Abstract
NELF and DSIF act together to inhibit transcription elongation in vitro, and are implicated in causing promoter proximal pausing on the hsp70 gene in Drosophila. Here, further characterization of Drosophila NELF is provided. Drosophila NELF has four subunits similar to subunits of human NELF. The amino acid sequences of NELF-B and NELF-D are highly conserved throughout their lengths, while NELF-A and NELF-E contain nonconserved regions inserted between conserved N- and C-terminal regions. Immunodepletion of NELF or DSIF from a nuclear extract desensitizes transcription in vitro to DRB. Immunodepletion of NELF also impairs promoter proximal pausing on the hsp70 promoter in vitro without affecting initiation. Chromatin immunoprecipitation analyses detect NELF at the promoters of the hsp70 and β1-tubulin genes where promoter proximal pausing has been previously detected. Heat shock induction of hsp70 results in a marked decrease in NELF at the hsp70 promoter. Immunofluorescence analysis of polytene chromosomes shows extensive colocalization of the NELF-B and NELF-D subunits at hundreds of interbands. Neither subunit appears to be recruited to puffs. These results provide a foundation for genetic and biochemical analysis of NELF in Drosophila.
INTRODUCTION
NELF and DSIF are two proteins that regulate transcription elongation by Pol II. They were first discovered in human cells during an investigation aimed at understanding how the nucleoside analog, DRB, inhibits transcription elongation (1–3). Biochemical analyses indicated that NELF and DSIF act together to inhibit elongation by Pol II (3). Hyperphosphorylation of Pol II by the kinase, P-TEFb, appears to alleviate this inhibition (2,4). Since DRB is a potent inhibitor of P-TEFb, it has been proposed that DRB blocks phosphorylation of Pol II, thus allowing DSIF and NELF to inhibit elongation (5).
The opposing actions of P-TEFb and the combination of NELF and DSIF could serve as an important mechanism for transcriptional control. P-TEFb associates with several genes during activation, and direct interaction between P-TEFb and transcriptional activators has been observed (6–9). A fusion protein composed of P-TEFb and the Gal4 DNA-binding domain was observed to stimulate transcription of a test promoter containing binding sites for Gal4 (6). We recently showed that NELF and DSIF cause Pol II to pause in the promoter proximal region of the hsp70 gene in Drosophila under normal growth conditions (10). Heat shock induction results in rapid association of P-TEFb with hsp70 (11). NELF but not DSIF appears to dissociate from the elongation complex during heat shock induction (10). In contrast to hsp70, NELF was found to be recruited to estrogen-stimulated genes where it attenuates the level of induction achieved by the addition of estrogen (12). Thus, studies so far indicate that NELF may play different roles on different genes.
Much remains to be learned about the roles of P-TEFb, NELF and DSIF. Although early studies indicated that hyperphosphorylation of the CTD of Pol II might be sufficient to overcome inhibition by NELF and DSIF (3), more recent results indicate that phosphorylation of NELF and DSIF may also be involved (13–15). Flavopiridol, a potent inhibitor of P-TEFb, did not significantly diminish the level of Pol II associated with the hsp70 gene after heat shock induction as would be expected if phosphorylation by P-TEFb was responsible for releasing paused Pol II (16). Biochemical data indicated that NELF and DSIF might provide a checkpoint during early elongation that ensures proper capping of nascent transcripts (17). The broad and overlapping distributions of NELF and DSIF observed on polytene chromosomes are consistent with these proteins affecting transcription of many genes (10).
Although DSIF and P-TEFb have homologs in eukaryotes ranging from yeast to human, no homologs of the four subunits of NELF identified in humans are evident in model organisms such as yeast or Caenorhabditis elegans (18). Thus, the regulatory potential provided by NELF could be restricted to a subset of eukaryotes. Our previous work focused on NELF-D and NELF-E from Drosophila and its role in promoter proximal pausing on the hsp70 gene (10). Here, we report on the characterization of the entire NELF complex from Drosophila, and provide a foundation for further study of this protein in Drosophila.
MATERIALS AND METHODS
Preparation of NELF-A and NELF-B antibodies
NELF-A and NELF-B subunits were identified through BLAST searches of the Drosophila genome using the sequences of human NELF subunits. dNELF-A has the gene identification CG5874 and dNELF-B has the gene identification CG32721. Two EST cDNA clones, SD09448 (NELF-A) and GH10333 (NELF-B) were obtained from the Berkeley Drosophila Genome Project.
NELF-A is predicted to encode a 1248 amino acid polypeptide. The region of cDNA clone SD09448 encoding amino acids 1150–1248 was amplified with the following primers: 5′-CGCGGATCCCGTGGACTCTCTCTATCGAA and 5′-CCGGAATTCGCGTATGACCCTTGTGGA. The resulting DNA fragment was digested with NheI and HindIII and subcloned into HindIII/NheI cut pET28a(+)vector (Novagen). His-tagged NELF-A protein was expressed in BL21(DE3) cells and purified with a HIS-Select™ Cartridge (Sigma) in the presence of urea. The isolated protein was dialyzed into 15 mM Tris–Cl, pH 8.0 and used to raise antibodies in guinea pigs (Pocono Rabbit Farm & Laboratory).
NELF-B is predicted to encode a 594 amino acid polypeptide. A primer set, NELF-B (5′)-5′-CGCAATCCATATGATAATGAGCACACCGGCGAAA and NELF-B (3′-1)-5′-TCGAAGCTTCTATTGGACATTATAGTGC, was used to amplify the region of GH10333 encoding full-length NELF-B. The amplified DNA fragment was digested with NdeI and HindIII and subcloned into HindIII/NdeI cut pET28a(+)vector. His-tagged NELF-B protein was expressed in BL21(DE3) cells and purified in the presence of urea with nickel-NTA agarose (Qiagen). Protein was dialyzed against 100 mM KCl/HEMG and used to produce antisera in guinea pigs. HEMG is 25 mM HEPES, pH 7.6, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol and 1 mM DTT.
