Abstract
The semi-enclosed nature of the Red Sea (20.2°N–38.5°N) makes it a natural laboratory to study the influence of environmental gradients on microbial communities. This study investigates the composition and structure of microbial prokaryotes and eukaryotes using molecular methods, targeting ribosomal RNA genes across different regions and seasons. The interaction between spatial and temporal scales results in different scenarios of turbulence and nutrient conditions allowing for testing of ecological theory that categorizes the response of the plankton community to these variations. The prokaryotic reads are mainly comprised of Cyanobacteria and Proteobacteria (Alpha and Gamma), with eukaryotic reads dominated by Dinophyceae and Syndiniophyceae. Periodic increases in the proportion of Mamiellophyceae and Bacillariophyceae reads were associated with alterations in the physical oceanography leading to nutrient increases either through the influx of Gulf of Aden Intermediate Water (south in the fall) or through water column mixing processes (north in the spring). We observed that in general dissimilarity amongst microbial communities increased when nutrient concentrations were higher, whereas richness (observed OTUs) was higher in scenarios of higher turbulence. Maximum abundance models showed the differential responses of dominant taxa to temperature giving an indication how taxa will respond as waters become warmer and more oligotrophic.
Introduction
Ecological and biogeochemical processes in the ocean are dependent on a diverse assemblage of microbes including members from Archaea, Bacteria and Eukarya. The diverse plankton assemblages comprising both prokaryotes and eukaryotes fulfill a wide variety of ecological roles in the marine system including carbon fixation (e.g., refs 1 and 2), biogeochemical cycling1, 3 and trophic energy transfer (e.g., ref. 4). Approximately half of the global primary production is carried out by oceanic microbes5, with contributions from the bacterial genera Synechococcus and Prochlorococcus, and diverse lineages of small eukaryotes, being especially important2. In the oligotrophic regions, this primary production is tightly recycled in the microbial loop, with a small proportion being transferred into higher trophic levels4.
Until the regular use of molecular techniques within the marine environment, studies generally relied on morphological characteristics (limiting investigations to those taxa which could be identified under a microscope) or through pigment analysis (limited to pigmented taxa). Therefore, until relatively recently, a detailed understanding of the biological processes occurring in the marine environment was impossible and an integrated assessment of diversity was missing. Indeed, the full extent of their diversity, especially among the eukaryotes, is still poorly understood with novel clades, even at the Kingdom level of classification, being discovered relatively recently (i.e., Picobiliphyta6 or Rappemonada7). Molecular techniques have been used to examine changes in the community of marine plankton across spatial (e.g., refs 8–10) and temporal (e.g., ref. 11) gradients and to assess the effects of environmental patterns on their composition. In the Red Sea, the number of molecular studies investigating the diversity of the microbial component is extremely limited (e.g., refs 12–16). Other methods have investigated the planktonic diversity in the Red Sea including fingerprinting techniques17, microscopy and HPLC18. Studies in the Red Sea tend to be restricted in their extent either spatially or temporally. For example, studies undertaken by Kürten et al.17, Pearman et al.16 and Kheireddine et al.19 have a wide spatial coverage but are limited to a single temporal period while Touliabah et al.20 and Al-Najjar et al.18 investigated seasonal effects in planktonic communities in a restricted geographic range.
Understanding the control mechanisms and resilience of communities is a critical topic in ecology, especially considering the ever increasing climatic and anthropogenic impacts the marine environment is subjected to. Physical processes can determine the predominant planktonic groups present in a pelagic environment and inherent patterns in size frequency distribution, size spectra and the community composition that are prevalent21. For example, it has been shown that the size structure of the phytoplankton community shows a relatively stable inter-annual trend in lake systems21 with Gin et al.22 showing less variability in oligotrophic marine waters compared with coastal waters. Here, stability is not suggested as being a constant but more oscillations around a central point with a known periodicity (i.e., seasonal cycles)21. Margalef23 and expanded on by Cullen et al.24 proposed that a combination of turbulence (descried as turbulent mixing of the water column by external forces, such as winds, tides, or upwelling) and nutrient concentrations, which can be affected by seasonal patterns, could determine the community structure. This concept has been further expanded to incorporate further effects or response traits to explain community structures in the plankton25.
It has been proposed that in low nutrient and low turbulence areas, typical of stratified tropical oceans the competition for nutrients and the retention of these nutrients, via recycling of organic nutrients, within the microbial loop is the dominant process resulting in a predominance of picophytoplankton (e.g., Synechococcus, Prochlorococcus and eukaryotic phytoplankton less than 2 μm) and slow growing groups with specialist strategies (e.g., mixotrophy)24, 25. Indeed, it has been found that in a stable stratified water column, oscillations in productivity26, abundance27, 28 and community structure29 are minor. As nutrients increase within the system it is proposed there is an increase in biomass and the size of cells but with a slower turnover24. It was further proposed that as turbulence increased toward a high nutrient and turbulence situation increased size and biomass would be favored, and there would be transient selection for taxa with rapid growth (e.g., diatoms)24, 25. One further category was proposed by Cullen et al.24 that of the high turbulence and low nutrient area where low biomass and turnover would be prevalent with a selection for organisms, which efficiently used light and nutrients.
In order to test whether responses of plankton in the Red Sea conform to current morphologic-based ecological theories we tested responses over large spatial and temporal gradients using molecular data. The Red Sea is a narrow, semi-confined basin, with limited exchange to other seas. Distinct gradients in temperature, salinity and nutrients are observed along its latitudinal axis. Temperature increases from north to south, salinity generally follows the opposite trend30, and nutrients are higher in southern regions compared with the north13, 16, 31. Satellite imaging in the main body of the Red Sea indicates a strong seasonality in surface chlorophyll a concentrations32. During the Arabian Sea northeast monsoon (winter), the prevailing winds over the Red Sea are SE to SSE in the south resulting in a two-layer exchange of water across the Bab el Mandab region33. The winds bring surface water into the Red Sea from the Gulf of Aden while Red Sea deep-water outflows into the Gulf34. In the northern Red Sea, vertical mixing of the water column can bring nutrients to the surface allowing for increased chlorophyll a concentrations35. During the Arabian Sea southwest monsoon (summer), the wind forcing is from the NW along the entire Red Sea. The shift to NW winds in the south leads to a three-layer flow pattern with the surface water being driven from the Red Sea into the Gulf of Aden, while the deep water continues to be exported from the Red Sea. The Gulf of Aden Intermediate Water (GAIW), which can occupy up to 70% of the water column, transports cool, lower salinity, nutrient-rich water into the southern Red Sea contributing to the productivity of the system. Despite the increasing body of literature focusing on the Red Sea circulation patterns, a better understanding of how they can affect the biogeochemical processes is still needed. Further, due to the effects of global warming a larger proportion of the world’s oceans is likely to become warmer and more oligotrophic. This makes the Red Sea, which has a gradient in its upper layer from lower temperature and low nutrients in the north to high temperature and higher nutrients in the south, a perfect natural laboratory to study the structure and diversity of microbial plankton communities and their main environmental drivers. Seasonal changes in not only circulation patterns and consequent alterations of water turbulence and nutrient availability but also temperature and salinity allow the investigation of the responses of microbial communities to alterations in prevailing environmental variables under different scenarios.
In accordance with the categories proposed by Cullen et al.24, we propose that the food web in the northern region of the Red Sea will be dominated by the microbial loop in the summer, while the more nutrient rich southern region will show an increase in the abundance of larger cells typical of fast growing taxa (e.g., diatoms). We do anticipate intra-region seasonal variability driven by changes in temperature and nutrient availability. We undertake species distribution modeling of some of the dominant taxa observed in the Red Sea to gain a greater understanding of the response of these groups to changes in their environment and so increasing the understanding of the limits to their distribution that can be of interest when addressing ecological trajectories under climate change scenarios.
Results
Physical structure
Mean profiles in the north in spring showed a predominantly mixed water column (mixed layer depth 84 m) with average chlorophyll maximum at approximately 70 m (Fig. 1) (Supplementary Table 1). In the fall, the mixed layer is considerably shallower at 10 m. In the south, the upper layer shows strong seasonality with average chlorophyll maximum at about 40 m. The mixed layer was approximately 50 m in the spring but only 15 m in the fall (Fig. 1) (Supplementary Table 1). In the south, an intrusion of low temperature, low salinity and low oxygen water is observed between 50 and 100 m depth in the water column during fall.
Figure 1.
