Abstract
The Ewings Sarcoma Oncoprotein (EWS) interacts with several components of the mammalian transcriptional and pre-mRNA splicing machinery and is also found in the cytoplasm and even on the cell surface. The apparently diverse cellular functions of EWS are, however, not well characterized. EWS harbours a potent N-terminal transcriptional activation domain (the EAD) that is revealed in the context of oncogenic EWS-fusion proteins (EFPs) and a C-terminal RNA-binding domain (RBD) that recruits pre-mRNA splicing factors and may couple transcription and splicing. In contrast to EFPs, the presumed transcriptional role of normal EWS remains enigmatic. Here, we report that multiple RGG-boxes within the RBD are necessary and sufficient for cis-repression of the EAD and that RGG-boxes can also repress in-trans, within dimeric partners. Lys can functionally substitute for Arg, indicating that the basic nature of the Arg side chain is the critical determinant of RGG-box-mediated repression. In addition to the EAD, RGG-boxes can repress a broad range of activation domains (including those of VP16, E1a and CREB), but repression can be alleviated by the simultaneous presence of more than one activation domain. We therefore propose that a key function of RGG boxes within native EWS is to restrict promiscuous activation by the EAD while still allowing EWS to enter functional transcription complexes and participate in other transactions involving pre-mRNAs.
INTRODUCTION
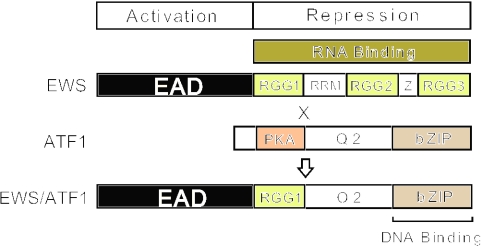
Much of what is known about the normal function of the Ewings Sarcoma oncogene (EWS) is derived from studies of oncogenic fusion proteins (EWS-Fusion-Proteins, EFPs). EFPs cause a variety of malignancies [reviewed in (1–3)] and arise from chromosomal fusion of the EWS gene with multiple partners. Our previous work has focused on EWS/ATF1 (Figure 1), an EFP which is a causative agent of soft tissue clear cell sarcoma (CCS) (4). EFPs are potent promoter-specific transcriptional activators, due to the presence of the N-terminal region of EWS (the EWS-Activation-Domain, EAD) and a DNA binding domain from the fusion partner. In addition to influencing transcriptional events in tumour cells, EFPs also perturb pre-mRNA splicing, suggesting that the mechanisms of tumorigenesis may be quite complex.
Figure 1.
Functional regions of EWS, ATF1 and EWS/ATF1. EWS contains an N-terminal transcriptional activation domain (the EWS-Activation Domain, EAD) and a C-terminal RNA-binding domain (RBD). The RBD contains two elements [an RNA-Recognition Motif (RRM) and RGG boxes] commonly found in RNA binding proteins (6–8) and a C2–C2 putative zinc finger (Z). The RRM (otherwise called the CS-RBD or the RNP motif) consists of ∼100 residues with a conserved three dimensional structure (6–8). The three RGG rich regions (RGG1, RGG2 and RGG3) contain 5, 3 and 12 tripeptide RGG motifs, respectively. The term RGG-box refers to clusters of closely spaced RGG tripeptides flanked by aromatic residues and a consensus RGG-box (containing 5 RGG tri-peptides) has been deduced (8). ATF1 is a PKA-inducible activator (47,48). The bZIP domain of ATF1 (amino acids 214–271) mediates dimerization and DNA-binding, Q2 is a glutamine-rich constitutive activation domain (48) and PKA represents the Kinase Inducible Domain (48) including a single PKA-phosphoacceptor site. EWS/ATF1 is an oncogenic EWS-Fusion-Protein (EFP) that is associated with soft tissue clear cell sarcoma (CCS) (4). The chromosomal cross-over point that produces EWS/ATF1 is shown with a black cross resulting in the EAD (residues 1–325) fused to the C-terminal region of ATF1 (residues 66–271). EWS/ATF1 lacks the PKA phosphoacceptor site of ATF1 and functions as a potent constitutive activator of ATF-dependent promoters (20,21) dependent on the EAD and the bZIP domain of ATF1.
The cellular role of the normal EWS protein is not well characterized. In addition to the EAD, EWS contains a C-terminal RNA-binding domain (RBD, Figure 1) and together with the highly related proteins TLS/FUS and TAF68, forms a sub-group [the TET family (5)] within the RNP family of RNA-binding proteins (6–8). The presence of the RBD points to a role for TETs in aspects of RNA biogenesis other than transcription and there is much evidence that TETs are directly involved in pre-mRNA splicing and transport or in coupling of the splicing and transcriptional machinery (9–13). Finally, TETs are targets for tyrosine kinases (14,15) and possibly other signalling pathways (16,17), and are also found to shuttle between several cellular locations (14,18,19) including the cell surface (19). Thus, it is apparent that TETs participate in a diverse array of cellular functions.
Similar to splicing, there is a significant body of evidence to implicate EWS/TETs in transcription. First, as mentioned above, the EAD functions as a potent transcriptional activation domain in the context of EFPs (1,3) and particularly in the case of EWS/ATF1 (20–22). Second, native EWS/TETs interacts with a gamut of transcription factors, including Pol II subunits (23,24), activators (25–27) and co-activators (28,29), and the TET fly homologue, SARFH, associates with many Pol II transcription units in vivo (30). Different TETs (including EWS) are present in distinct TFIID sub-populations however (5), indicating that, despite making contact with multiple transcriptional components, TETs are not general transcription factors. Third, native EWS (28,29) and TLS (27) can modestly stimulate transcription in a cell or promoter-specific manner (27–29) further pointing to a specialized transcriptional role of TETs. The finding that a cis-linked EWS RBD can block trans-activation by the EAD in the context of EFPs (31) or Gal4-fusions (28,31,32) indicates that the RBD can profoundly influence the transcriptional properties of EWS. The RBD contains features common to several other RNA-binding proteins [(6–8) and see Figure 1] although the natural RNA ligands are not yet known. The RBD binds to synthetic ribopolymers and to ssDNA (33,34) and the recent demonstration of sequence-specific RNA binding by TLS (35) may point to a more specific regulatory role. Significantly, the isolated RGG3 region of EWS [(33), Figure 1] and the homologous RGG rich regions of TLS (34,35) and hnRNPU protein (36) are sufficient for the interactions with RNA mentioned above. Besides a role in RNA binding, RGG-boxes are also able to mediate protein–protein interactions (7,8) and particular RGG-box interacting proteins include splicing components, namely the SR family of splicing factors (37) and the multifunctional SMN protein (38). Finally, the RGG-boxes of G3BP and nucleolin are implicated in RNA/DNA helicase activity (39) and RGG-boxes in hnRNPK (40) and hnRNPD and nucleolin (41) are required for transcriptional activation. Together, the above findings suggest that RGG-boxes, like TETs, may perform several functions.