Isolation of FLAG-NELF-E complexes
NELF-E cDNA was obtained by RT–PCR from total RNA of S2 cells. A DNA fragment encoding full-length NELF-E and encoding FLAG peptide (DYKDDDDK) at its N-terminus was inserted into the plasmid pA5CΔP, which contains the Drosophila actin 5C promoter and polyadenylation signals (19). The resulting plasmid was called pA5CΔP-FLAG-NELF-E. To generate pA5CΔP-neo, the neomycin phosphotransferase gene cassette was excised from the plasmid pKO SelectNeo (Lexicon Genetics) and inserted into pA5CΔP.
Drosophila S2 cells were grown at 25°C in Drosophila S2 medium supplemented with 10% fetal calf serum (Gibco BRL). Two micrograms of pA5CΔP–FLAG–NELF–E and 0.5 μg of pA5CΔP-neo were cotransfected into 1 ml of cells (5 × 105) using SuperFect reagent (Qiagen). Cells transfected with pA5CΔP-neo alone served as a negative control. Cells were cultured for 2 days in the absence of gentamicin and then cultured with regular passages for 1 month in media containing gentamicin (500 μg/ml; Invitrogen) to establish stably transformed cell lines.
To purify NELF complexes from transformed cells, nine 150 cm2 T-flasks, each containing 40 ml of Flag-NELF-E expressing cells were cultured in the presence of gentamicin (100 μg/ml) for 3 days. Cells were harvested by centrifugation at 1000 g at 4°C, and washed once with cold phosphate-buffered saline. The cell pellet was rapidly frozen in liquid nitrogen and stored at −80°C. All subsequent steps were carried out at 4°C. Cell pellets were resuspended in 15 ml of modified TBS buffer (50 mM Tris–HCl, pH 7.4; 150 mM NaCl; 10% glycerol; 1 mM DTT; and 1× complete proteinase inhibitor) and sonicated four times for 8 s intervals. Supernatant was collected by centrifugation at 8000 r.p.m. for 15 min in a Sorvall SS34 rotor, and incubated with 1.5 ml of anti-FLAG M2 beads (Sigma) for 1 h. The beads were poured into a column and washed with 15 ml of modified TBS buffer. NELF was eluted with FLAG peptide (100 μg/ml) in TBS.
Preparation of antibody resins for immunodepletions
Antibody resins were generated by incubating 500 μl of serum with 250 μl of protein A-sepharose followed by covalent coupling with dimethyl pimelimidate as described by Harlow and Lane (20). Immunoaffinity sepharose was prepared that contained antibody against NELF-B (described above), a mixture of antibodies against both NELF-D and NELF-E (10), or antibody against the largest subunit of DSIF (21). Control antibody-sepharose was prepared from preimmune antibody. To reduce nonspecific binding during depletions, the antibody-sepharose was first incubated with 500 μl of nuclear extract for 4 h. Antibody-sepharose was washed extensively with 0.15 M KCl/HEMG buffer, followed by two washes with 0.1 M glycine, pH 2.5, and extensive washing with 0.15 M KCl/HEMG buffer. Antibody-sepharose was stored at 4°C.
Immunodepletion of NELF and DSIF from nuclear extracts and evaluation of DRB sensitive transcription
Nuclear extracts were prepared from Drosophila embryos as described by Biggin and Tjian (22), but dialyzed at the final step to a conductivity that matched 0.15 M KCl/HEMG. To deplete DSIF or NELF, 500 μl of nuclear extract was incubated for 1.5 h with 100 μl of the appropriate antibody-sepharose. This step and all subsequent steps for the depletion were done at 4°C. Supernatants were collected through a Handee Spin Cup (Pierce) and then incubated with a fresh 100 μl portion of antibody-sepharose. While the second cycle of depletion was occurring, the first portion of antibody-sepharose was washed three times with 0.15 M KCl/ HEMG, two times with 0.1 M glycine, pH 2.5 and then three times with 0.15 M KCl/HEMG. Two more rounds of depletion were performed by alternating between regenerated batches of antibody-sepharose. Depleted extracts were stored at −80°C.
The DNA template used for in vitro transcription consisted of the Drosophila hsp70 promoter region spanning from −194 to +1 fused upstream of a 410 bp G-less cassette from p(C2AT) (23). Transcription reactions were performed by first preparing a 20 μl solution of 7.5 ng/μl supercoiled G-less DNA template, 27.5 mM HEPES, pH 7.6, 0.125 U/μl RNase T1, 45 mM KCl, 30 μM Na2EDTA, 3.75 mM MgCl2, 3% glycerol, 0.5 mM DTT and 6 μg/μl nuclear extract protein. The mixture was incubated at 25°C for 25 min to allow formation of preinitiation complexes. Transcription was started by adding a 10 μl solution of 1 mM ATP, 1 mM UTP, 0.1 mM CTP, 0.5 μCi/μl [32P]CTP (6000 Ci/mmol), 0.02 mM 3′ O-methyl GTP, 22.5 mM HEPES, pH 7.6, 75 mM KCl, 1.25 mM MgCl2, 0.5 mM DTT and 1% glycerol. When added, DRB was present in the 10 μl solution at 150 μM so that the final concentration in the 30 μl transcription reaction was 50 μM. Transcription was allowed to occur at 25°C for 20 min. Transcription was stopped by adding 120 μl of 20 mM EDTA, pH 8, 0.2 M NaCl, 1% SDS and 0.1 μg/μl yeast RNA. Samples were digested for 30 min with proteinase K followed by phenol/chloroform extraction. RNA was ethanol precipitated and analyzed on a 6% polyacrylamide gel containing 7 M urea.
Evaluation of promoter proximal pausing in vitro
Transcription reactions were performed with 10 μl of NELF-depleted or mock-depleted extracts in 40 μl reaction as described previously (24). The permanganate footprinting analysis was performed as described previously (24,25).