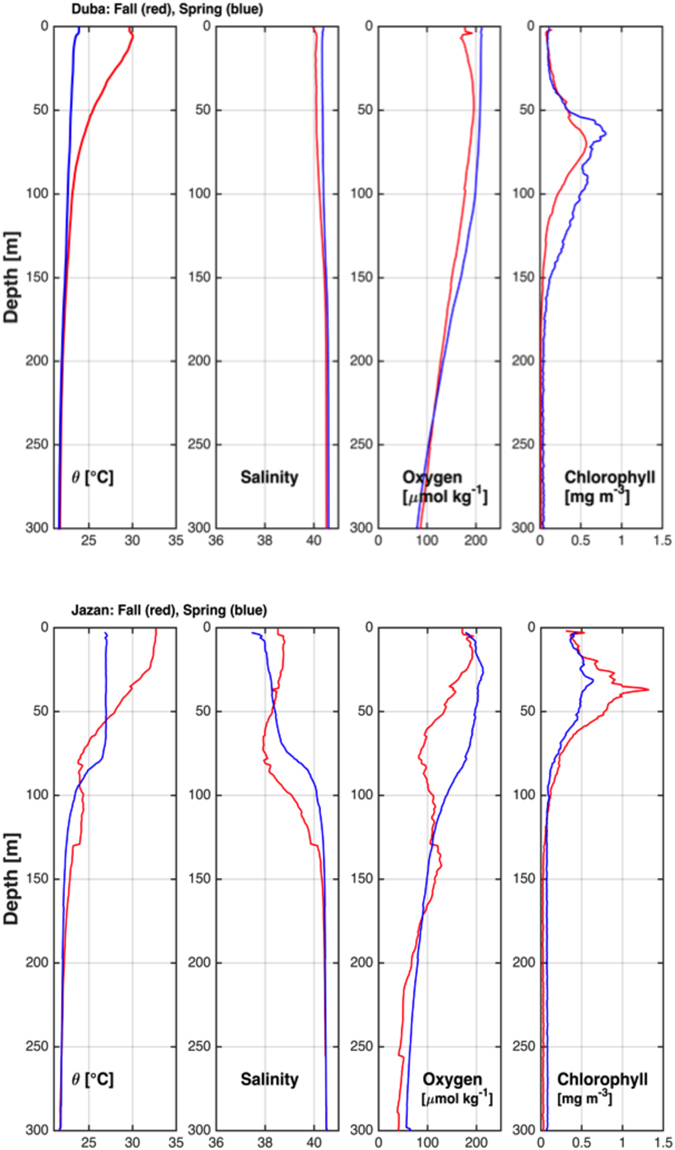
Mean vertical profiles of the hydrographic properties in the northern Red Sea (above) and the southern Red Sea (below).
Biological data
After quality checking of the sequencing reads there were 9442538 prokaryotic reads and 17665062 eukaryotic reads and these reads were used for the clustering as described in the methods. After reference sequences had been taxonomically assigned, OTUs not taxonomically relevant to the current study were removed (Eukaryotes: non-identified phyla or metazoan, 1808 OTUs; Prokaryotes: non-identified phyla or chloroplasts, 1653 OTUs). After multiple rarefactions at an even depth this resulted in 4211 (prokaryotes) and 3054 (eukaryotes) OTUs across all regions and both seasons.
There were no cosmopolitan eukaryotic OTUs with only 36 OTUs (1.2%) being observed in >90% of the samples. 1453 OTUs (47.6%) were present in less than 10% of the samples. In contrast, the prokaryotic component presented 11 OTUs (0.3%), which were observed in all samples, with 77 (1.8%) occurring in at least 90% of the samples. Similar to the eukaryotes, a substantially higher proportion of OTUs were found in fewer than 10% of samples (1497 OTUs, 35.5%). Only 477 prokaryotic (11.3%) and 518 eukaryotic (17.0%) OTUs were shared between all regions/depths/season. For both the eukaryotic and prokaryotic components the central region had a lower number of shared OTUs with either northern or southern regions than those extreme regions did with each other (Supplementary Figure 1).
Large-scale patterns of variability
Regional variability – three regions, spring period
Prokaryotic and eukaryotic fractions varied significantly among regions in terms of Faith’s diversity (PD) (ANOVA; F = 10.374; p < 0.001 and F = 5.545; p = 0.006, respectively). Faith’s diversity of prokaryotes also changed significantly with “Depth” (F = 4.411; p = 0.041). Post-hoc tests (Tukey HSD) revealed that the southern region had the highest diversity with the lowest observed in the central region.
Seasonal variability – north and south regions, spring and fall periods
The prokaryotic fraction had a significantly different Faith’s diversity across “Region” (F = 6.492; p = 0.0121), “Depth” (F = 32.158; p < 0.001) and “Season” (F = 9.758; p = 0.002). For the eukaryotic fraction, differences across regions and season were inconsistent, i.e., there was a significant interaction between “Region” and “Season” (F = 6.932; p = 0.010). Post-hoc tests revealed that sampling in the north in the fall had significantly higher values of Faith’s diversity than either region in the spring as well as the south in the fall.
Four scenarios (North_Fall, North_Spring, South_Fall and South_Spring) were established based on turbulence and nutrient conditions. In general, higher nitrate concentrations are registered in the South_Fall whilst the South_Spring had the highest levels of phosphate and silicate (Supplementary Figure 2). In terms of community dissimilarity (measured with unweighted and weighted UniFrac), higher levels of dissimilarity were observed in the South_Fall (weighted UniFrac) and South_Spring (unweighted UniFrac) (Supplementary Table 2 and Supplementary Figure 2). A similar trend was observed for the prokaryote structure (unweighted UniFrac) where both the South_Spring and South_Fall (no significant difference between the two) were higher than either the North_Spring or North_Fall.
Variability in taxonomic groups
The main dominant eukaryotic groups in the Red Sea in both seasons were the Alveolata classes Dinophyceae and Syndiniophyceae (mainly group I clade 1, 4 and 5 and group II clade 6, 7 and 10+11) with an average percentage of eukaryotic reads of 20.2% and 28.7%, respectively (Fig. 2a). Bacillariophyta increased in dominance in the south during the fall, reaching a peak of 23.7% of total number of reads. The eukaryotes Mamiellophyceae also showed increased numbers of reads in the southern region during the fall (average of 36.1% of reads at the Deep Chlorophyll Maximum (DCM)) and in the north during the spring (average of 30.5% at the DCM). The three main Mamiellophyceae genera (Ostreococcus, Micromonas and Bathycoccus) showed distinct patterns of distribution: Ostreococcus was more prevalent in the spring in the north while Micromonas and Bathycoccus presented higher proportional abundances in the south in the fall (Supplementary Figure 3). The Prasinophyte-Clade VII only had a high number of reads in the central region during the spring.
Figure 2.
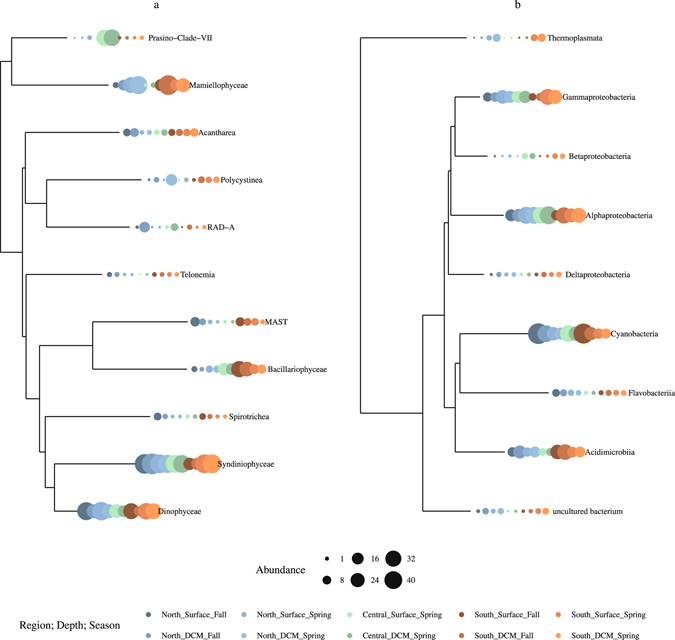
Phylogenetic tree of the main classes of (a) eukaryotes and (b) prokaryotes based on alignments of the 18 S rRNA and 16 S rRNA genes, respectively. The proportional abundance of specific classes in each regions (north, south), depths (surface, deep chlorophyll maximum – DCM), and seasons (spring, fall) (color coded) is denoted by the size of the circle at the end of each branch.
In terms of the prokaryotes, only four classes accounted for on average 87% of the reads: Cyanobacteria (28.6%), Alphaproteobacteria (27.8%), Gammaproteobacteria (16%), and Acidimicrobiia (14.6%) (Fig. 2b). Among the Cyanobacteria, Synechococcus, and to a lesser extent Prochlococcus, were the dominant genera (Supplementary Figure 4). SAR11 and SAR86 were the main contributors of Alphaproteobacteria and Gammaproteobacteria, respectively. Finally, the Acidimicrobiia mainly consisted of clade OM1. Although never accounting for a high number of reads, the archaeal class Thermoplasmata showed increased abundances in the spring period in both the northern and southern regions.