Here, we demonstrate that RGG-boxes within the EWS RBD are necessary and sufficient for repression of several transcriptional activation domains, including the EAD. We also describe additional features of RGG-box mediated repression which point to a significant role for RGG boxes in determining the transcriptional properties of native EWS.
MATERIALS AND METHODS
Plasmids and constructions
The reporters pΔ(−71)SomCAT (42), pG1E4TCAT, pG5E4TCAT, pZ7E4TCAT (43) and pBS2CAT (44) are as previously described. The protein expression vectors pSVEWS/ATF1 (20), pΔ87C (45), p87R, pNC and pG4NC (31), pBSAPCREB (44) and pbZ3EA/pbZ12EA (45) are as described previously. pRGG contains EAD residues 1–86 fused to the RGG3 region of EWS (residues 545–656) and was obtained by insertion of a BglII/XhoI fragment from pRM2 (31) into BglII and XhoI digested p87R. pLR1 was obtained by inserting a BglII/EcoRI ended PCR product containing EWS residues 571–604 (from RGG3) into BglII/EcoRI digested pRGG. To construct pLR2 and pLR4, a BglII/XbaI fragment from pLR1 or pLR2 respectively, was inserted into BamHI/XbaI digested pLR1 or pLR2. pSR1 was obtained by inserting oligonucleotides encoding EWS residues 587–604 (from RGG3) into BglII/EcoRI digested pRGG. pSR2 and pSR4 were constructed by cloning a BglII/XbaI fragment from pSR1 or pSR2 into BamHI/XbaI digested pSR1 or pSR2. pSRm1, pSRm2, pSRm3 and pSRm4 were constructed in the same manner as pSR4, using oligonucleotides containing the desired mutations. All inserts were sequenced to ensure the absence of extraneous mutations. pEASR4 and pEASRm1 were obtained by inserting a HindIII/BglII fragment from pNC into HindIII/BglII digested pSR4 and pSRm1 respectively. pBCREBvec was obtained by inserting an oligonucleotide containing the desired restriction sites into XbaI digested pBSAPCREB. pBCREBR was obtained by inserting an XbaI fragment from pG4NC into NheI/XbaI digested pBCREBvec. pG4E1a and pG4VP16 were obtained by inserting SalI/BglII ended PCR products containing the activation domains of E1a (residues 121–223) and VP16 (residues 413–490), into SalI/BglII digested pG4NC. pE1aR and pVP16R were obtained by insertion of a BglII/XbaI fragment from pG4NC into BglII/XbaI digested pG4E1a and pG4VP16, respectively. pVPSRm2 and pVPSR4 were obtained by inserting a BglII/BamHI fragment from pSRm2 and pSR4 respectively into BglII digested pG4VP16. pVPvec was obtained by cloning an oligonucleotide containing an in-frame EcoRI site, into pVP16R digested with BglII and XbaI. pVPNC was obtained by inserting in a EcoRI fragment from pVPvec into EcoRI digested pG4NC. pVPE was obtained by digestion of pVPNC with XbaI and religation to remove the RBD. VPESRm2 and pVPESR4 were obtained by inserting a BglII/XbaI fragment from pVPSRm2 and pVPSR4 into BglII/XbaI digested pVPNC. pbZ3 and pbZ12 were obtained by inserting oligonucleotides containing a functional initiation codon into BglII/NdeI digested pbZ3EA and pbZ12EA. pbZ3LR4 (or pbZ12LR4), pbZ3SR4 (or pbZ12SR4) and pbZ3SRm1 (or pbZ12SRm1) were obtained by inserting the BglII/BamHI fragment from pLR4, pSR4 and pSRm1 into pbZ3EA or pbZ12EA digested with BglII. pbZ3RBD was obtained by inserting a BglII/NdeI fragment from pNC into BglII/NdeI digested pbZ3EA, followed by removal of the ATF1 N-terminal sequence by digestion with BglII and NdeI, and insertion of an oligonucleotide to maintain the reading frame. pbZ3VPSR4 was obtained by inserting a blunt-ended PCR fragment containing the VP16 activation domain into BglII digested blunt-ended pbZ3SR4.
Transfections, CAT assays and western blotting
JEG3 cells were grown in DMEM containing 10% FCS. Transfections, CAT assays and western blotting of epitope-tagged proteins using monoclonal antibody KT3 (46) were carried out as previously described (31). For quantitation, percentage conversion of unacetylated to acetylated 14C-chloramphenicol under linear assay conditions was determined by excision of spots from the TLC plate and quantitation of radioactivity using a liquid scintillation counter.