Chromatin immunoprecipitation (ChIP) analysis
ChIP analysis was performed as described by Park et al. (26). Cross-linked chromatin from 107 Drosophila S2 cells was immunoprecipitated with the following amounts of sera: 7 μl of NELF-B preimmune serum or antiserum, 7 μl of NELF-E preimmune serum or antiserum, 4 μl of DSIF preimmune serum or antiserum, 4 μl of GAGA factor preimmune serum or antiserum. DNA recovered from immunoprecipitates and from 10, 1 and 0.1% input DNA was analyzed by PCR as described previously (10). The following primers were used to amplify 5′ and 3′ regions of each gene: hsp70 −48F (AAAAGAGCGCCGGAGTATAAATAGA) and hsp70 +250R (GCAGGCATTGTGTGTGAGTTCT); hsp70 +1357F (CTGTGCAGGCCGCTATCC) and hsp70 +1730R (GGCCTTGCCCGTACTCATCTC); β1-tubulin −40F (ACTGCGGCCATCGTATAAAAG) and β1-tubulin +222R (TGGCAACTGTACTCTGGATGG); β1-tubulin +3890F (CCACCTTCATCGGCAACTC) and β1-tubulin +4104R (GAACTCGGCGTCCTCGTC).
RESULTS
Identification of the Drosophila NELF complex
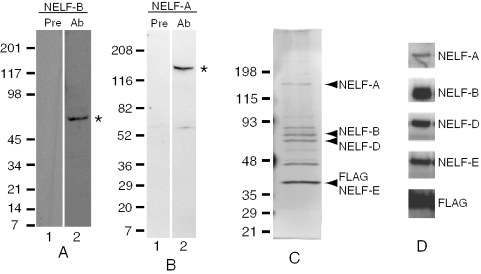
BLAST analyses identified Drosophila candidates for each of the four subunits of NELF. Antibodies against Drosophila NELF-D and NELF-E were previously shown to recognize proteins of the expected sizes in Drosophila nuclear extracts, and co-immunoprecipitation analyses showed that NELF-D and NELF-E associated with each other (10). cDNA clones putatively encoding Drosophila NELF-A and NELF-B were obtained from the Drosophila Genome Resource Center and portions encoding each protein were subcloned into bacterial expression vectors. Antibodies were raised against portions of NELF-A and NELF-B isolated from Escherichia coli. These antibodies were observed to react with proteins of sizes matching those predicted by the Drosophila genome (Figure 1A and B, asterisks). The size of Drosophila NELF-A is notable as the protein is approximately twice the size of the human ortholog. Nevertheless, this size is consistent with the current annotation of the Drosophila genome.
Figure 1.
Identification of subunits of the NELF complex. (A) Immunoblot analysis of Drosophila nuclear extract with preimmune antiserum (lane 1) and anti-NELF-B antiserum (lane 2). The polypeptide marked by an asterisk matches the predicted size of NELF-B. (B) Immunoblot analysis of Drosophila nuclear extract with preimmune antiserum (lane 1) and anti-NELF-A antiserum (lane 2). The polypeptide marked by an asterisk matches the predicted size of NELF-A. (C) SDS–PAGE analysis of immunoaffinity purified FLAG-NELF-E complex. Polypeptides were separated on a 4–15% polyacrylamide gradient gel and visualized by silver staining. NELF subunits designated on the right were identified based on co-migration with polypeptides detected on immunoblots with respective antibodies. (D) Immunoblot analysis of immunoaffinity purified NELF. Separate blots were probed with antiserum against different subunits of NELF or antibody against the FLAG-tag. Only the portion of each blot showing signals is presented. The variation in the molecular weight markers designated by the numbers flanking (A, B and C) are due to batch to batch variation in the calibration of pre-stained markers (BioRad).
To test if the four putative NELF subunits formed a complex, we generated a Drosophila cell line that expressed a FLAG-tagged version of NELF-E. Analysis of the immunoaffinity purified sample on SDS–PAGE stained with silver revealed prominent polypeptides corresponding to the predicted sizes of the four subunits of NELF (Figure 1C). The prominent polypeptide migrating at ∼48 kDa is also recovered from Drosophila cells that do not express FLAG-tagged NELF-E so it appears to be unrelated to NELF (data not shown). The identities of the remaining polypeptides have not been determined. Immunoblotting with antibodies against each subunit of NELF verified that the silver stained polypeptides comigrated with NELF subunits (Figure 1D).
Immunodepletion of NELF or DSIF diminishes the sensitivity of transcription in vitro to DRB
NELF and DSIF were originally isolated from human cell extracts because they were involved in rendering transcription reactions sensitive to DRB (1,3). To determine if Drosophila NELF and DSIF functioned like their human counterparts, we assessed whether depletion of either protein from a Drosophila nuclear extract would affect the sensitivity of transcription towards DRB. Transcription was performed with nuclear extracts from nonheat shocked embryos on a plasmid that contained the hsp70 promoter fused at +1 to a 410 bp G-less cassette.
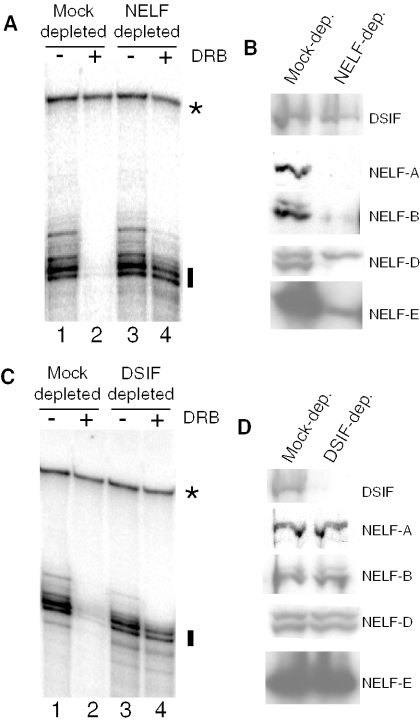
As shown in Figure 2A, immunodepletion of NELF from the Drosophila nuclear extract desensitized the transcription reaction to DRB. Fifty micromolar DRB markedly inhibited the level of transcription that occurred in extract subjected to a mock depletion with preimmune antibody (Figure 2A, compare lanes 1 and 2). In contrast, extract subjected to depletion by a mixture of antibodies against NELF-E and NELF-D was significantly less sensitive to DRB inhibition (Figure 2A, compare lanes 3 and 4). Immunoblots confirmed the specificity of the immunodepletion: the levels of all four subunits of NELF were significantly less in the NELF-depleted extract than the mock-depleted extract (Figure 2B). In contrast, comparable amounts of DSIF were present in both extracts.
