vent (exo(-)) polymerase. Org. Lett. 2004;6:489–492. doi: 10.1021/ol0360290. [DOI] [PubMed] [Google Scholar]
- 26.Cook A.F., Vuocolo E., Brakel C.L. Synthesis and hybridization of a series of biotinylated oligonucleotides. Nucleic Acids Res. 1988;16:4077–4095. doi: 10.1093/nar/16.9.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lermer L., Roupioz Y., Ting R., Perrin D.M. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- 28.Hobbs F.W. Palladium-catalyzed synthesis of alkynylamino nucleosides—a universal linker for nucleic-acids. J. Org. Chem. 1989;54:3420–3422. [Google Scholar]
- 29.Dey S., Sheppard T.L. Ketone-DNA: a versatile postsynthetic DNA decoration platform. Org. Lett. 2001;3:3983–3986. doi: 10.1021/ol016626r. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.E., Vyle J.S., Williams D.M., Grasby J.A. Novel syntheses of (Z)-alkene and alkane base-modified nucleosides. Tetrahedron Lett. 2000;41:267–270. [Google Scholar]
- 31.Rosemeyer H., Toth G., Golankiewicz B., Kazimierczuk Z., Bourgeois W., Kretschmer U., Muth H.P., Seela F. Syn anti-conformational analysis of regular and modified nucleosides by 1D H-1 NOE difference spectroscopy—a simple graphical-method based on conformationally rigid molecules. J. Org. Chem. 1990;55:5784–5790. [Google Scholar]
- 32.Rinkel L.J., Altona C. Conformational-analysis of the deoxyribofuranose ring in DNA by means of sums of proton–proton coupling-constants—a graphical-method. J. Biomol. Struct. Dyn. 1987;4:621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- 33.Perrin D.D. Dissociation Constants of Organic Bases in Aqueous Solution. London: Butterworths; 1965. [Google Scholar]
- 34.Hall H.K., Jr Correlation of the base strengths of amines. J. Am. Chem. Soc. 1957;79:5441–5443. [Google Scholar]
- 35.Asensio J.L., Dosanjh H.S., Jenkins T.C., Lane A.N. Thermodynamic, kinetic, and conformational properties of a parallel intermolecular DNA triplex containing 5′ and 3′ junctions. Biochemistry. 1998;37:15188–15198. doi: 10.1021/bi980057m. [DOI] [PubMed] [Google Scholar]
- 36.Froehler B.C., Wadwani S., Terhorst T.J., Gerrard S.R. Oligodeoxynucleotides containing C-5 proplyne analogs of 2′- deoxyuridine and 2′-deoxycytidine. Tetrahedron Lett. 1992;33:5307–5310. [Google Scholar]
- 37.Phipps A.K., Tarkoy M., Schultze P., Feigon J. Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)- 2′-deoxyuridine residues in the third strand. Biochemistry. 1998;37:5820–5830. doi: 10.1021/bi972811u. [DOI] [PubMed] [Google Scholar]
- 38.Ahmadian M., Zhang P.M., Bergstrom D.E. A comparative study of the thermal-stability of oligodeoxyribonucleotides containing 5-substituted 2′-deoxyuridines. Nucleic Acids Res. 1998;26:3127–3135. doi: 10.1093/nar/26.13.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gold B. Effect of cationic charge localization on DNA structure. Biopolymers. 2002;65:173–179. doi: 10.1002/bip.10215. [DOI] [PubMed] [Google Scholar]
- 40.Dande P., Liang G.N., Chen F.X., Roberts C., Nelson M.G., Hashimoto H., Switzer C., Gold B. Regioselective effect of zwitterionic DNA substitutions on DNA alkylation: evidence for a strong side chain orientational preference. Biochemistry. 1997;36:6024–6032. doi: 10.1021/bi962602u. [DOI] [PubMed] [Google Scholar]
- 41.Liang G.N., Encell L., Nelson M.G., Switzer C., Shuker D.E.G., Gold B. Role of electrostatics in the sequence-selective reaction of charged alkylating-agents with DNA. J. Am. Chem. Soc. 1995;117:10135–10136. [Google Scholar]
- 42.Poznanski J., Felczak K., Kulikowski T., Remin M. H-1 NMR conformational study of antiherpetic C5-substituted 2′-deoxyuridines: insight into the nature of structure–activity relationships. Biochem. Biophys. Res. Commun. 2000;272:64–74. doi: 10.1006/bbrc.2000.2725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.