RESULTS
A repression assay for small regions of the EWS RBD
Our previous studies of EFPs and RBD-mediated repression of the EAD have focused on EWS/ATF1 (Figure 1), the EFP which causes soft tissue clear cell sarcoma (4). Unlike ATF1 which is a cAMP-inducible activator (47,48), EWS/ATF1 is a potent constitutive activator of ATF-dependent promoters (20,21), dependent on multiple elements dispersed throughout the EAD (45) including a consensus SYGQQS repeat with a critical Tyr residue (22). Our previous findings (31) suggested that RGG-boxes within the EWS RNA-binding domain (RBD, Figure 1) and the RGG tripeptides therein, might be necessary for cis-repression of the EAD. Testing this hypothesis is hindered by the number of RGG tripeptides (a total of 20) in the RBD which renders mutational analysis technically difficult. Realistically, the above task can only be achieved via total synthesis of mutated oligonucleotides and reiterative subcloning. This in turn requires establishment of a functional repression assay for smaller regions of the RBD (in practice, ∼40 residues or 120 nt). In light of the contribution of the RGG3 region to repression (31) and due to the high density of RGG tripeptides within RGG3, we attempted to exploit the RGG3 region to achieve the above objective.
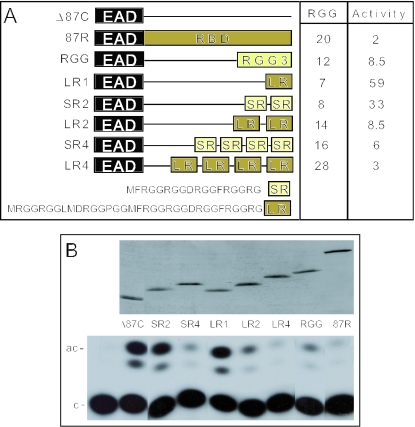
Despite the significant effect of deleting RGG3 on repression of the intact EAD (31) RGG3 alone only moderately represses the EAD, perhaps due to the magnitude of activation by the EAD. To establish a more workable assay for RGG3-mediated repression (Figure 2), we tested the ability of RGG3 to repress the lower levels of trans-activation by the N-terminal region of the EAD (present in Δ87C) using a previously described transient assay (31,45). Briefly, an expression vector for the test protein and an ATF-dependent reporter [pΔ(−71)SomCAT] were introduced into JEG3 cells and trans-activation monitored by CAT assay. As previously shown (31), the RBD strongly represses Δ87C (50-fold repression, compare Δ87C and 87R, Figure 2B) and RGG3 alone is indeed able to repress reasonably well (12-fold repression, compare Δ87C and RGG).
Figure 2.
An assay for repression by small regions of the EWS RBD. (A) Structure of experimental proteins. For reference, the structure of EWS, ATF1 and EWS/ATF1 are described in Figure 1. Δ87C (45) is a derivative of the EWS/ATF1 oncogene containing only the N-terminal 86 residues of the EAD fused to ATF1 (residues 66–271) including the bZIP domain that allows dimerization and DNA binding. Δ87C is a constitutive activator of ATF-dependent promoters dependent on the EAD region (45). RGG3 corresponds to residues 545–656 of intact EWS. All of the novel proteins tested are derived from Δ87C by insertion of the indicated sequence between the EAD and ATF1 regions. The ATF1 region is thus present in all proteins but is not shown in the diagram. The amino acid sequence of a short repeat (SR, light green box) and a long repeat (LR, dark green box) are present within the RGG3 region of EWS (LR contains EWS residues 571–604 and SR contains EWS residues 587–604). The number of RGG-tripeptides in each protein and the relative transcription activity are shown to the right. (B) Transcription assays. JEG3 cells were transfected with 5 μg of ATF-dependent reporter [pΔ−(71)SomCat] and 5 μg of plasmid expressing the activator indicated. Transcriptional activity was monitored by CAT assay and a representative TLC result is shown at the bottom (c = chloramphenicol; and ac = acetylated chloramphenicol). The corresponding western blot, using monoclonal antibody KT3 (45) to monitor epitope-tagged activator levels, is shown above the CAT assay.
We next created a series of synthetic proteins (Figure 2A) containing one (LR1), two (LR2) or four (LR4) copies of a long repeat (LR, EWS residues 571–605, including seven RGG tripeptides), or two (SR2) or four (SR4) copies of a shorter repeat (SR, EWS residues 587–605, including four RGG tripeptides) present with RGG3. All the above proteins are expressed at comparable levels in transfected cells (Figure 2B). While the presence of seven (LR1, 1.7-fold) or eight RGG tripeptides (SR2, 3-fold) results in very modest repression, 14 (LR2, 12-fold) or 16 RGG tripeptides (SR4, 17-fold) gave strong repression and 28 RGG tripeptides (LR4, 32-fold) repress more or less as efficiently as the intact RBD (Figure 2B). Overall, the above experiments reveal a clear inverse correlation between trans-activation and the number of RGG tripeptides present, suggesting that RGG tripeptides are necessary for repression. Furthermore, given that RGG tripeptides account for ∼70% of the residues present in LR and SR, it seems probable that they are sufficient for repression (see below). Although the above analysis is not extensive, the results also indicate quite clearly that RGG tripeptides act synergistically above a certain threshold. For example, 7 RGG tripeptides (LR1) give very a modest effect (∼2-fold repression) while 14 (LR2) give much more than an additive effect (12-fold repression).
Mutational analysis of SR4
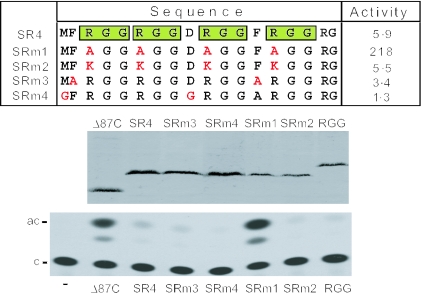
The assay described above enabled creation and testing of mutants to precisely define the residues within SR4 that mediate repression (Figure 3). Simultaneous substitution of Arg by Ala in all four RGG tripeptides present (SRm1) eliminated repression and even resulted in modest stimulation (2-fold), thus providing direct evidence that the Arg residues within RGG tripeptides are required for repression. We note that while SRm1 and in addition SRm2 (Arg to Lys) proteins are expressed at lower levels relative to other mutants tested, the high transcriptional activity of SRm1 demonstrates that this level of expression is quite sufficient to reflect activity. Thus, Lys is able to substitute for Arg (SRm2) indicating that the basic nature of the Arg side chain is the important repression determinant. The RGG tripeptides in SR4 are flanked by Phe residues which are a conserved feature of RGG-boxes in other RNA-binding proteins (8,49). However, substitution of the Phe residues in SR4 by Ala (SRm3) had no obvious effect on the magnitude of repression. Similarly, substitution of the two remaining residues (Met and Asp) present in SR4 by Gly (SRm4) had no effect on repression. Because Gly residues constitute 50% of SR and even substitution by Ala would dramatically alter the structure of SR, we did not test the effect of substituting Gly residues. Together, the above results demonstrate that the RGG tripeptides within SR4 are necessary and sufficient for repression, with Arg residues playing a crucial role.