Non-metric multidimensional scaling (NMDS) indicated that both the structure (weighted) and composition (unweighted) of the eukaryotic and prokaryotic communities changed according to region, season and depth (Fig. 3). For the eukaryotes, both unweighted and weighted, significant interactions were observed between the factors, suggesting inconsistent variations in the trends. Pairwise comparisons (Supplementary Table 3) showed that, in general, community composition differed significantly with region and season. In general the surface community was also different from that at the DCM, with exception of those stations in the south. Community structure of eukaryotes showed similar patterns for the region and season. For the depths it was found that in the spring the surface and DCM were similar, differing in the fall. Interactions were also observed in both the weighted and unweighted matrices for prokaryotes. Pairwise, comparisons showed general significant differences between the north and the south, seasons and depth (Supplementary Table 3).
Figure 3.
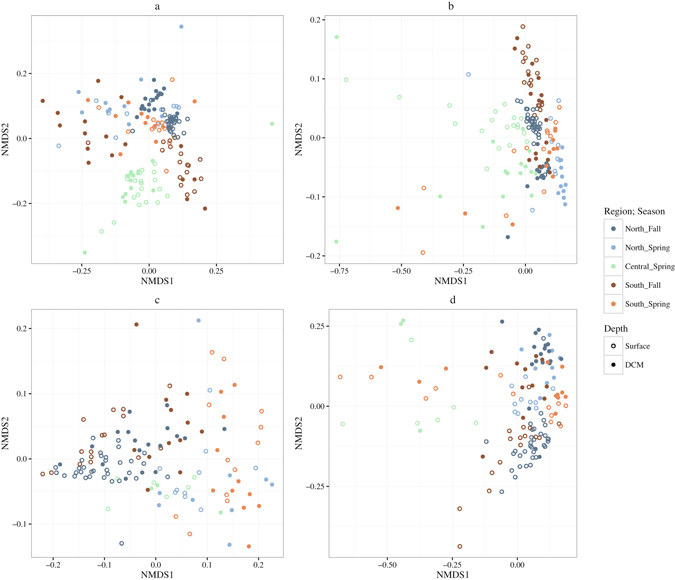
NMDS for eukaryotes – (a) weighted UniFrac (b) unweighted UniFrac and prokaryotes (c) weighted UniFrac and (d) unweighted UniFrac.
Overall, CCA models (removing the central region due to missing nutrient data) for both eukaryotes and prokaryotes were significant (p < 0.001 and p = 0.01 respectively). Surface samples from the southern fall stations were typified by higher temperatures and the increase in abundance of Micromonas, as well as raphid pennate diatoms (Fig. 4a). The Chlorophyte genus Bathyococcus was associated with the higher nutrient concentrations observed in the DCM of the southern fall stations while the abundance of the genus Ostreococcus responded positively to depth. For the prokaryotic component, the environmental variables measured could not account for the distribution of the most abundant groups (Fig. 4b).
Figure 4.
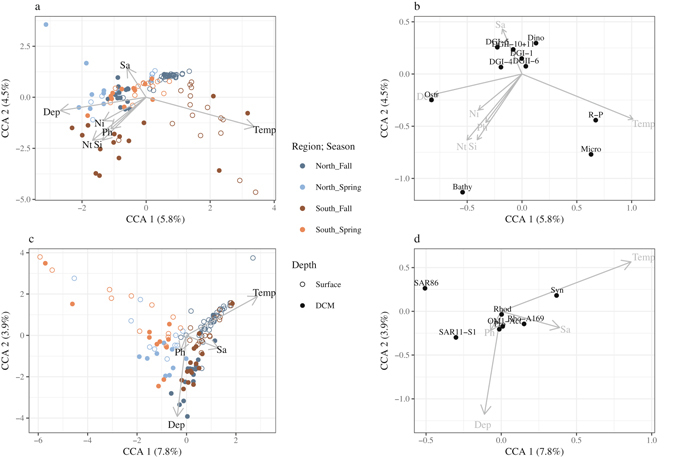
CCA analysis of the eukaryotic stations (a) and most abundant eukaryotic groups (b) and for the prokaryote stations (c) and the prokaryotic groups (d) with the environmental data. The seasons and regions are color coded while open circles denote samples taken at the surface with closed circles denoting the DCM. For (b) the taxa codes are: Dino = unclassified Dinophyceae; DG-II-10+11 = Syndinophyceae Group II clade 10 and 11; DG-II-6 = Syndinophyceae Group II clade 6; DG-I-1 = Syndinophyceae Group I clade 1; DG-I-4 = Syndinophyceae Group I clade 4; DG-I-5 = Syndinophyceae Group I clade 5; R-P = Raphid pennate diatoms; Ostr = Ostreococcus; Bathy = Bathycoccus and Micro = Micromonas. For (d) the taxa codes are: OM1-Act = Actinomarina OM1 clade; Pro = Prochlorococcus; Syn = Synechococcus; Rho-A169 = Rhodospirillaceaea AEGEAN-169 group; Rhod = uncultured Rhodobacteraceae; SAR11-S1 = SAR11 surface 1; SAR86 = SAR86. For all plots the environmental variables are: Ni = Nitrate; Nt = Nitrite; Ph = Phosphate; Si = Silicate; Temp = Temperature and Sa = Salinity. Please note the differences in the scales.
Mantels tests between showed that, with the exception of the prokaryotic unweighted comparison, the communities showed similar patterns to the environmental data (Table 1).
Table 1.
Mantel test results for the OTU dissimilarity matrices resulting from the different datasets (Eukaryotic and Prokaryotic) with the environmental distance matrix.
| R | p | |
|---|---|---|
| Eukaryote_weighted | 0.2352 | 0.002 |
| Eukaryote_unweighted | 0.3217 | 0.001 |
| Prokaryote_weighted | 0.292 | 0.001 |
| Prokaryote_unweighted | 0.095 | 0.062 |
Based on the 95th percentile maximum abundance models, we observed that the Syndiniophyce groups (Group I clades 1, 4 and 5 and Group II clade 10+11), the Mamiellophyceae genus Ostreococcus and the Alphaproteobacteria SAR11 responded negatively to the increase in temperature towards the maximum of 34 °C. In contrast, as temperature increased, Synechococcus (Cyanobacteria), Neoceratium (Alveolata) and Micromonas (Mamiellophyceae) became more abundant. Bathycococcus (Mamiellophyceae), Gonyaulax (Alveolata) and SAR86 (Gammaproteobacteria) had intermediate peaks in abundance while Prochlorococcus (Cyanobacteria) showed a bimodal distribution (Fig. 5). What is more, even within the same genus, different responses were observed. For example, the two most abundant OTUs assigned to Synechococcus showed different responses to changes in temperature (Supplementary Figure 5).
Figure 5.
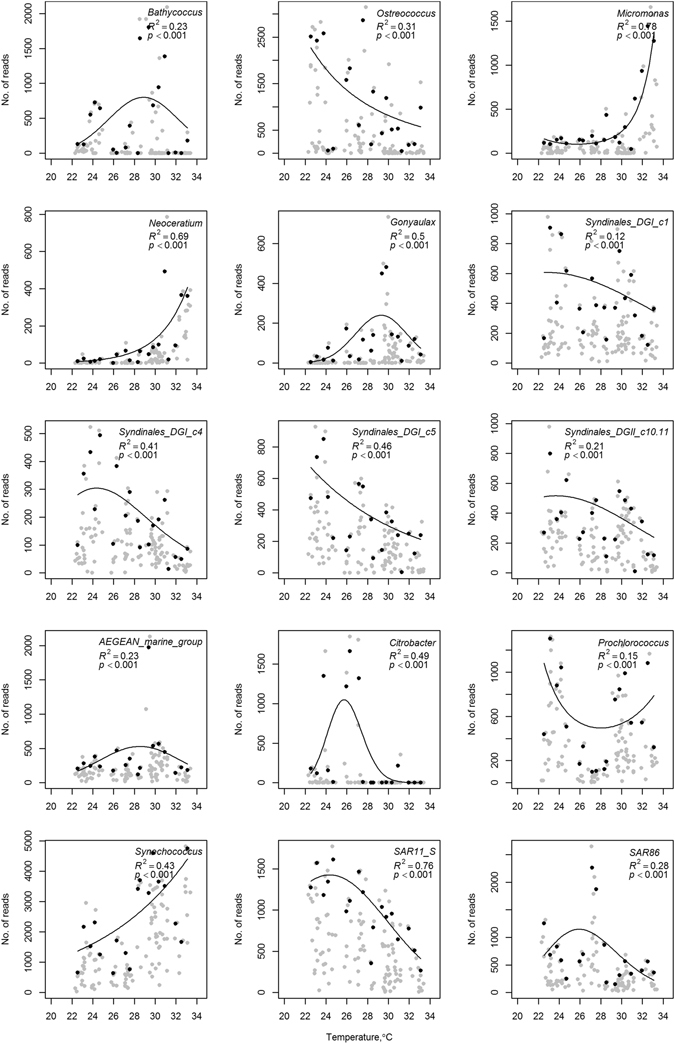
Maximum abundance models for various taxa against temperature. Grey points represent the data while black dots are the 95th percentile of the maximum abundance.