Figure 2.
Immunodepletion of NELF or DSIF desensitizes transcription in vitro to DRB. (A) Mock-depleted (lanes 1 and 2) or NELF-depleted (lanes 3 and 4) extracts were tested for transcription in vitro in the presence or absence of DRB. NELF was depleted with a mixture of antibodies against NELF-D and NELF-E. Transcription was measured using an hsp70-G-less cassette substrate. The radiolabeled G-less transcription products are marked by a vertical line. The identity of the band marked with an asterisk is not known, but this band is evident even when transcription reactions are done in the presence of alpha amanitin (data not shown). It serves as a convenient control for RNA recovery. (B) Immunoblot analysis of proteins in mock-depleted and NELF-depleted extracts. (C) Mock-depleted (lanes 1 and 2) or DSIF-depleted (lanes 3 and 4) extracts were tested for transcription in vitro in the presence or absence of DRB. Antibody against the largest subunit of DSIF was used to deplete DSIF. (D) Immunoblot analysis of proteins in mock-depleted and DSIF-depleted extracts.
Interestingly, two polypeptides were detected on the western blot with antibody against NELF-D, but only the more rapidly migrating polypeptide was depleted with NELF antibodies (Figure 2B, NELF-D). Both polypeptides were detected with crude NELF-D antiserum or with affinity-purified NELF-D antibody (data not shown). More work is required to assess whether the polypeptide that is refractory to depletion in Figure 2B is a form of NELF-D that is independent of the rest of NELF or a cross-reacting polypeptide that is detected on the immunoblots. NELF-B also appeared as a doublet. In this case, both polypeptides were depleted suggesting that both are NELF-B and associated with the other subunits of NELF.
Immunodepletion of DSIF from the Drosophila nuclear extract also desensitized the transcription reaction to DRB (Figure 2C). Fifty micromolar DRB markedly inhibited the level of transcription that occurred in extract subjected to a mock depletion with preimmune antibody (Figure 2C, compare lanes 1 and 2). In contrast, extract subjected to depletion by DSIF antibody was significantly less sensitive to DRB inhibition (Figure 2C, compare lanes 3 and 4). Immunoblot analysis confirmed that the DSIF depletion had indeed removed DSIF but not the four subunits of NELF (Figure 2D).
Immunodepletion of NELF from a Drosophila nuclear extract impairs promoter proximal pausing
Previously, we determined that reducing the level of NELF-E using RNA interference impaired promoter proximal pausing on the hsp70 gene in salivary glands (10). It remained possible, however, that the depletion of NELF-E was affecting initiation rather than pausing. To address this possibility, we evaluated the impact of depleting NELF from the nuclear extract on promoter proximal pausing in vitro. Previously, we showed that promoter proximal pausing can be reconstituted in Drosophila nuclear extracts and that depletion of DSIF from the extract with antibody inhibited pausing but not initiation (10,25).
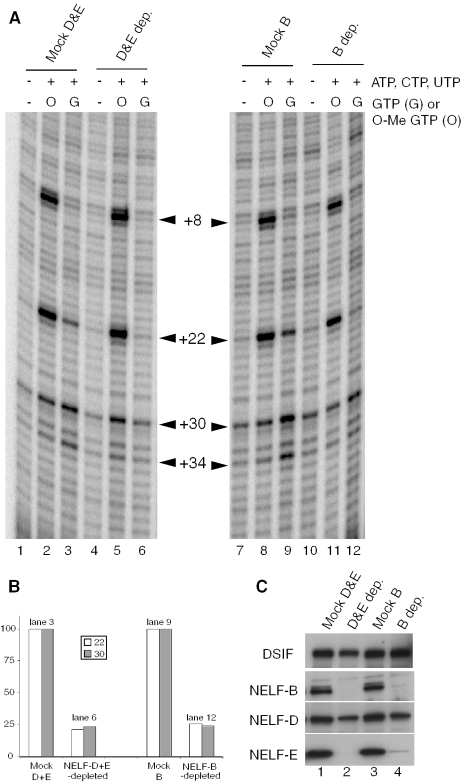
Depletion of NELF was achieved with a mixture of NELF-D and NELF-E antibodies or with NELF-B antibody alone. Antibodies from respective preimmune sera served as negative controls. After four successive incubations of extract with antibody conjugated to sepharose, immunoblots showed that NELF subunits were substantially depleted from the extract while DSIF was mostly unaffected (Figure 3C). Approximately half the protein detected with the NELF-D antibody on the immunoblot was refractory to depletion. This is in accord with the result in Figure 2B showing that there is a second protein that migrates closely to NELF-D but is not depleted by NELF antibodies.
Figure 3.
Immunodepletion of NELF impairs promoter proximal pausing in vitro. (A) Permanganate footprinting analysis of mock-depleted and NELF-depleted extracts. Transcription reactions were done in the absence of nucleotides (lanes 1, 4, 7 and 10), the presence of ATP, CTP, UTP and 3′ O-methyl GTP (lanes 2, 5, 8 and 11), or the presence of ATP, CTP, UTP and GTP (lanes 3, 6, 9, and 12). At the end of 30 min, Pol II was detected on the DNA by permanganate footprinting. Permanganate reactivity at +8 and +22 in lanes 2, 5, 8 and 11 was due to Pol II arresting at +15 following incorporation of the 3′ O-methyl GTP. Permanganate reactivity at +22, +30 and +34 in lanes 3 and 9 was due to Pol II stably paused in the promoter proximal region of hsp70. (B) Quantification of band intensities detected at +22 and +30 in lanes 3, 6, 9 and 12. Background intensities detected in lanes 1, 4, 7 and 10 have been subtracted. (C) Immunoblot analysis showing the specificity of the NELF depletions. Depleted extracts were run-on a 4–15% gradient gel and blotted to nitrocellulose. The largest subunit of DSIF and the indicated subunits of NELF were detected with antibodies.