Figure 3.
Mutational analysis of SR4. Residues present in each mutant are aligned with the SR4 amino acid sequence. RGG tri-peptides are highlighted by green boxes and point mutations are in red. The relative transcription activity for each mutant (compared with Δ87C set at 100) is indicated to the right. For experiments, JEG3 cells were transfected with 5 μg of ATF-dependent reporter [pΔ−(71)SomCat] and 5 μg of plasmid expressing the indicated activator. Transcriptional activity was monitored by CAT assay. Middle panel. western blot analysis of KT3 epitope-tagged proteins in transfected cells. Lower panel: representative TLC of CAT assay (c = chloramphenicol; and ac = acetylated chloramphenicol).
Fusion of the synthetic RGG-containing elements present in SR4 and SRm1 to the intact EAD (EASR4 and EASRm1, Figure 4A) also resulted in effective repression in the case of EASR4 (10-fold) but not in the case of EASRm1 (2-fold). Thus, the RGG tripeptides present within SR4 can repress the intact EAD (see also Figure 5) although less efficiently than can the intact RBD.
Figure 4.
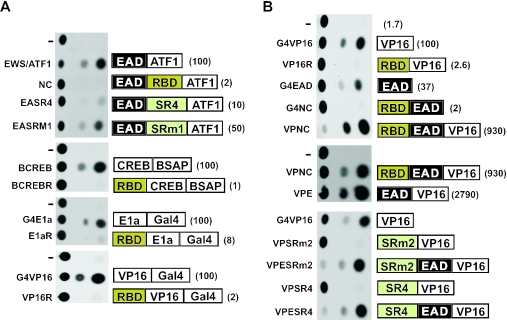
Specificity and dominance of RGG-box mediated repression. (A) Effect of the RBD on single activation domains (EAD, VP16, E1a and CREB). (B) Effect of RBD on two activation domains (EAD and VP16) present together. JEG3 cells were transfected with plasmids expressing the proteins indicated (as described in Materials and Methods) together with the following reporter constructs: p(Δ− 71)SomCat (for activators containing the ATF1 DNA-binding domain); pBS2CAT (for activators containing the BSAP DNA-binding domain); pG5E4TCAT (for G4E1a and E1aR) and pG1E4TCAT (for VP16 and derivatives). CAT assays were performed 40 h post-transfection and a representative CAT assay is shown. Relative transcription activity is shown in parenthesis to the right of each protein tested. For single activation domains (A) activity in the absence of the RBD is set at 100. For the joint activity of VP16 and the EAD (B) the activity of G4VP16 is set at 100.
Figure 5.
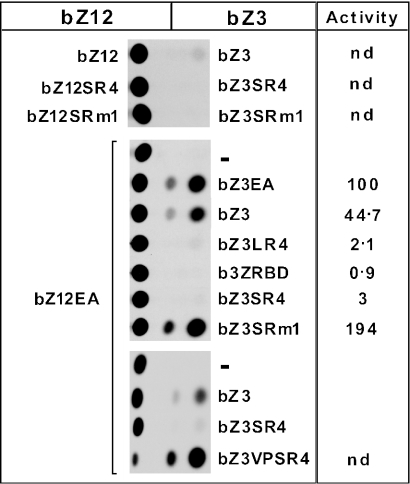
Effect of the RBD in obligatory heterodimers. JEG3 cells were transfected with a Zta reporter plasmid (pZ7E4TCAT) together with two other plasmids expressing either a bZ12 derivative (as indicated to the left) or bZ3 derivative (indicated to the right). Thus, the resultant dimeric proteins in each case will consist of a heterodimer containing one bZ12 partner and one bZ3 partner. CAT assays were performed 40 h post-transfection and a representative CAT assay is shown. Relative transcription activity for each heterodimer is shown in the right hand column with the bZ3EA/bZ12EA combination set at 100. nd, not determined.
Repression of a range of activation domains by the RGG-box
To assess repression specificity, we examined the effect of the RBD on multiple distinct activation domains (Figure 4A). G4E1a and G4VP16 contain the Gal4 DNA-binding domain linked to the adenovirus E1a and VP16 activation domains respectively and BCREB contains the BSAP DNA-binding domain linked to the glutamine-rich activation domains of CREB (44). E1aR, VP16R and BCREBR are the corresponding proteins containing the EWS RBD. Following co-transfection of JEG3 cells using the appropriate reporter constructs (see legend, Figure 4A) each of the above activation domains was strongly repressed by the RBD, although the magnitude of repression did vary (E1a, 12.6-fold repression; VP16, 54-fold; BCREB, 125-fold). We also tested the capacity of the synthetic element in SR4 to repress VP16 (Figure 4B) and found that the magnitude of repression of VP16 in this case (73-fold, compare G4VP16 with VPSR4) is comparable with that achieved by the intact RBD (54-fold).