Discussion
Overall distributions
Autotrophs
Increased temperatures in nutrient-limited conditions are primarily thought to influence plankton distribution through physical mechanisms (e.g., stratification and nutrient supply) favoring the picoplankton36. The dominance of picoplankton has been observed in several oligotrophic regions including the Mediterranean Sea (e.g., refs 37 and 38), the Sargasso Sea39, and the Atlantic40. In the Red Sea, picoplankton has been shown to account for over 90% of the primary production in ref. 4. The picoplankton comprises cyanobacteria (e.g., genera Prochlorococcus and Synechococcus), as well as a diverse assortment of small eukaryotic algal taxa. Synechococcus is ubiquitous in the marine environment although it is more abundant in more nutrient-rich regions while Prochlorococcus is more restricted to oligotrophic tropical and sub-tropical waters41, 42. In tropical and sub-tropical regions, High performance liquid chromatography (HPLC) analysis indicated that Prochlorococcus could account for a large proportion of the phototrophs (e.g., refs 43–45) and it has previously been shown, using molecular methods, to account for up to 91% of the picocyanobacteria in the Red Sea13. Surprisingly, in the current study, except in the northern region in the spring, where Prochlorococcus reads numbers equaled those of Synechococcus, the latter was the dominant cyanobacterial genus in the Red Sea. The study conducted by Ngugi et al.13 was conducted in the northern Red Sea and during the spring, attenuating the differences observed between our and their study. Veldhuis & Kraay46 found that at the surface, Synechococcus abundance was higher than that of Prochlorococcus.
While cyanobacteria were large contributors to the 16 S rRNA gene libraries, and undoubtedly played a substantial role in the primary production, in the eukaryotic fraction, Dinophyceae were the dominant plastid-containing taxa throughout the Red Sea, in agreement with previous findings for this8, 20, and other oligotrophic regions (e.g., refs 8, 47 and 48). Being mixotrophic protists, they gain energy from sunlight and acquire inorganic nutrient requirements and essential organic nutrients, such as amino acids and vitamins, via bacterivory49, 50. This may be especially important in oligotrophic regions where high light levels are present, which could selectively favor mixotrophic grazers over heterotrophs51. The mixotrophic nature of Dinophyceae may allow for the propagation of dinoflagellates in oligotrophic conditions, especially in warmer conditions where grazing is reported at higher rates52 but in more nutrient replete regions they are likely to be outcompeted by other phototrophs53.
In the central Red Sea, although seasonal differences were unable to be determined with the current dataset, the eukaryotic community in this region was generally distinct from either the north or south with the predominance of prasinophyte clade VII A reads. This clade has previously been observed in mesotrophic regions in the Pacific54, 55 as well as in the Red Sea during the Tara Ocean cruise56. The dominant clade in the Red Sea during the Tara Oceans project (A4) was proposed to be a more coastal strain and although in the current study it was observed in open water this could be due to influence from the water column mixing, which can result from eddy structures that are present in this area creating the conditions for the upwelling of nutrients into the photic zone. However, inter annual variability or the effect of using different filter types, cannot be ruled out as the reason for the differences between the regions in this study.
Heterotrophs
The most dominant heterotrophic class in the prokaryotic fraction was Alphaproteobacteria. As is typical of other open ocean environments, the dominant group in this class is SAR11 (e.g., refs 57–59), specifically SAR11 - Surface 1, typical of the photic zone60. This group was dominant throughout the sampling irrespective of region, depth and season. The apparent cosmopolitan behavior observed in the present study is probably due to the inability of the 16 S rRNA gene to detect ecological differentiation within the clade that is known to have a high level of diversity in phylotypes13. SAR86 of the class Gammaproteobacteria, which has previously been reported in oligotrophic waters in the Pacific61, was prevalent in the spring when nutrients are limited but detectable. These groups with their small size and genome streamlining have a selective advantage in nutrient limited regions due to resource specialization62. It has been suggested that the specialization on different carbon compounds of SAR11 and SAR86 allows them to limit competition and thus have high abundances in the open ocean62.
Syndiniophyceae, parasitic members of the Alveolata are known to account for a large proportion of Alveolata reads in marine systems63, 64. The dominant groups in the euphotic zone belong to Group I (clades 1, 4 and 5) and Group II (clades 6, 7 and 10+11)63. Syndiniophyceae are likely to be highly opportunistic and infect a variety of hosts including dinoflagellates65 and radiolarians66. The proportion and diversity of parasites in the Red Sea may have a substantial impact on biogeochemical cycles. As well as releasing dissolved organic material into the environment, due to the destruction of host cells, the production of dinospores provides a nutrition source for crustacean zooplankton. This activity also returns a proportion of the energy to higher trophic levels, which could otherwise be lost due to the sinking of larger phytoplankton cells67, 68.
Seasonal and regional variations in the distributions of taxa
The lowest average richness observed in the current study was observed in the southern region in the fall (Fig. 6), a period of stratification, i.e. low turbulence, and high nutrients input through the GAIW (a relatively cold, nutrient rich and low salinity water mass; 31). With the increase in nutrients, small cells are no longer selected for, as fast growing larger taxa such as diatoms can effectively compete for the available nutrients. Also, in a stratified environment these larger cells may be able to escape grazing pressure as the rates of contact between predator and prey are lower in less turbulent regions69. The dominance of diatoms in these conditions deviates slightly from previous concepts23, 24 where bloom-forming dinoflagellates are proposed to dominate. However, the N form prevalent in the water column may determine whether bloom forming dinoflagellates or diatoms are present with inorganic forms of N favoring the latter25. The low richness observed under a scenario of high nutrient availability is also in agreement with Irigoien et al.70 who showed a reduction of biodiversity at high levels of phytoplankton biomass. The highest levels of richness were driven by high turbulence and low nutrients. Barton et al.71 proposed that turbulence can increase the flux of nutrients towards the cell and so increase the cell’s resource affinity. Also, as turbulence can increase the number of predator prey interactions69, zooplankton can prevent smaller sizes from consuming all the available resources. Therefore, larger cells may be able to compete with smaller cells in turbulent conditions, reducing competitive exclusion and increasing richness.
Figure 6.
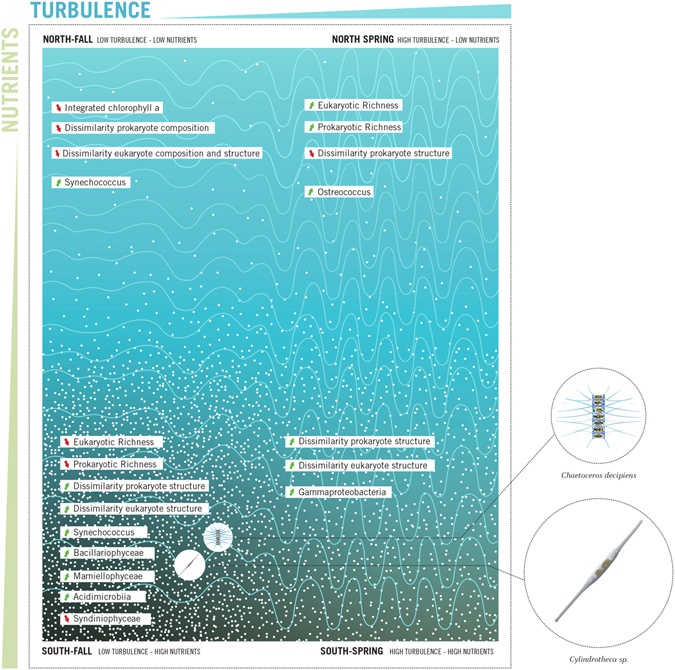
Generalized depiction of the responses of the microbial community in the Red Sea to variations in nutrients and turbulence.
The northern Red Sea in the fall is characterized by being highly stratified (low turbulence) and having low nutrient concentrations. In regimes where nutrients and turbulence are both low the microbial loop or specialist groups such as mixotrophs would dominate the food web24, 25. Indeed, the current study showed that cyanobacteria dominated the prokaryotic fraction while Dinophyceae, which contain a large proportion of mixotrophic taxa, and the parasitic Syndiniophyceae were the dominant eukaryotic groups. The highest average similarity within a region/season was observed here. This is not surprising, as when perturbations are kept at a low frequency similarity will increase72. In general, in the current study, increases in dissimilarity were observed as nutrient concentrations increase especially for the eukaryotes. The relationship between higher nutrients and dissimilarity has been shown previously (e.g., ref. 73) and could be due to the fact that high productivity tends to favor instability and compositional turnover74.
Determination of niches
As sea surface temperatures increase and the size of oligotrophic regions expand75, 76, taxa will respond to the changes in the environmental conditions depending on their genomic characteristics77. Understanding how taxa will respond to these climatic changes is important to understand how biogeochemical cycles will be affected.