Transcription reactions were performed in the presence of all four nucleotides with the NELF-depleted and mock-depleted extracts. After 30 min, the presence of Pol II on the promoters was monitored with permanganate (25). Permanganate reacts preferentially with thymines in single-stranded regions, and promoter proximal pausing on hsp70 causes thymines at +22, +30 and +34 on the nontranscribed strand to be hyper-reactive (25,27,28). Extracts depleted of NELF exhibited less permanganate reactivity at these positions than the mock-depleted extracts (Figure 3A, compare lanes 3 to 6 and lanes 9 to 12). Quantification of band intensities at +22 and +30 after subtracting background intensities in lanes 1, 4, 7 and 10 are provided in Figure 3B. The intensities of the +22 and +30 bands for the NELF-depleted samples were ∼4-fold less than the preimmune controls. Importantly, similar results were obtained when the depletions were done with two different NELF affinity resins—one composed of a mixture of NELF-D and NELF-E antibodies and the other composed of NELF-B antibodies.
To test if depletion of NELF impaired initiation, we did transcription reactions with 3′ O-methyl GTP instead of GTP. 3′ O-methyl GTP causes the Pol II to arrest at +15 where the first guanylate nucleotide is incorporated, and this arrest results in permanganate hyper-reactivity at +8 and +22 (10). Since initiation must occur for Pol II to reach +15, the intensities of the bands at +8 and +22 reflect the level of initiation. As shown in Figure 3A (lanes 2, 5, 8 and 11), permanganate analysis detects little difference in the reactivity of thymines at +8 and +22. Thus, depletion of NELF is affecting promoter proximal pausing but not initiation.
Unfortunately, we were unable to assess the fate of the Pol II that failed to pause as a consequence of depleting NELF. It may continue to elongate on the template or it might disengage. The efficiency of pausing in our extract is never complete. Consequently, read-through products are always generated even in the presence of NELF, so these obscure detection of read-through transcripts that might be generated as a consequence of NELF depletion.
ChIP analyses of hsp70 in nonheat shocked and heat shocked cells
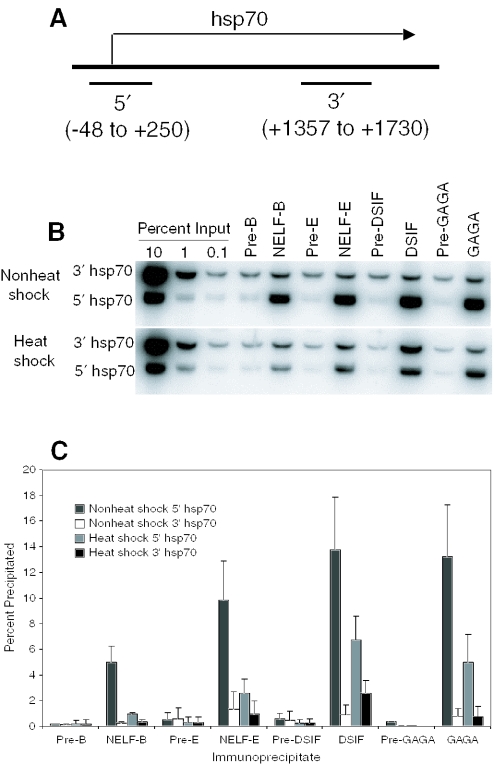
Previously, we observed that NELF-D and NELF-E cross-linked to the promoter region but not the 3′ region of the hsp70 gene in nonheat shocked cells (10). We were interested in determining if any changes in the association of NELF would be apparent during heat shock. Results in Figure 4 indicate that there was ∼5-fold less NELF-B and NELF-E cross-linking to the hsp70 promoter region in heat shocked cells than in nonheat shocked cells. The level of each NELF subunit cross-linking to the 3′ region of hsp70 was low and essentially the same for heat shocked and nonheat shocked cells. The residual cross-linking of NELF subunits to the promoter region in heat shocked cells appears to be specific because the amount of DNA recovered in NELF precipitates from the 5′ region exceeded the amount from the 3′ region or the amounts of either 5′ or 3′ regions recovered in the preimmune precipitates. Some evidence indicates that promoter proximal pausing persists transiently during heat shock induction (29), so this could account for the NELF detected on the hsp70 promoter region during heat shock.
Figure 4.
ChIP analysis of proteins associated with the hsp70 gene before and after heat shock. Cross-linked chromatin was prepared from nonheat shocked cells and cells that had been heat shocked for 15 min. The cross-linked chromatin was subjected to immunoprecipitation with antisera against NELF-B, NELF-E, DSIF or GAGA factor, or with corresponding preimmune sera. The immunoprecipitated DNA and the indicated amounts of input DNA were analyzed by PCR to determine the amount of DNA from the promoter region and the 3′ region cross-linked to each protein. (A) Schematic of the hsp70 gene showing regions that were amplified by PCR (not drawn to scale). (B) Representative results obtained from a ChIP analysis. (C) Quantification of results obtained from three independent ChIP analyses. Error bars represent standard deviations.
We also analyzed DSIF and GAGA factor as both have been implicated in promoter proximal pausing (10,35). As seen before, both proteins cross-link to the hsp70 promoter region but not the 3′ region prior to heat shock induction. The level of DSIF cross-linking to the promoter region decreased ∼2-fold following heat shock. In contrast, the level of DSIF cross-linking to the 3′ region of hsp70 increased ∼2-fold following heat shock. These results are in good agreement with rapid cross-linking results reported by Saunders et al. (30). The level of GAGA factor cross-linking to the promoter region also diminished following heat shock, but the level of cross-linking to the 3′ region remained low and similar to the level seen in nonheat shocked cells. The apparent absence of GAGA factor from the downstream region of hsp70 in heat shocked cells is not in agreement with previously reported UV cross-linking experiments (31) but is in agreement with recent immunofluorescence data showing that GAGA factor does not colocalize with elongation complexes on polytene chromosomes (16). The heat shock-dependent reduction in GAGA factor cross-linking to the promoter region could be due to displacement by HSF, whose binding sites are interdigitated and overlapping with GAGA factor binding sites in the promoter region (32).