The above experiments do not indicate whether repression by the RGG boxes is direct or indirect. To address this issue, we asked whether the synthetic element in SR4 or the RBD can simultaneously repress two distinct activation domains (VP16 and the EAD) present in the same molecule (Figure 4B). In the experiment shown, the RBD strongly represses the EAD (18-fold) and VP16 (39-fold) activation domains when the latter are present alone but when the EAD and VP16 are together (VPNC) potent transcriptional activation is restored (compare VPNC with VP16R or G4NC). Quantitation (in the linear range of the CAT assay) reveals that activation by VPNC (550-fold) is about 7-fold higher than the sum of VP16 (60-fold) and the EAD (22-fold) alone. Thus, even when the RBD is present (VPNC) VP16 and the EAD together give very potent synergistic activation, indicating that two activation domains can work together to counteract RGG-mediated repression. Deletion of the RBD from VPNC results in only a modest increase in trans-activation (3-fold, compare VPNC and VPE) confirming that repression is indeed largely overcome by the joint action of VP16 and the EAD. Similar to the effect of the RBD, while the EAD and VP16 alone are repressed by either SR4 or SR4m2 (Figure 4B), activation is restored in both cases when the EAD and VP16 are present together (compare VPESR4 with VPSR4 and VPESRm2 with VPSRm2). The above results demonstrate that RGG-boxes (or the RBD) do not prevent entry of cis-linked activation domains (the EAD and VP16) into functional transcription complexes. In addition, while all activation domains tested can be repressed, repression is not dominant but can (at least in the case of VP16 and the EAD) be counteracted by the presence of an additional activation domain.
Trans-repression by the RGG-box within heterodimeric complexes
While the RBD does not repress the EAD in trans (i.e. when not recruited to the promoter) (data not shown), we wanted to determine whether the RBD (or SR4) could repress the EAD when present in heterodimeric complexes. To achieve this, we exploited obligatory dimerization partners (Figure 5) and a Zta DNA-binding domain to avoid interference from endogenous ATF1/CREB as previously described (45,47). bZIP domain proteins bZ3 and bZ12 contain the Zta basic domain (b) and mutated ATF1/CREB leucine zippers (Z3 and Z12) that do not homodimerize but do form heterodimers (47,50). Thus, bZ3 and bZ12 do not homodimerize or bind to Zta binding sites, while bZ3/bZ12 heterodimers form efficiently and can therefore be exploited to produce and assay any desired heterodimer.
As expected (Figure 5), a combination of bZ3 and bZ12 gave little trans-activation of a reporter (pZ7E4TCAT) containing Zta binding sites, while a bZ3EA/bZ12EA heterodimer (essentially representing homodimeric EWS/ATF1 with each partner containing the intact EAD and ATF1) strongly activates (set at 100%) as previously shown (45). We analysed a series of heterodimers obtained by combining bZ12EA with various derivatives of bZ3. bZ3 alone combines with bZ12EA to give significant activation (45% of bZ3EA/bZ12EA), indicating that the presence of the EAD in only one partner of a dimer is sufficient for relatively high levels of activation on a multi-site reporter (pZ7E4TCAT). bZ3RBD (containing the intact RBD, 0.9%), bZ3LR4 (containing the LR4 synthetic element, 2.1%) and bZ3SR4 (containing the SR4 synthetic element, 3%) all failed to significantly activate bZ12EA. This lack of activation strongly suggested that the RBD and SR4 are able to repress the EAD within heterodimeric complexes. To substantiate this conclusion, bZ3SR4m1 (containing Arg to Ala substitutions within SR4) and bZ3VPSR4 (containing the VP16 activation domain in addition to SR4) were both able to strongly activate in combination with bZ12EA. A combination of bZ3SRm1 and bZ12SRm1 is inactive, thus ruling out the possibility that these proteins activate by themselves. Identical results were obtained, in each case, in complementary experiments using a combination of bZ3EA and all of the derivatives described above in a bZ12 background (data not shown). To summarize, there is a correlation between the ability to repress the EAD in-cis and the inability to support activation by the EAD in-trans, and we therefore conclude that the EWS RBD or RGG-boxes, directly repress the EAD in heterodimeric complexes.
DISCUSSION
Structural requirements for RGG-box-mediated repression
We have shown that the RGG-box present within the RBD of a TET family member (EWS) can repress transcriptional activation by the EAD and a range of other activation domains. The arginine residues present within multiple RGG tripeptide motifs are necessary for repression and our data indicate that the minimal repression element is a reiterated RGG tripeptide of approximately 12 copies (Figure 2). Lys can functionally substitute for Arg, indicating that the previously demonstrated Arg methylation of RGG-boxes within EWS (19) is not critical for repression. This is similar to the requirement for nucleolar localization of nucleolin (51) and indicates that the basic nature of the Arg side chain is the primary repression determinant of RGG-boxes. There is little conservation of spacing for the intact RGG3 region compared with the synthetic elements LR2 and SR4, all of which have a similar number of RGG tripeptides and repress to similar degrees. Thus our results indicate that the spatial arrangement of RGG tripeptides is not highly constrained. RGG-boxes form a flexible β-spiral structure (7,8) but beyond that, a high resolution structure/function relationship for the RGG-box (in relation to transcriptional repression, RNA binding or other functions) has yet to be determined and represents an important task for the future.
Mechanism of repression by the RGG-box
Our finding that repression can be effectively counteracted by the synergistic action of two transcriptional activation domains, indicates that the RGG-box does not act indirectly, e.g., via inhibition of nuclear export of nascent transcripts. In addition, the likely presence of native EWS in a high proportion of cellular transcription complexes (5,24) is also incompatible with a generally repressive effect of the RBD on events distal to transcription. We therefore interpret our results to indicate that the individual activation domains tested are directly repressed by the RGG-box/RGG-tripeptides.
We have previously suggested (31) that repression by the EWS RBD might be quite selective for the EAD and result from the RBD blocking interaction between the EAD and the Pol II subunit RPB7 (23,52). However, the findings reported here show that a range of distinct activation domains are susceptible to repression. A priori, the ability of two activation domains to alleviate repression, might most readily be explained by direct physical, and hence competing, interactions between the activation domains and the RBD. However, the unrelated primary structure of the activators in question (EAD, E1a, VP16 and the Q-rich activation domains of CREB) does not support the above possibility. A more likely explanation is that a net increase in activation domains results in strong synergy and thus a switch from repression to activation. Striking synergistic effects (more than an order of magnitude) are indeed observed upon a minimal increase in the number of activation domains in other systems (43,44,53,54). Thus, we suggest that repression (by the RBD/RGG-boxes) and activation (by the EAD/VP16) may target different steps in transcription complex assembly and that the joint positive effect of the EAD and VP16 overrides the negative effect of the RBD. Such ‘kinetic synergism’ has been proposed previously for the multi-step process of transcription complex assembly (55).