Raising temperatures are predicted to favor small groups, such as cyanobacteria over large phytoplankton (e.g., diatoms)78. Our study suggested that, as temperatures increase toward 34 °C, the abundance of reads of Synechococcus increased while Prochlorococcus showed a bimodal distribution with peaks around 24 and 30 °C. The bimodal distribution of Prochlorococcus suggests strain-level differences in their distribution. Low light and high light clades have been previously described for Prochlorococcus 79, 80. Further, and specifically in the Red Sea, Shibl et al.81, 82 showed differences in the composition of Prochlorococcus in the water column, with strains adapted to conditions lower in the water column possibly having a different thermal tolerance. The peak in abundances around 30 °C is in agreement with culture tests on high light II cultures of Prochlorococcus whose growth rate declines rapidly around 28–29 °C83. The increased abundances of Synechococcus in warmer temperatures is also in agreement with Moore et al.84 who showed that the growth optimum of Synechococcus is higher than that of Prochlorococcus. With higher growth optimum, it is possible that Synechococcus are better adapted to take advantage of the nutrient inputs into the southern Red Sea (which is also warmer) in the summer and so increase their abundances relative to Prochlorococcus. This is especially true as Synechococcus is able to take advantage of the influxes of nitrate, such as those from the GAIW, whereas most strains of Prochlorococcus seem unable to80, 85. Temperature related growth optima are likely to be clade-specific84, which is further supported by the current findings.
Three of the most important eukaryotic players in the photic zone of Red Sea waters14, the genera of picoeukaryotic green algae (Bathyococcus, Micromonas and Ostreococcus; class Mamiellophyceae), showed distinct responses to depth and temperature. Bathycoccus was observed predominantly at the DCM, which seems to be typical of its distribution in other oceans86, 87 and is likely to be linked to the increased availability of nutrients at this depth86. In contrast, Micromonas has a higher affinity to surface waters, particularly near the coast, in line with reports in other biogeographical studies16, 64, 88, 89. Meanwhile, Ostreococcus showed a more even distribution in the photic zone of the water column. Its abundance was in general higher in the DCM than at the surface86, 90. Increased abundances of Ostreococcus were recorded in the colder waters of the northern Red Sea during the spring although this may be associated with an increase in the amount of nutrients available in the photic zone due to convective mixing of the water column, which has previously been shown to support high levels of phytoplankton until stratification occurs91.
Limited knowledge is available concerning the distribution and habitat preferences of the Syndiniophyceae. While it seemed that the major clades identified in the current study were generally observed throughout the water column, a preference toward lower surface seawater temperature was apparent. While temperature may play a role in the distribution of these parasitic clades, other studies have suggested that the abundance of the host92 and nutrient availability65 were driving factors in the abundance patterns of for example Amoebophyra spp. Further work will have to be undertaken to determine the host species of this diverse collection of parasites to fully understand how the distribution of these groups alters throughout the Red Sea, and in general how they respond to changes in temperature or depth. A greater understanding of the role of parasites in the marine system is required as they play an important role in energy transfer in the marine environment and this is especially important in oligotrophic regions where recycling of organic matter is vital. They may play a role in the control of harmful algal blooms92, which are becoming more prevalent93.
The prevalence of SAR11, a typical oligotrophic group in Atlantic waters, and a substantial contributor to the Red Sea planktonic community, has been proposed to increase its prevalence as oceans become warmer94 and for some strains more oligotrophic95. Field data from the warm Red Sea do not fully support this theory as maximum abundance modeling suggests that at the higher temperatures observed in the Red Sea they are not at their growth optima and abundances decline. The warmer water conditions in the Red Sea were also associated with increased nutrients and consequently more mesotrophic groups are likely to be able to take advantage of the nutrient conditions and to selectively out-compete SAR11.
Conclusion
The present study reinforces the relevance of the Red Sea as a natural laboratory for a better understanding of the responses of the plankton to changes in the physio-chemical environment. By looking at different regions and seasons, we were able to identify different scenarios aligned with the categories proposed by Cullen et al.24, testing their theory based on field data. Current results show that increasing nutrient levels tended to increase community dissimilarity within a region, especially for the eukaryotes, while higher turbulence, indicated by a deeper mixed layer, was associated with higher OTU richness. Phytoplankton communities in the Red Sea responded similarly to the framework proposed by Cullen et al.24, with the mixotrophic eukaryotes and cyanobacteria predominant in low turbulence and nutrient conditions being replaced by mesotrophic groups (Bacillariophyceae and Mamiellophyceae) as nutrient levels increased. Picophytoplankton (e.g. Ostreococcus), which are specialized to efficiently utilize light and nutrients, dominated the high turbulence low nutrient regimes as proposed by Cullen et al.24. The use of molecular techniques also allowed a broader assessment of the planktonic community to be assessed compared with Cullen et al.24 who focused predominantly on phytoplankton.
The present study shows different distributional patterns for the three main groups of Mamiellophyceae (Bathycoccus, Ostreococcus and Micromonas), which were dominant in various parts of the Red Sea. Based on this field-based dataset and on molecular data, we were also able to identify increases in Synechococcus, the photosynthetic cyanobacteria, with temperature although this response was shown to be specific to different OTUs. Further investigations of the distributional patterns of taxa and their relationships with other groups will give a more comprehensive understanding of the trophic structure in oligotrophic regions where mixotrophy and parasiticism are likely to be important nutritional modes.
Methods
Sample collection
Samples were collected during four research cruises undertaken in August and September 2014 (as detailed in 16) and February and April 2015 at both the surface (5 m) and the deep chlorophyll maximum (DCM). A total of 33 stations were sampled in the north of the Red Sea in August and 10 in February while 20 and 13 were sampled in the south of the Red Sea in September and April respectively (Fig. 7). A fifth cruise was undertaken during March/April 2013 in the central region of the Red Sea (21 stations). The coordinates of the stations for each sampling point are presented in Supplementary Table 1 (alongside ancillary information such as sampling depth, temperature, salinity, chlorophyll a, nitrate, nitrite, phosphate and silicate concentrations, mix layer depth).
Figure 7.
Sampling stations in the north, central and south of the Red Sea. Green triangles represent fall sampling points while orange circles are spring sampling. The map was made in ArcGIS (version: 10.3.1; https://www.arcgis.com/).
Sampling was undertaken at 5 m (described as the surface) and the DCM, with the DCM being determined during the downcast of the CTD/rosette profiler. During the upcast, 10 L Niskin bottles were triggered for closure at the desired depth.
From the 10 L Niskin bottles, 5 L of water (with no prefiltration) was filtered through either 0.22 µm membrane filters (Millipore) (used during the cruises to the north and south of the Red Sea) or through a 0.22 µm CellTrap (MemTeq) during the cruise in the central region. Individual filters were stored in 15 mL tubes with ~5 mL lysis buffer and frozen at −20 °C until analysis. The concentrated cell samples from the CellTrap were immediately frozen at −20 °C.
Water, for nutrient analysis and chlorophyll a, was collected at the same depths, which were sampled for DNA analysis. The method for nutrient analysis and chlorophyll a is as described in Pearman et al.16.
The mixed layer depth was ascertained from density measurements calculated from temperature and salinity values obtained from the rosette profiler. The mixed layer depth was used as a proxy for turbulence (e.g. A deep mixed layer equates to high turbulence whilst a small mixed layer equals a stratified and stable water column).
DNA extraction and PCR amplification
Concentrated cells from the CellTraps were centrifuged at 21 000 × g for 30 min prior to being re-suspended in 180 μL ATL buffer (Qiagen) and 20 μL proteinase K (20 mg mL−1). The filters were removed from the falcon tubes, placed in 2 mL eppendorfs with 540 μL ATL buffer (Qiagen) and 60 μL proteinase K (20 mg mL−1) added to them. Sample tubes from both methods were incubated at 55 °C for 30 min and DNA extraction and PCR amplification followed the same procedure as described in Pearman et al.16. The primers used for amplification targeted the v3 and v4 regions of the 16S rRNA gene96 and the v4 region of the 18S rRNA gene97. Subsequent to the PCR amplification (where a no negative control was also run) samples were cleaned and normalized using a SeqPrep Normalization plate prior to MiSeq library preparation. The library was prepared following the Illumina 16S metagenomic sequencing library preparation protocol. Before sequencing the samples were cleaned and normalized a second time and tagged samples were pooled for sequencing on a MiSeq sequencing platform at the King Abdullah University Core Laboratory. Raw reads were submitted to the NCBI SRA archive and can be accessed under the project accessions: SRP060785 and SRP081162.
Data analysis
Automatic demultiplexing of samples occurred during the MiSeq sequencing. The forward and reverse raw reads were joined using the join_seqs.py script in QIIME98 and quality filtered (phred = 25). Quality filtered reads were concatenated and the forward and reverse primers removed in mothur99. Any reads not containing the forward or reverse primers (with no errors) were removed from the analysis. Clustering of reads in OTUs followed a two-step process in QIIME. Firstly CD-HIT100 was implemented with the trie function before representative sequences were selected (first sequence of cluster was selected). A second round of clustering was undertaken using USEARCH (version 5.2.236)101 at 97% similarity with a minimum cluster size of two. During the USEARCH clustering chimeras were detected using both a denovo approach and against a reference database (SILVA 123102). Reference sequences obtained from the USEARCH clustering were taxonomically identified using the PR2103 database for eukaryotes and the SILVA 123 database for prokaryotes using the uclust algorithm. Sequences not assigned to at least the phylum level were removed from the analysis as were sequences relating to Metazoa in the eukaryotic fraction (due to these organisms not being representatively sampled using Niskin bottles) and chloroplasts (due to being from eukaryotic organisms) from the prokaryotic dataset.