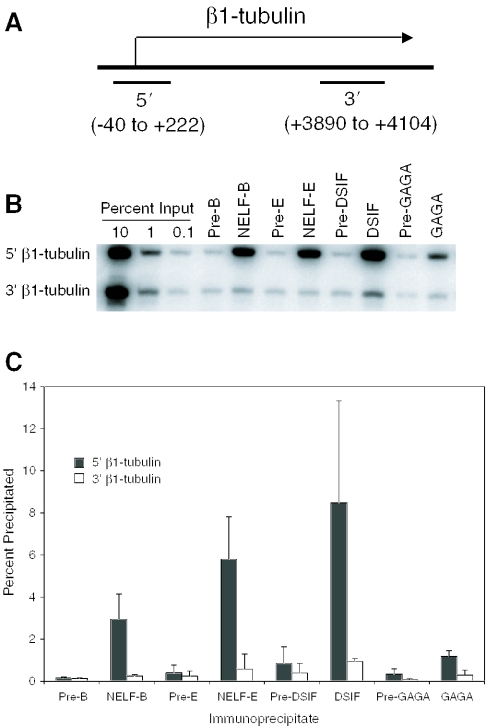
ChIP analysis of the β1-tubulin gene
Previous analyses detected paused Pol II on the promoter proximal region of the β1-tubulin gene in Drosophila cells (27,33), so we tested whether NELF and DSIF were present at this gene. ChIP analysis revealed that both NELF and DSIF were present in the promoter region but not the 3′ region (Figure 5). Interestingly, significantly less GAGA factor was detected on the β1-tubulin promoter than on the hsp70 promoter. This is consistent with visual inspection of the sequences constituting these two promoters (data not shown)—multiple GAGA elements are evident in hsp70 but only two short elements are evident in the region from +1 to −250 of β1-tubulin. Nevertheless, the signal for GAGA factor on the β1-tubulin promoter region is above the background shown by the preimmune control. GAGA factor might be involved in setting up the paused Pol II on β1-tubulin, since GAGA factor is involved in setting up paused Pol II on hsp70 (34,35).
Figure 5.
ChIP analysis of proteins associated with the β1-tubulin gene in nonheat shocked cells. Cross-linked chromatin from nonheat shocked cells was subjected to immunoprecipitation with antisera against NELF-B, NELF-E, DSIF or GAGA factor and with corresponding preimmune sera. (A) Schematic of β1-tubulin gene showing regions that were amplified by PCR (not drawn to scale). (B) Representative results obtained from a ChIP analysis. (C) Quantification of results obtained from three independent ChIP analyses.
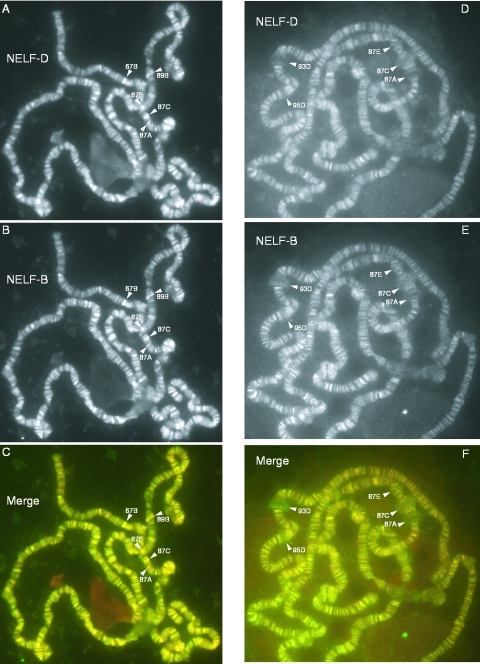
Immunofluorescence analysis of NELF-B and NELF-D on polytene chromosomes
Immunofluorescence microscopy was used to compare the distributions of NELF-B and NELF-D on polytene chromosomes. Previously, we observed that antibody against NELF-D stained hundreds of loci on polytene chromosomes (10). Figure 6A–C show that antibody against NELF-B stains hundreds of loci and that there is extensive overlap in the distributions of NELF-B and NELF-D on polytene chromosomes. Both proteins are concentrated in regions known as interbands, which are regions of decondensed chromatin. Control experiments showed that the preimmune serum for each antibody did not stain the chromosomes. Moreover, preincubation of either antibody with its respective antigen but not with an unrelated protein blocked staining of the chromosomes.
Figure 6.
Immunofluorescence analyses of NELF-D and NELF-B on polytene chromosomes derived from nonheat shocked and heat shocked larvae. Images in (A, B and C) are of chromosomes from nonheat shocked larvae. Images in (D, E and F) are of chromosomes from heat shocked larvae. NELF-D was detected with affinity-purified antibody made in rabbit. NELF-B was detected with antiserum made in guinea pig. NELF-D antibody on the chromosomes was detected with Alexa 568-tagged secondary, giving red fluorescence [converted to grayscale in (A and D)]. NELF-B antibody bound to the chromosomes was detected with Alexa 488-tagged secondary, giving green fluorescence (converted to grayscale in panels B and E). Merged images for the two antibodies are shown in (C and F).
We were particularly interested to see staining at 67B, 87A, 87C and 87E because all of these locations are known to harbor Pol II paused in the promoter proximal regions of particular genes (27,36). 67B contains several small heat shock genes, and 87A and 87C contain copies of the hsp70 heat shock gene. 87E contains an hsp70 transgenic promoter previously shown to interact with NELF-D and to have paused Pol II (10). A fly line lacking the transgene at 87E showed reduced staining of 87E with antibodies against NELF-B or NELF-D (data not shown).
The NELF-D antiserum detected two polypeptides in Drosophila nuclear extracts raising concern that immunofluorescence could be due to a polypeptide that was not part of the NELF complex (Figure 2C). This is unlikely for two reasons. First, the immunodepletion results in Figure 2B are consistent with the majority of NELF-B being associated with NELF-D. Second, the slowly migrating polypeptide that appears to be independent of NELF seems to be unable to associate with the NELF-D antibody in its native state, since it fails to be immunodepleted with NELF-D antibody (Figure 2B).