The RGG-box within the RGG3 region of EWS (33), TLS (34) and the Fmrp protein (56), is sufficient for synthetic ribopolymer binding. In addition, TLS (and the RGG2/3 region therein) is able to recognize GGUG containing RNAs of synthetic origin (35). This raises the possibility that RGG-boxes might mediate repression through their intrinsic RNA-binding function. In light of the susceptibility of several different promoters to repression (Figure 4A), we suggest that sequence-specific binding of RGG-boxes to nascent mRNA transcripts is unlikely to be involved in repression. An alternative possibility is that RGG-boxes might interact with a putative RNA-component of the transcriptional machinery, precedent for which comes from identification of an RNA that participates in estrogen inducible transcription (58,59). It has been proposed that RGG-boxes bind to RNA in a manner similar to the HIV TAT protein (7) in which case an Arg residue is critical for binding to the HIV TAR element and Lys cannot substitute for Arg (57). In the above eventuality, the ability of Lys to allow transcriptional repression by RGG-boxes would again suggest that RNA-binding by EWS is not involved in repression.
The RGG-boxes of TETs bind to SR family of splicing factors (37) and the multifunctional Survival of Motor Neuron (SMN) protein (38) that may be involved in coupling of transcription and splicing. Binding of EWS to SMN is reduced by Arg to Lys mutations in the RGG-boxes however (38), indicating that SMN is not likely to be involved in repression. Another potential feature of repression is suggested by the DNA/RNA helicase and ATPase activity residing within the RGG boxes of G3BP and nucleolin (39). Future progress in uncovering the molecular mechanism of repression will require biochemical analysis and may also arise from identification of novel RGG-box interacting proteins and the physiological RNA ligands for EWS.
Function of EWS
The ability of the isolated RBD to repress transcription by an array of activation domains raises the possibility that native EWS could act as repressor. The existence of several other RNA-binding proteins that repress transcription, including Nrd1 (60), hnRNP-U (61) and NELF-E (62), ssDBF (63), SPEN proteins (64), TDP-43 (65) and Sam68 (66), provides ample precedent for this. In the event that EWS is a repressor, the involvement of RGG boxes in repression would represent a novel mode of action for vis a vis the other repressors mentioned above. However, based on the finding that the EWS RBD does not act as a dominant repressor, we suggest that native EWS is unlikely to act as a general repressor within mammalian transcription complexes that, commonly, contain multiple synergistic activators.
The finding that the EAD is such a potent activation domain in the context of EFPs and yet is effectively repressed (50-fold) by the RBD, suggests that EAD function within EWS may be quite distinct to that uncovered in the context of EFPs. Consistent with this suggestion, the EAD and intact EWS can have apparently opposite transcriptional effects (28,29,67). Binding of the EAD (in EWS/ATF1) to the co-activator CBP represses p53-mediated trans-activation (67) while in contrast, binding of intact EWS to CBP allows transcriptional activation (28,29). It is also of significance that trans-activation by EWS (28,29) and TLS (27) is very modest compared with the EAD and in one case the N-terminal 246 residues of EWS (including most of the EAD) is dispensable for activation by EWS (28). The latter observation indicates that trans-activaton by EWS does not reflect residual EAD activity but instead reveals a novel activation capacity of the EWS RBD. Finally, whatever the transcriptional role of EWS might be, the EAD presumably plays a positive role in one or more of the wide range of potential EWS functions alluded to in the introduction (9–19).
Given the presence of EWS in a wide range of transcription complexes and the fact that a single, promoter-bound EFP can activate transcription with such potency (20), it is apparent that promiscuous activation by the EAD, and hence gross de-regulation of cellular transcription, must somehow be averted. Repression of the EAD by RGG-boxes provides a mechanism to achieve this. Thus, we suggest that one function of the EWS RBD is to serve as a negative control element for the EAD, while still allowing other activators present in particular transcription complexes to activate. This idea is strongly supported by the finding that additional activation domains can overcome repression by the RBD (Figures 4 and 5). If EWS is, in many cases, neutral for transcription, then the presence of EWS in most transcription complexes may reflect a role in coupling of transcription to other RNA transactions in the cell. The abundant evidence that EWS functionally interacts with several splicing factors (9–13) supports this idea.
Repression and oncogenesis
The work of several groups (2) has indicated that EFPs are involved in tumour maintenance, raising the possibility that EFP/EAD inhibitors might serve as therapeutic leads. Indeed this objective stimulated the study reported here. The finding that RGG-boxes can repress a broad range of activation domains, however, argues that RGG-boxes will have limited potential as specific EAD inhibitors. In contrast, the ability of RGG-boxes to strongly repress the EAD in heterodimeric complexes (Figure 5) predicts that RGG-boxes fused to the ATF1 bZIP domain should function as highly specific and effective EWS/ATF1 inhibitors. Thus, it will be of interest to utilize such molecules to downregulate EWS/ATF1 and observe the biological effect on CCS cells.
Acknowledgments
This work was supported by an Association for International Cancer Research grant (AICR grant # 03-131) to K.A.W.L. We are grateful to Dr Mingjie Zhang for helpful comments on the manuscript. The Open Access publication charges for this article were paid by K.A.W.L.