To ensure an even depth of sequencing per sample reads were rarefied to 5000 reads per sample for the eukaryotes and 5700 reads for the prokaryotes multiple times (n = 100). Samples not reaching this threshold were subsequently removed from the analysis. The composition of the taxonomic groups was assessed using the R104 package phyloseq 105 using the average number of reads per region/depth/season and visualized with ggplot 106. Faith’s phylogenetic distance (PD) measure of alpha diversity was calculated in QIIME and analysis of variance (ANOVA) statistics were calculated in R. To achieve this the data was subset into a regional subset and a seasonal subset. The regional subset contained all the regions (north, central and south) but only for the spring. A two-way ANOVA for region (three levels: North, Central and South) and depth (two levels: Surface and DCM) was undertaken on this subset. For the analysis of seasons the subset contained data for both seasons (fall and spring) and the regions north and south. A three-way ANOVA was undertaken with the factors region (two levels: North and South), depth (two levels: Surface and DCM) and season (two levels: Spring and Fall). ANOVA was used to test the dissimilarity of the communities (measured with weighted and unweighted Unifrac) with four levels to the factor (North_Fall, North_Spring, South_Fall and South_Spring).
Non-metric multidimensional scaling (NMDS) ordination was undertaken using weighted and unweighted UniFrac107 distance matrices within the framework of the package phyloseq. Using the PRIMER v6 package108 with the PERMANOVA + add on109 a three-way PERMANOVA was undertaken. This assessed the significance of the factors “region” (orthogonal three levels, North, Central and South), “season” (orthogonal two levels, fall and spring) and “depth” (orthogonal two levels, surface and DCM). The statistical significance was tested using 9999 permutations of the residuals under a reduced model with a significance level of α = 0.05. Those effects, which were significant, were further investigated through a series of pairwise comparisons. Constrained Correspondence Analysis (CCA) was undertaken on the OTU tables merged at the level of genus. The CCA model was tested for significances using the permutational ANOVA (permutations = 999) within the vegan 110 package of R. Comparative (Mantel-type) tests were undertaken on the community dissimilarity matrices and an environmental distance matrix (Eucilidean) in vegan using the Pearson’s rank method.
Species Distribution Models based on maximum abundances across environmental gradients of depth, temperature and salinity were modeled using the method proposed by Blackburn et al.111. For these models, each of the three variables (depth, temperature, salinity) was divided into categories. Each category included no more than 20 observations, and there were roughly equal numbers of observations in at least three categories. For each category, the 95th percentile abundance was calculated for each taxon. For all taxa, regressions were conducted using the number of observations in each category as a weighting. Generalized Linear Models (GLMs) were fitted to the data, using the 95th percentile in each category as the dependent variable, a log link function and a Poisson likelihood function. The independent variable offered to the GLM included up to three-degree polynomials. The number of higher degree terms entered into the final model was based on the Akaike Information Criterion (AIC) and only higher degree terms that increased the proportion of deviance explained by more than 1% were included. The final model used for each taxon was that function which explained the most variability. All analyses were conducted using the R software package.
Electronic supplementary material
Acknowledgements
The authors would like to thank the crews of the RV Aegaeo and the RV Thuwal who enabled the sampling to be undertaken. We would like to thank Saskia Kürten for undertaking the nutrient analysis, Ute Langner for producing the map figure and Maria Ferri Sanz for producing Fig. 6. Also the help from the Coastal and Marine Resources Core Laboratory in obtaining permits and assistance in the field was deeply appreciated. The authors would also like to thank two anonymous reviewers for their valuable comments. The research reported in this publication was supported by funding from King Abdullah University of Science and Technology (KAUST) as well as from collaboration between KAUST and Saudi Aramco within the framework of the Saudi Aramco – KAUST Center for Marine Environmental Observations.
Author Contributions
J.K.P. and S.C. designed the experiment while J.K.P., J.E., Y.V.B.S. and S.C. analyzed the data. X.I. and B.H.J. provided reagents and equipment. J.K.P. and S.C. wrote the paper with input from J.E. and X.I. and all co-authors read and commented on the various drafts of the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06928-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Strom SL. Microbial ecology of ocean biogeochemistry: A community perspective. Science. 2008;320:1043–1045. doi: 10.1126/science.1153527. [DOI] [PubMed] [Google Scholar]
- 2.Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- 3.Falkowski PG, Fenchel T, DeLong EF. The Microbial Engines That Drive Earth’s Biogeochemical Cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 4.Weisse T. The microbial loop in the Red Sea: dynamics of pelagic bacteria and heterotrophic nanoflagellates. Marine Ecology Progress Series. 1989;55:241–250. doi: 10.3354/meps055241. [DOI] [Google Scholar]
- 5.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 6.Not F, et al. Picobiliphytes: A Marine Picoplanktonic Algal Group with Unknown Affinities to Other Eukaryotes. Science. 2007;315:253. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- 7.Kim E, et al. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. PNAS. 2011;108:1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vargas, C. et al. Eukaryotic plankton diversity in the sunlit ocean. Science348 (2015). [DOI] [PubMed]
- 9.Kirkham AR, et al. Basin-scale distribution patterns of photosynthetic picoeukaryotes along an Atlantic Meridional Transect. Environmental Microbiology. 2011;13:975–990. doi: 10.1111/j.1462-2920.2010.02403.x. [DOI] [PubMed] [Google Scholar]
- 10.Pommier T, et al. Spatial patterns of bacterial richness and evenness in the NW Mediterranean Sea explored by pyrosequencing of the 16S rRNA. Aquatic Microbial Ecology. 2010;61:221–233. doi: 10.3354/ame01484. [DOI] [Google Scholar]
- 11.Gilbert JA, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian P-Y, et al. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 2011;5:507–518. doi: 10.1038/ismej.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngugi DK, Antunes A, Brune A, Stingl U. Biogeography of pelagic bacterioplankton across an antagonistic temperature–salinity gradient in the Red Sea. Mol Ecol. 2012;21:388–405. doi: 10.1111/j.1365-294X.2011.05378.x. [DOI] [PubMed] [Google Scholar]
- 14.Acosta F, Ngugi DK, Stingl U. Diversity of picoeukaryotes at an oligotrophic site off the Northeastern Red Sea Coast. Aquatic Biosystems. 2013;9:16. doi: 10.1186/2046-9063-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari MI, Harb M, Jones B, Hong P-Y. Molecular-based approaches to characterize coastal microbial community and their potential relation to the trophic state of Red Sea. Scientific Reports. 2015;5:9001. doi: 10.1038/srep09001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearman JK, Kürten S, Sarma YVB, Jones B, Carvalho S. Biodiversity patterns of plankton assemblages at the extremes of the Red Sea. FEMS Microbiol Ecol. 2016;92:fiw002. doi: 10.1093/femsec/fiw002. [DOI] [PubMed] [Google Scholar]
- 17.Kürten B, et al. Ecohydrographic constraints on biodiversity and distribution of phytoplankton and zooplankton in coral reefs of the Red Sea, Saudi Arabia. Marine Ecology. 2014;36(4):1195–1214. doi: 10.1111/maec.12224. [DOI] [Google Scholar]
- 18.Al-Najjar T, Badran MI, Richter C, Meyerhoefer M, Sommer U. Seasonal dynamics of phytoplankton in the Gulf of Aqaba, Red Sea. Hydrobiologia. 2007;579:69–83. doi: 10.1007/s10750-006-0365-z. [DOI] [Google Scholar]
- 19.Kheireddine, M. et al. Assessing Pigment-Based Phytoplankton Community Distributions in the Red Sea. Frontiers in Marine Science4(32), doi:10.3389/fmars.2017.00132 (2017).
- 20.Touliabah HE, Abu El-Kheir WS, Kuchari MG, Abdulwassi NIH. Phytoplankton Composition at Jeddah Coast–Red Sea, Saudi Arabia in Relation to some Ecological Factors. JKAU: Sci. 2010;22:115–131. doi: 10.4197/Sci.22-1.9. [DOI] [Google Scholar]
- 21.Kamenir Y, Dubinsky Z, Zohary T. Phytoplankton size structure stability in a meso-eutrophic subtropical lake. Hydrobiologia. 2004;520:89–104. doi: 10.1023/B:HYDR.0000027729.53348.c7. [DOI] [Google Scholar]
- 22.Gin KYH, Chisholm SW, Olson RJ. Seasonal and depth variation in microbial size spectra at the Bermuda Atlantic time series station. Deep Sea Research Part I. 1999;46:1221–1245. doi: 10.1016/S0967-0637(99)00004-7. [DOI] [Google Scholar]
- 23.Margalef R. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanologica Acta. 1978;1:493–509. [Google Scholar]
- 24.Cullen, J. J., Franks, P. J. S., Karl, D. M. & Longhurst, A. Physical Influences on marine ecosystem dynamics. In, Robinson, A. R., McCarthy, J. J. & Rothschild, B. J. (eds) The Sea. John Wiley & Sons, Inc.: New York (2002).