Figure 6D–F show the distributions of NELF-B and NELF-D on polytene chromosomes from heat shocked larvae. As was the case for the nonheat shocked samples, there is extensive overlap in the distributions of NELF-B and NELF-D. In accord with our previous observation (10), staining of the puffs with antibody against NELF-D was low. Heat shock induced genes that cause puffs in the specimen shown in Figure 6 reside at 87A, 87C, 87E, 93D and 95D. Although staining of puffs with antibody against NELF-B was slightly elevated relative to NELF-D, the level of staining was markedly less than the level of staining observed for DSIF at puffs (10) (data not shown).
We have also stained polytene chromosomes with antibodies against NELF-A and NELF-E (data not shown). There was considerable overlap in the staining patterns with NELF-B and NELF-D at interbands. As was the case with NELF-B antibody, the staining of puffs with NELF-A and NELF-E antibodies were variable and much less intense than the staining observed with DSIF antibody. More analysis is required to determine the significance of the weak staining at puffs as this staining could be due to nonspecific binding by the NELF antibodies.
DISCUSSION
Comparison of human and Drosophila NELF
Using immunoaffinity techniques, we have determined that Drosophila NELF contains four polypeptides with similarity to the four subunits of human NELF. All four polypeptides copurify with a FLAG-tagged version of NELF-E expressed in Drosophila cells. Moreover, immunodepletions done with antibodies against subsets of NELF subunits result in the removal of other subunits. The amino acid sequences of the four Drosophila polypeptides have significant similarity to their human counterparts indicating that they might have similar structures. In addition, Drosophila and human NELF appear to be functionally similar because depletion of NELF from human (1,3) and Drosophila nuclear extracts desensitizes transcription to DRB.
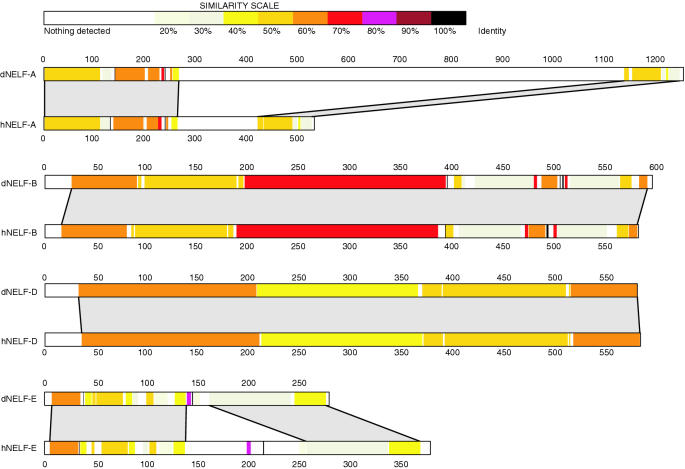
We aligned the amino acid sequences of the various NELF subunits to learn more about NELF (Figure 7). The sequences of NELF-B and NELF-D orthologs exhibit several regions with >50% identity distributed over their entire lengths. Analysis of subunit interactions lead Narita et al. (18) to propose that NELF-B and NELF-D form a central core that brings together NELF-A and NELF-E. NELF-A associates with NELF-D while NELF-E associates with NELF-B. The high degree of homology between NELF-B and NELF-D orthologs is consistent with constraints that might be dictated by the requirements that these two proteins interact with each other and with other subunits of NELF. The extensive colocalization of NELF-B and NELF-D on polytene chromosomes is also consistent with these two subunits forming the core of the NELF complex.
Figure 7.
Schematic representation of amino acid sequence alignments between human and Drosophila subunits of NELF. Regions of homology between Drosophila and human NELF subunits were determined using LFasta and illustrated using LalnView. The nonhomologous region of ∼800 amino acids in dNELF-A spans from approximately amino acids 300 to 1100. The RNA recognition motif in dNELF-E corresponds to amino acids 167–232.
When NELF-A was initially identified through a BLAST search, we were concerned about the genome annotation because dNELF-A was over twice the size of human NELF-A. Our immunoblot analysis verifies that dNELF-A is approximately twice the size of hNELF-A. The alignment in Figure 7 shows that the size difference is due to a large nonconserved region located approximately between amino acids 300 and 1100. Inspection of the amino acid sequence in this region of Drosophila NELF-A reveals numerous tracts of poly-glutamine, poly-asparigine, poly-threonine and poly-alanine that are absent from hNELF-A. This lack of conservation raises the possibility that this region serves as a linker between two distinct functional domains defined by the regions of homology spanning the first 250 amino acids and the last 100 amino acids of NELF-A. The N-terminal region of homology in hNELF-A has been shown to interact with Pol II and NELF-D (37). Moreover, this region is required for NELF-mediated repression in vitro (18). Deletion of amino acids 431–528 of hNELF-A, which corresponds to the last 100 amino acids of dNELF-A, does not impair the ability of the NELF complex to inhibit transcription in vitro (18). Future analysis using genetic and molecular genetic approaches in Drosophila may help elucidate the function of the large nonconserved region and the C-terminal domain.
hNELF-E binds RNA, and the region responsible for this activity exhibits 30% identity to amino acids 167–232 in dNELF-E. This RNA recognition motif has been shown to be required for NELF inhibition in human extracts (18). Two features found in hNELF-E are notably absent in dNELF-E. A 46 amino acid stretch spanning from 196 to 242 in hNELF-E that is composed primarily of alternating arginine and aspartic acid residues is absent—the function of this region in hNELF-E is not known. Also absent from dNELF-E are serines that correspond to amino acids 181, 185, 187 and 191 in hNELF-E. These serines in hNELF-E are phosphorylated in vitro by P-TEFb, and these modifications reduce the binding between hNELF-E and RNA (15). It will be interesting to learn if phosphorylation of dNELF-E regulates its RNA binding activity.
NELF is involved in promoter proximal pausing
Previously, we provided two results that implicated NELF in promoter proximal pausing on the hsp70 heat shock gene (10). Depleting salivary glands of NELF using RNA interference diminished the level of promoter proximal pausing occurring on hsp70 in salivary glands. Also, NELF was found to cross-link to the hsp70 promoter region prior to heat shock induction. Here, we provide additional data strengthening the conclusion that NELF is involved in promoter proximal pausing.