REFERENCES
- 1.Kim J., Pelletier J. Molecular genetics of chromosome translocations involving EWS and related family members. Physiol. Genomics. 1999;1:127–138. doi: 10.1152/physiolgenomics.1999.1.3.127. [DOI] [PubMed] [Google Scholar]
- 2.Kovar H., Aryee D., Zoubek A. The Ewing family of Tumors and the search for the Achilles heel. Curr. Opin. Oncol. 1999;11:275–284. doi: 10.1097/00001622-199907000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Arvand A., Denny C.T. Biology of EWS/ETS fusions in Ewing's family tumors. Oncogene. 2001;20:5747–5754. doi: 10.1038/sj.onc.1204598. [DOI] [PubMed] [Google Scholar]
- 4.Zucman J., Delattre O., Desmaze C., Epstein A.L., Stenman G., Speleman F., Fletchers C.D.M., Aurias A., Thomas G. EWS and ATF1 gene fusion induced by t(12:22) translocation in malignant melanoma of soft parts. Nature Genet. 1993;4:341–345. doi: 10.1038/ng0893-341. [DOI] [PubMed] [Google Scholar]
- 5.Bertolotti A., Lutz Y., Heard D.J., Chambon P., Tora L. hTAFII68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes S.R. The RNP Motif Protein Family. The New Biologist. 1992;4:421–429. [PubMed] [Google Scholar]
- 7.Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 8.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 9.Yang L., Chansky H.A., Hickstein D.D. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine–arginine protein-mediated RNA splicing. J. Biol. Chem. 2000;275:37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 10.Knoop L.L., Baker S.J. The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem. 2000;275:24865–24871. doi: 10.1074/jbc.M001661200. [DOI] [PubMed] [Google Scholar]
- 11.Knoop L.L., Baker S.J. EWS/FLI alters 5′-splice site selection. J. Biol. Chem. 2001;276:22317–22322. doi: 10.1074/jbc.M008950200. [DOI] [PubMed] [Google Scholar]
- 12.Chansky H.A., Hu M., Hickstein D.D., Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- 13.Ohkura N., Yaguchi H., Tsukada T., Yamaguchi K. The EWS/NOR1 fusion gene product gains a novel activity affecting pre-mRNA splicing. J. Biol. Chem. 2002;277:535–543. doi: 10.1074/jbc.M109018200. [DOI] [PubMed] [Google Scholar]
- 14.Felsch J.S., Lane W.S., Peralta E.G. Tyrosine kinase Pyk2 mediates G-protein-coupled receptor regulation of the Ewing sarcoma RNA-binding protein EWS. Curr. Biol. 1999;9:485–488. doi: 10.1016/s0960-9822(99)80214-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee H.J., Kim S., Pelletier J., Kim J. Stimulation of hTAFII68(NTD)-mediated trans-activation by v-Src. FEBS Lett. 2004;564:188–198. doi: 10.1016/S0014-5793(04)00314-X. [DOI] [PubMed] [Google Scholar]
- 16.Deloulme J.C., Prichard L., Delattre O., Storm D.R. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J. Biol. Chem. 1997;272:27369–27377. doi: 10.1074/jbc.272.43.27369. [DOI] [PubMed] [Google Scholar]
- 17.Olsen R.R., Hinrichs S.H. Phosphorylation of the EWS IQ domain regulates transciptional activity of the EWS/ATF1 and EWS/FLI1 fusion proteins. Oncogene. 2001;20:1756–1764. doi: 10.1038/sj.onc.1204268. [DOI] [PubMed] [Google Scholar]
- 18.Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 19.Belyanskaya L.L., Gehrig P.M., Gehring H. Exposure on cell surface and extensive arginine methylation of Ewing sarcoma (EWS) protein. J. Biol. Chem. 2001;276:18681–18687. doi: 10.1074/jbc.M011446200. [DOI] [PubMed] [Google Scholar]
- 20.Brown A.D., Lopez-Terrada D., Denny C.T., Lee K.A.W. Promoters containing ATF-binding sites are de-regulated in tumour-derived cell lines that express the EWS/ATF1 oncogene. Oncogene. 1995;10:1749–1756. [PubMed] [Google Scholar]
- 21.Fujimura Y., Ohno T., Siddique H., Lee L., Rao V.N., Reddy E.S.P. The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene. 1996;12:159–167. [PubMed] [Google Scholar]
- 22.Feng L., Lee K.A.W. A repetitive element containing a critical tyrosine residue is required for transcriptional activation by the Ewings sarcoma oncogene. Oncogene. 2001;20:4161–4168. doi: 10.1038/sj.onc.1204522. [DOI] [PubMed] [Google Scholar]
- 23.Petermann R., Mossier B.M., Aryee D.N., Khazak V., Golemis E.A., Kovar H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17:603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 24.Bertolotti A., Melot T., Acker J., Vigneron M., Delattre O., Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallier M., Lerga A., Barnache S., Tavitian A., Moreau-Gachelin F. The transcriptional factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- 26.Powers C.A., Mathur M., Raaka B.M., Ron D., Samuels H.H. TLS (Translocated-in-Liposarcoma) is a high-affinity interactor for steroid, thyroid hormone and retinoid receptors. Mol. Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 27.Uranishi H., Tetsuka T., Yamashita M., Asamitsu K., Shimizu M., Itoh M., Okamoto T. Involvement of the pro-oncoprotein TLS (Translocated in Liposarcoma) in nuclear factor-κB p65-mediated transcription as a coactivator. J. Biol. Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- 28.Rossow K.I., Janknecht R. The Ewing's sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- 29.Araya N., Hirota K., Shimamoto Y., Miyagishi M., Yoshida E., Ishida J., Kaneko S., Kaneko M., Nakajima T., Fukamizu A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003;278:5427–5432. doi: 10.1074/jbc.M210234200. [DOI] [PubMed] [Google Scholar]
- 30.Immanuel D., Zinszner H., Ron D. Association of SARFH (sarcoma associated RNA-binding fly homologue) with regions of chromatin transcribed by RNA polymerase II. Mol. Cell. Biol. 1995;15:4562–4571. doi: 10.1128/mcb.15.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K.K.C., Lee K.A.W. Transcriptional activation by the EWS oncogene can be cis-repressed by the EWS RNA-binding domain. J. Biol. Chem. 2000;275:23053–23058. doi: 10.1074/jbc.M002961200. [DOI] [PubMed] [Google Scholar]