- 25.Gilbert PM. Margalef revisited: A new phytoplankton mandala incorporating twelve dimensions, including nutritional physiology. Harmful Algae. 2016;55:25–30. doi: 10.1016/j.hal.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Dunbar MJ. The evolution of stability in marine environments. Natural selection at the level of the ecosystem. The American Naturalist. 1960;94:129–136. [Google Scholar]
- 27.Landry MR, Kirchman DL. Microbial community structure and variability in the tropical Pacific. Deep Sea Research Part II: Topical Studies in Oceanography. 2002;49:2669–2693. doi: 10.1016/S0967-0645(02)00053-X. [DOI] [Google Scholar]
- 28.Venrick EL. Phytoplankton in an oligotrophic ocean: Species structure and interannual variability. Ecology. 1990;71:1547–1563. doi: 10.2307/1938291. [DOI] [Google Scholar]
- 29.Cermeño P, de Vargas C, Abrantes F, Falkowski PG. Phytoplankton biogeography and community stability in the ocean. PLoS ONE. 2010;5:e10037. doi: 10.1371/journal.pone.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sofianos SS, Johns WE. Observations of the summer Red Sea circulation. Journal of Geophysical Research – Oceans. 2007;112(C6):C06025. doi: 10.1029/2006JC003886. [DOI] [Google Scholar]
- 31.Churchill JH, Bower AS, McCorkle DC, Abualnaja Y. The transport of nutrient-rich Indian Ocean water through the Red Sea and into coastal reef systems. Journal of Marine Research. 2014;72:165–181. doi: 10.1357/002224014814901994. [DOI] [Google Scholar]
- 32.Raitsos DE, Pradhan Y, Brewin RJW, Stenchikov G, Hoteit I. Remote Sensing the Phytoplankton Seasonal Succession of the Red Sea. PloS ONE. 2013;8(6):e64909. doi: 10.1371/journal.pone.0064909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sofianos SS, Johns WE. An Oceanic General Circulation Model (OGCM) investigation of the Red Sea circulation: 2. Three-dimensional circulation in the Red Sea. Journal of Geophysical Research – Oceans. 2003;108(C3):3066. doi: 10.1029/2001JC001185. [DOI] [Google Scholar]
- 34.Murray SP, Johns W. Direct observations of seasonal exchange through the Bab el Mandab Strait. Geophysical Research Letters. 1997;24:2557–2560. doi: 10.1029/97GL02741. [DOI] [Google Scholar]
- 35.Acker J, Leptoukh G, Shen S, Zhu T, Kempler S. Remotely-sensed chlorophyll a observations of the northern Red Sea indicate seasonal variability and influence of coastal reefs. Journal of Marine Systems. 2008;69:191–204. doi: 10.1016/j.jmarsys.2005.12.006. [DOI] [Google Scholar]
- 36.Lewandowska AM, et al. Effects of sea surface warming on marine plankton. Ecology Letters. 2014;17(5):614–623. doi: 10.1111/ele.12265. [DOI] [PubMed] [Google Scholar]
- 37.Siokou-Rrangou I, et al. Plankton in the open Mediterranean Sea: a review. Biogeosciences. 2010;7:1543–1586. doi: 10.5194/bg-7-1543-2010. [DOI] [Google Scholar]
- 38.Ignatiades L, et al. Phytoplankton size-based dynamics in the Aegean Sea (Eastern Mediterranean) Journal of Marine Systems. 2002;36:11–28. doi: 10.1016/S0924-7963(02)00132-X. [DOI] [Google Scholar]
- 39.Caron DA, Peele ER, Lim EL, Dennett MR. Picoplankton and nanoplankton and their trophic coupling in surface waters of the Sargasso Sea south of Bermuda. Limnology and Oceanography. 1999;44:259–272. doi: 10.4319/lo.1999.44.2.0259. [DOI] [Google Scholar]
- 40.Teira E, et al. Variability of chlorophyll and primary production in the Eastern North Atlantic Subtropical Gyre: potential factors affecting phytoplankton activity. Deep-Sea Research Part I-Oceanographic Research Papers. 2005;52:569–588. doi: 10.1016/j.dsr.2004.11.007. [DOI] [Google Scholar]
- 41.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance. Microbiology and Molecular Biology Reviews. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouman HA, et al. Water-column stratification governs the community structure of subtroptical marine picophytoplankton. Environmental Microbiology Reports. 2011;3(4):473–482. doi: 10.1111/j.1758-2229.2011.00241.x. [DOI] [PubMed] [Google Scholar]
- 43.Veldhuis MJW, Kraay GW. Vertical distribution and pigment composition of a picoplanktonic prochlorophyte in the subtropical North Atlantic: a combined study of HPLC- analysis of pigments and flow cytometry. Marine Ecology Progress Series. 1990;68:121–127. doi: 10.3354/meps068121. [DOI] [Google Scholar]
- 44.Goericke R, Repeta DJ. Chlorophylls a and b and divinyl chlorophylls a and b in the open subtropical North Atlantic Ocean. Marine Ecology Progress Series. 1993;101:307–313. doi: 10.3354/meps101307. [DOI] [Google Scholar]
- 45.Letelier RM, et al. Temporal variability of phytoplankton community structure based on pigment analysis. Limnology and Oceanography. 1993;38:1420–1437. doi: 10.4319/lo.1993.38.7.1420. [DOI] [Google Scholar]
- 46.Veldhuis MJW, Kraay GW. Cell abundance and fluorescence of picophytoplankton in relation to growth irradiance and nitrogen availability in the Red Sea. Netherlands Journal of Sea Research. 1993;31:135–145. doi: 10.1016/0077-7579(93)90003-B. [DOI] [Google Scholar]
- 47.Estrada M. Phytoplankton assemblages across a NW Mediterranean front: Changes from winter mixing to spring stratification. Oecologia Aquatica. 1991;10:157–185. [Google Scholar]
- 48.Estrada M, et al. Phytoplankton across Tropical and Subtropical Regions of the Atlantic, Indian and Pacific Oceans. PLoS ONE. 2016;11(3):e0151699. doi: 10.1371/journal.pone.0151699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unrein F, Massana R, Alonso-Saez L, Gasol JM. Significant year-round effect of small mixotrophic flagellates on bacterioplankton in an oligotrophic coastal system. Limnology and Oceanography. 2007;52:456–469. doi: 10.4319/lo.2007.52.1.0456. [DOI] [Google Scholar]
- 50.Hartmann M, et al. Mixotrophic basis of Atlantic oligotrophic ecosystems. PNAS. 2012;109:5756–5760. doi: 10.1073/pnas.1118179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ptacnik R, et al. A light-induced shortcut in the planktonic microbial loop. Scientific Reports. 2016;6:29286. doi: 10.1038/srep29286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilken S, Huisman J, Naus-Wiezer S, Van Donk E. Mixotrophic organisms become more heterotrophic with rising temperature. Ecology Letters. 2013;16:225–233. doi: 10.1111/ele.12033. [DOI] [PubMed] [Google Scholar]
- 53.Litchman E, Klausmeier CA, Miller JR, Schofield O, Falkowski PG. Multi-nutrient, multi-group model of present and future oceanic phytoplankton communities. Biogeosciences. 2006;3:585–606. doi: 10.5194/bg-3-585-2006. [DOI] [Google Scholar]
- 54.Lepère C, Vaulot D, Scanlan DJ. Photosynthetic picoeukaryote community structure in the South East Pacific Ocean encompassing the most oligotrophic waters on Earth. Environmental Microbiology. 2009;11:3105–3117. doi: 10.1111/j.1462-2920.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 55.Shi XL, Lepère C, Scanlan DJ, Vaulot D. Plastid 16S rRNA Gene Diversity among Eukaryotic Picophytoplankton Sorted by Flow Cytometry from the South Pacific Ocean. PloS ONE. 2011;6(4):e18979. doi: 10.1371/journal.pone.0018979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lopes dos Santos, A. et al. Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters. ISME J, doi:10.1038/ismej.2016.120 (2016). [DOI] [PMC free article] [PubMed]
- 57.Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappe MS. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol. 2009;11:2291–2300. doi: 10.1111/j.1462-2920.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- 58.Carlson CA, et al. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- 59.Morris RM, Frazar CD, Carlson CA. Basin-scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the South Atlantic. Environ Microbiol. 2012;14:1133–1144. doi: 10.1111/j.1462-2920.2011.02694.x. [DOI] [PubMed] [Google Scholar]
- 60.Ngugi DK, Stingl U. Combined Analyses of the ITS Loci and the Corresponding 16S rRNA Genes Reveal High Micro- and Macrodiversity of SAR11 Populations in the Red Sea. PLoS ONE. 2012;7:e50274. doi: 10.1371/journal.pone.0050274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West NJ, et al. Distinct Spatial Patterns of SAR11, SAR86, and Actinobacteria Diversity along a Transect in the Ultra-oligotrophic South Pacific Ocean. Frontiers in Microbiology. 2016;7:234. doi: 10.3389/fmicb.2016.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillou L, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environmental Microbiology. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 64.Shi XL, Marie D, Jardillier L, Scanlan DJ, Vaulot D. Groups without Cultured Representatives Dominate Eukaryotic Picophytoplankton in the Oligotrophic South East Pacific Ocean. PloS ONE. 2009;4(10):e7657. doi: 10.1371/journal.pone.0007657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siano R, et al. Distribution and host diversity of Amoebophryidae parasites across oligotrophic waters of the Mediterranean Sea. Biogeosciences. 2011;8:267–278. doi: 10.5194/bg-8-267-2011. [DOI] [Google Scholar]