Immunodepletion of NELF from nuclear extracts impaired formation of the paused Pol II. Importantly, the cell-free transcription reactions with 3′ O-methyl GTP indicated that NELF does not contribute to steps in the transcription process encompassing initiation and elongation to +15. We have also determined that NELF is associated with the β1-tubulin promoter. Genomic footprinting with permanganate and nuclear run-on analysis indicate that promoter proximal pausing occurs on the β1-tubulin gene (27,36).
Changes in NELF and DSIF associations at hsp70 during heat shock induction
Based on strong immunofluorescence staining detected for DSIF and the weak staining for NELF at heat shock puffs, we proposed that NELF dissociates from elongation complexes during activation but DSIF remains associated with the elongation complexes (10). Our observation that the level of NELF cross-linking to the hsp70 promoter region after heat shock is ∼5-fold less than before heat shock is consistent with this model. However, we also observed a 2-fold decrease in the level DSIF cross-linking to the hsp70 promoter region after heat shock, in agreement with a recent report from Saunders et al. (30). The decrease in DSIF cross-linking near the promoter was unexpected since heat shock puffs on polytene chromosomes stain very strongly with DSIF antibody. However, much of the signal detected on the polytene chromosomes probably originates from DSIF associated with elongation complexes in the body of the gene. In addition, the interaction of DSIF with Pol II in the promoter proximal region might be transient or not occur at all on some elongation complexes in the promoter proximal region. The rate of reinitiation at the hsp70 promoter is exceptionally high, once every few seconds, so a significant portion of the Pol II molecules might escape into the body of the gene before associating with DSIF.
NELF and its role in gene expression
The broad distribution of NELF-B and NELF-D on polytene chromosomes could mean that hundreds of genes harbor paused Pol II similar to that found on hsp70. Alternatively, NELF could have additional functions. mRNA capping is coupled to transcription (38), and recent results lead to the hypothesis that pausing induced by NELF might be important for coupling transcription and mRNA capping (17). A subset of genes like hsp70 and β1-tubulin might use cofactors such as GAGA factor to generate a stably paused state.
Acknowledgments
We thank Maria Horvat-Gordon for preliminary evaluation of antibodies on polytene chromosomes. This work was supported by Grant GM47477 from the National Institutes of Health to D.S.G. and by a Grant for Research and Development Projects in Cooperation with Academic Institutions from the New Energy and Industrial Technology Development Organization to H.H. Funding to pay the Open Access publication charges for this article was provided by grant GM47477 from the National Institutes of Health.
REFERENCES
- 1.Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wada T., Takagi T., Yamaguchi Y., Watanabe D., Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi Y., Inukai N., Narita T., Wada T., Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi Y., Narita T., Inukai N., Wada T., Handa H. SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem. 2001;129:185–191. doi: 10.1093/oxfordjournals.jbchem.a002842. [DOI] [PubMed] [Google Scholar]
- 6.Lis J.T., Mason P., Peng J., Price D.H., Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 7.Kanazawa S., Okamoto T., Peterlin B.M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 8.Barboric M., Nissen R.M., Kanazawa S., Jabrane-Ferrat N., Peterlin B.M. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 9.Eberhardy S.R., Farnham P.J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 10.Wu C.H., Yamaguchi Y., Benjamin L.R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehm A.K., Saunders A., Werner J., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aiyar S.E., Sun J.L., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H., et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renner D.B., Yamaguchi Y., Wada T., Handa H., Price D.H. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 14.Ivanov D., Kwak Y.T., Guo J., Gaynor R.B. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujinaga K., Irwin D., Huang Y., Taube R., Kurosu T., Peterlin B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- 17.Mandal S.S., Chu C., Wada T., Handa H., Shatkin A.J., Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl Acad. Sci. USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D.K., Hasegawa J., Omori M., Inukai N., et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thummel C.S., Boulet A.M., Lipshitz H.D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E., Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor NY: Cold Spring Harbor Press; 1999. [Google Scholar]
- 21.Zhang Z., Wu C.H., Gilmour D.S. Analysis of Pol II elongation complexes by native gel electrophoresis: evidence for a novel CTD-mediated termination mechanism. J. Biol. Chem. 2004;279:23223–23228. doi: 10.1074/jbc.M402956200. [DOI] [PubMed] [Google Scholar]
- 22.Biggin M.D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 23.Sawadogo M., Roeder R.G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc. Natl Acad. Sci. USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamin L.R., Gilmour D.S. Nucleosomes are not necessary for promoter-proximal pausing in vitro on the Drosophila hsp70 promoter. Nucleic Acids Res. 1998;26:1051–1055. doi: 10.1093/nar/26.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B., Weber J.A., Chen Y., Greenleaf A.L., Gilmour D.S. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol. Cell. Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J.M., Werner J., Kim J.M., Lis J.T., Kim Y.J. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 27.Giardina C., Perez-Riba M., Lis J.T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 28.Weber J.A., Taxman D.J., Lu Q., Gilmour D.S. Molecular architecture of the hsp70 promoter after deleting the TATA box or the upstream regulatory region. Mol. Cell. Biol. 1997;17:3799–3808. doi: 10.1128/mcb.17.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien T., Lis J.T. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol. Cell. Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien T., Wilkins R.C., Giardina C., Lis J.T. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 1995;9:1098–1110. doi: 10.1101/gad.9.9.1098. [DOI] [PubMed] [Google Scholar]
- 32.Gilmour D.S., Thomas G.H., Elgin S.C.R. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science. 1989;245:1487–1490. doi: 10.1126/science.2781290. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen E.B., Lis J.T. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J. Mol. Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 34.Shopland L.S., Hirayoshi K., Fernandes M., Lis J.T. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 35.Lee H., Kraus K.W., Wolfner M.F., Lis J.T. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- 36.Rougvie A.E., Lis J.T. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi Y., Filipovska J., Yano K., Furuya A., Inukai N., Narita T., Wada T., Sugimoto S., Konarska M.M., Handa H. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science. 2001;293:124–127. doi: 10.1126/science.1057925. [DOI] [PubMed] [Google Scholar]
- 38.Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]