- 32.Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 2000;18:8000–8010. doi: 10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 33.Ohno T., Ouchida M., Lee L., Gatalica Z., Rao V.N., Reddy E.S.P. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 34.Prasad D.D.K., Ouchida M., Lee L., Rao V.N., Reddy E.S.P. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 35.Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 36.Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through the RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., Embree L., Tsai S., Hickstein D.D. Oncoprotein TLS interacts with serine–arginine proteins involved in RNA splicing. J. Biol. Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 38.Young P.J., Francis J.W., Lince D., Coon K., Androphy E.J., Lorson C.L. The Ewing's sarcoma protein interacts with the Tudor domain of the survival motor neuron protein. Mol. Brain Res. 2003;119:37–49. doi: 10.1016/j.molbrainres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Costa M., Ochem A., Staub A., Falaschi A. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the Ras transduction pathway. Nucleic Acids Res. 1999;27:817–821. doi: 10.1093/nar/27.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee M.-H., Mori S., Raychaudhuri P. trans-Activation by the hnRNP K protein involves an increase in RNA synthesis from the reporter genes. J. Biol. Chem. 1996;271:3420–3427. doi: 10.1074/jbc.271.7.3420. [DOI] [PubMed] [Google Scholar]
- 41.Dempsey L.A., Hanakahi L.A., Maizels N. A specifc isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specifc duplex DNA binding protein. J. Biol. Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 42.Montminy M.R., Sevarino K.A., Wagner J.A., Mandel G., Goodman R.H. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl Acad. Sci. USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emami K.H., Carey M. A synergistic increase in potency of a multimerised VP16 transcriptional activation domain. EMBO J. 1992;11:5005–5012. doi: 10.1002/j.1460-2075.1992.tb05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krajewski W., Lee K.A.W. A monomeric derivative of the cellular transcription factor CREB functions as a constitutive activator. Mol. Cell. Biol. 1994;14:7204–7210. doi: 10.1128/mcb.14.11.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan S., Koh Y.M., Dunn T.A., Li K.K.C., Lee K.A.W. The EWS/ATF1 fusion protein contains a dispersed activation domain that functions directly. Oncogene. 1998;16:1625–1631. doi: 10.1038/sj.onc.1201671. [DOI] [PubMed] [Google Scholar]
- 46.MacArthur H., Walter G. Monoclonal antibodies specific for the carboxy terminus of Simian virus 40 large T antigen. J. Virol. 1984;52:483–491. doi: 10.1128/jvi.52.2.483-491.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribeiro A., Brown A.D., Lee K.A.W. An in vivo assay for members of the CREB family of transcription factors. J. Biol. Chem. 1994;269:31124–31128. [PubMed] [Google Scholar]
- 48.Lee K.A.W., Masson N. Transcriptional regulation by CREB and its relatives. Biochem. Biophys. Acta. 1993;1174:221–233. doi: 10.1016/0167-4781(93)90191-f. [DOI] [PubMed] [Google Scholar]
- 49.Kim S., Merrill B.M., Rajpurohit R., Kumar A., Stone K.L., Papov V.V., Schneiders J.M., Szer W., Wilson S.H., Paik W.K., Williams K.R. Identification of NG-Methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36:5185–5192. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- 50.Loriaux M.M., Rehfuss R.R., Brennan R.G., Goodman R.H. Engineered leucine zippers show that hemiphosphorylated CREB complexes are transcriptionally active. Proc. Natl Acad. Sci. USA. 1993;90:9046–9050. doi: 10.1073/pnas.90.19.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pellar G.J., DiMario P.J. Deletion and site-specific mutagenesis of nucleolin's carboxy GAR domain. Chromosoma. 2003;111:461–469. doi: 10.1007/s00412-003-0231-y. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H.-Q., Lee K.A.W. An hsRPB4/7-dependent yeast assay for trans-activation by the EWS oncogene. Oncogene. 2001;20:1519–1524. doi: 10.1038/sj.onc.1204135. [DOI] [PubMed] [Google Scholar]
- 53.Oliviero S., Struhl K. Synergistic transcriptional enhancement does not depend on the number of acidic activation domains bound to the promoter. Proc. Natl Acad. Sci. USA. 1991;88:224–228. doi: 10.1073/pnas.88.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carey M., Kolman J., Katz D.A., Gradoville L., Barberis L., Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J. Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herschlag D., Johnson F.B. Synergism in transcriptional activation: a kinetic view. Genes Dev. 1993;7:173–179. doi: 10.1101/gad.7.2.173. [DOI] [PubMed] [Google Scholar]
- 56.Brown V., Small K., Lakkis L., Feng Y., Gunter C., Wilkinson K.D., Warren S.T. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 57.Tao J., Frankel A.D. Specific binding of arginine to TAR RNA. Proc. Natl Acad. Sci. USA. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanz R.B., McKenna N.J., Onate S.A., Albrecht U., Wong J., Tsai S.Y., Tsai M-J., O'Malley B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 59.Freedman L.P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 60.Steinmetz E.J., Brow D.A. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl Acad. Sci. USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim M.K., Nikodem V.M. hnRNP U inhibits carboxy-terminal domain phosphorylation by TFIIH and represses RNA polymerase II elongation. Mol. Cell. Biol. 1999;19:6833–6844. doi: 10.1128/mcb.19.10.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi Y., Inukai N., Narta T., Wada T., Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smidt M.P., Russchen B., Snippe L., Wijnholds J., AB G. Cloning and characterisation of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res. 1995;23:2389–2395. doi: 10.1093/nar/23.13.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ariyoshi M., Schwabe J.W.R. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–1920. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H-Y, Wang I-F., Bose J., Shen C.K. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 66.Hong W., Resnick R.J., Rakowski C., Shalloway D., Taylor S.J., Blobel G.A. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol. Cancer Res. 2002;1:48–55. [PubMed] [Google Scholar]
- 67.Fujimura Y., Siddique H., Leo L., Rao V.N., Reddy E.S.P. EWS-ATF-1 chimeric protein in soft tissue clear cell sarcoma associates with CREB-binding protein and interferes with p53-mediated trans-activation function. Oncogene. 2001;20:6653–6659. doi: 10.1038/sj.onc.1204684. [DOI] [PubMed] [Google Scholar]