- 66.Bråte J, et al. Radiolaria Associated with Large Diversity of Marine Alveolates. Protist. 2012;163:767–777. doi: 10.1016/j.protis.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Lefèvre E, Roussel B, Amblard C, Sime-Ngando T. The Molecular Diversity of Freshwater Picoeukaryotes Reveals High Occurrence of Putative Parasitoids in the Plankton. PLoS ONE. 2008;3:e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jephcott TG, et al. Ecological impacts of parasitic chytrids, syndiniales and perkinsids on populations of marine photosynthetic dinoflagellate. Fungal Ecology. 2016;19:47–58. doi: 10.1016/j.funeco.2015.03.007. [DOI] [Google Scholar]
- 69.Kiørboe TT. Phytoplankton cell size, and the structure of pelagic food webs. Advances in Marine Biology. 1993;29:1–72. doi: 10.1016/S0065-2881(08)60129-7. [DOI] [Google Scholar]
- 70.Irigoien X, Huisman J, Harris RP. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature. 2004;429:863–867. doi: 10.1038/nature02593. [DOI] [PubMed] [Google Scholar]
- 71.Barton AD, Ward BA, Williams RG, Follows MJ. The impact of fine-scale turbulence on phytoplankton community structure. Limnology and Oceanography: Fluids & Environments. 2014;4:34–49. [Google Scholar]
- 72.Lindenschmidt K-E, Chorus I. The effect of water column mixing on phytoplankton succession, diversity and similarity. Journal of Plankton Research. 1998;20:1927–1951. doi: 10.1093/plankt/20.10.1927. [DOI] [Google Scholar]
- 73.Chase JM. Stochastic community assembly causes higher biodiversity in more productive environments. Science. 2010;328:1388–1391. doi: 10.1126/science.1187820. [DOI] [PubMed] [Google Scholar]
- 74.Steiner CF, Leibold MA. Cyclic assembly trajectories and scale-dependent productivity-diversity relationships. Ecology. 2004;85:107–113. doi: 10.1890/03-3010. [DOI] [Google Scholar]
- 75.Polovina JJ, Dunne JP, Woodworth PA, Howell EA. Projected expansion of the subtropical biome and contraction of the temperate and equatorial upwelling biomes in the North Pacific under global warming. Ices Journal of Marine Science. 2011 [Google Scholar]
- 76.Polovina JJ, Howell EA, Abecassis M. Ocean’s least productive waters are expanding. Geophysical Research Letters. 2008;35:L03618. doi: 10.1029/2007GL031745. [DOI] [Google Scholar]
- 77.DeLong EF, Karl DM. Genomic perspectives in microbial oceanography. Nature. 2005;437:336–342. doi: 10.1038/nature04157. [DOI] [PubMed] [Google Scholar]
- 78.Paerl HW, Huisman J. Blooms like it hot. Science. 2008;320:57. doi: 10.1126/science.1155398. [DOI] [PubMed] [Google Scholar]
- 79.Martiny AC, Tai APK, Veneziano D, Primeau F, Chisholm SW. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- 80.Scanlan DJ, et al. Ecological Genomics of Marine Picocyanobacteria. Microbiology and Molecular Biology Reviews. 2009;73:249. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shibl AA, Thompson LR, Ngugi DK, Stingl U. Distribution and diversity of Prochlorococcus ecotypes in the Red Sea. FEMS Microbiology Letters. 2014;356:118–126. doi: 10.1111/1574-6968.12490. [DOI] [PubMed] [Google Scholar]
- 82.Shibl AA, Haroon MF, Ngugi DK, Thompson LR, Stingl U. Distribution of Prochlorococcus ecotypes in the Red Sea basin based on analyses of rpoC1 sequences. Frontiers in Marine Science. 2016;3:104. doi: 10.3389/fmars.2016.00104. [DOI] [Google Scholar]
- 83.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nature Reviews Microbiology. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 84.Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Marine Ecology Progress Series. 1995;116:259–275. doi: 10.3354/meps116259. [DOI] [Google Scholar]
- 85.Berube PM, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2015;9:1196–1207. doi: 10.1038/ismej.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Choi DH, et al. Dynamic changes in the composition of photosynthetic picoeukaryotes in the northwestern Pacific Ocean revealed by high-throughput tag sequencing of plastid 16S rRNA genes. FEMS Microbiol Ecol. 2016;92(2):fiv170. doi: 10.1093/femsec/fiv170. [DOI] [PubMed] [Google Scholar]
- 87.Monier A, Worden AZ, Richards TA. Phylogenetic diversity and biogeography of the Mamiellophyceae lineage of eukaryotic phytoplankton across the oceans. Environmental Microbiology Reports. 2016;8:461–469. doi: 10.1111/1758-2229.12390. [DOI] [PubMed] [Google Scholar]
- 88.Not F, et al. Protistan assemblages across the Indian Ocean, with a specific emphasis on the picoeukaryotes. Deep-Sea Research Part I-Oceanographic Research Papers. 2008;55:1456–1473. doi: 10.1016/j.dsr.2008.06.007. [DOI] [Google Scholar]
- 89.Foulon E, et al. Ecological niche partitioning in the picoplanktonic green alga Micromonas pusilla: evidence from environmental surveys using phylogenetic probes. Environmental Microbiology. 2008;10:2433–2443. doi: 10.1111/j.1462-2920.2008.01673.x. [DOI] [PubMed] [Google Scholar]
- 90.Countway PD, Caron DA. Abundance and Distribution of Osteococcus sp. in the San Pedro Channel, California, as Revealed by Quantitative PCR. Applied and Environmental Microbiology. 2006;72:2496–2506. doi: 10.1128/AEM.72.4.2496-2506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kürten B, et al. Carbon and nitrogen stable isotope ratios of pelagic zooplankton elucidate ecohydrographic features in the oligotrophic Red Sea. Progress in Oceanography. 2016;140:69–90. doi: 10.1016/j.pocean.2015.11.003. [DOI] [Google Scholar]
- 92.Coats DW, Adam EJ, Gallegos CL, Hedrick S. Parasitism of photosynthetic dinoflagellates in a shallow subestuary of Chesapeake Bay, USA. Aquatic Microbial Ecology. 1996;11:1–9. doi: 10.3354/ame011001. [DOI] [Google Scholar]
- 93.Hallegraeff GM. Ocean Climate change, phytoplankton community responses and harmful algal blooms: A formidable predictive challenge. J Phycol. 2010;46:220–235. doi: 10.1111/j.1529-8817.2010.00815.x. [DOI] [Google Scholar]
- 94.Morán XAG, et al. More, smaller bacteria in response to ocean’s warming? Proceedings of the Royal Society B-Biological Sciences. 2015;282:20150371. doi: 10.1098/rspb.2015.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salter I, et al. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J. 2015;9:347–360. doi: 10.1038/ismej.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stoeck T, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19(Suppl 1):21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 98.Caporaso JG, et al. QIIME allows analysis of high throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schloss IR, et al. Picophytoplankton and nanophytoplankton abundance and distribution in the southeastern Beaufort Sea (Mackenzie Shelf and Amundsen Gulf) during Fall 2002. Journal of Marine Systems. 2008;74:978–993. doi: 10.1016/j.jmarsys.2008.01.004. [DOI] [Google Scholar]
- 100.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 101.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 102.Pruesse E, et al. Nucleic Acids Research. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB; pp. 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guillou L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Research. 2013;41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. ISBN 3-900051-07-0 (2016).
- 105.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PloS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wickham, H. ggplot2: Elegant Graphics for Data Analysis Springer-Verlag New York (2009).
- 107.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Applied and Environmental Microbiology. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clarke, K.R. & Gorley, R.N. Primer v6: User Manual/Tutorial. Plymouth, UK: Primer-E (2006).
- 109.Anderson, M. J., Gorley, R. N. & Clarke, K. R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth, U.K.: PRIMER-E
- 110.Oksanen, J. et al. (2016) vegan: Community Ecology Package. R package version 2.3–5. http://CRAN.R-project.org/package=vegan (2008).
- 111.Blackburn TM, Lawon JH, Perry JN. A Method of Estimating the Slope of Upper Bounds of Plots of Body Size and Abundance in Natural Animal Assemblages. Oikos. 1992;65:107–112. doi: 10.2307/3544892. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


