Abstract
The metabolic SearcH And Reconstruction Kit (metaSHARK) is a new fully automated software package for the detection of enzyme-encoding genes within unannotated genome data and their visualization in the context of the surrounding metabolic network. The gene detection package (SHARKhunt) runs on a Linux system and requires only a set of raw DNA sequences (genomic, expressed sequence tag and/or genome survey sequence) as input. Its output may be uploaded to our web-based visualization tool (SHARKview) for exploring and comparing data from different organisms. We first demonstrate the utility of the software by comparing its results for the raw Plasmodium falciparum genome with the manual annotations available at the PlasmoDB and PlasmoCyc websites. We then apply SHARKhunt to the unannotated genome sequences of the coccidian parasite Eimeria tenella and observe that, at an E-value cut-off of 10−20, our software makes 142 additional assertions of enzymatic function compared with a recent annotation package working with translated open reading frame sequences. The ability of the software to cope with low levels of sequence coverage is investigated by analyzing assemblies of the E.tenella genome at estimated coverages from 0.5× to 7.5×. Lastly, as an example of how metaSHARK can be used to evaluate the genomic evidence for specific metabolic pathways, we present a study of coenzyme A biosynthesis in P.falciparum and E.tenella.
INTRODUCTION
The genomics revolution continues to produce biological sequence data at an ever-increasing rate. At the time of writing, the GOLD database (http://www.genomesonline.org) lists 233 published and 1000 ongoing genome projects. However, the completion of genome sequencing only marks the beginning of a long and difficult process of deriving biological knowledge from these data. The huge task of ‘genome annotation’ includes both the identification of the set of coding regions in the genomic DNA and the assignment of function to those genes.
While there exist many computer packages to aid in this work, the complete annotation of a genome still requires a tremendous amount of effort from a dedicated team of professionals, and hence a very large investment of time and money. The sheer number of ongoing genome projects and their range of scales and budgets indicate that there is an urgent need for fully automated tools, both to generate preliminary annotations and to derive new knowledge as the genome sequencing is completed. Organisms that might benefit include numerous fungi of environmental, agricultural and medical importance, parasites of medical and veterinary importance in developing countries, such as trypanosomes and worms, and members of the kingdom Stramenopiles (or Chromista), including diatoms and important plant pathogens.
One such use of genome data is in the preparation of a preliminary annotation of enzyme-encoding genes, and hence the elucidation of the network of reactions they catalyse within an organism (1,2). Differences in metabolism between host and pathogen species may highlight novel drug targets to feed into the drug development pipeline (3). New methods for the qualitative analysis of metabolic networks, such as elementary mode analysis (4) or the closely related extreme pathway analysis (5), can reveal novel pathways and hence new insights into metabolic diseases. Enzyme annotation is a good candidate for automation owing to the large amount of knowledge that has been assembled in metabolic databases, such as KEGG (6), BioCyc (7) and BRENDA (8). These databases include references to protein sequence data from known enzymes in a wide variety of organisms, which can be used as models in searches of newly sequenced genomes.
Existing semi-automated enzyme annotation software (2,9–11) starts with a set of predicted proteins from an annotated genome and, by a variety of text mining and/or sequence comparison methods, constructs a list of the enzymatic functions that are asserted to be present. Our software (SHARKhunt) differs in that it requires only a set of DNA sequences [finished chromosomes, contigs, genome survey sequences or expressed sequence tags (ESTs)] as input, and hence can be applied to extract new knowledge of metabolic capabilities from preliminary data produced by unannotated and ongoing genome sequencing projects. Models derived from sets of known enzymes are used to search through the DNA sequences to find regions with significant similarity to the model sequences. The confidence of each functional assertion is measured by an E-value score, and the full set of predictions is output in various formats for consultation, human curation or further automated network analysis. Results may be uploaded to an online visualization tool (SHARKview), which permits users to browse freely around the KEGG metabolic network, run BLAST (12) searches on their predicted gene sequences and compare data from different organisms or different sources of annotation.
MATERIALS AND METHODS
Preparation of data
The SHARKhunt search protocol described below requires both a set of PSI-BLAST (12) polypeptide profiles and a set of associated HMMER profile hidden Markov models (HMMs) (13). We use the set of PRIAM PSI-BLAST profiles (11) (July 2004 version) and the protein sequence data from which they were derived as the basis for our profile HMMs. This set constitutes 2562 PSI-BLAST profiles covering a total of 1967 enzymatic functions, defined by Enzyme Commission (EC) number. There are more profiles than functions in the PRIAM data because some functions are represented by more than one homologous family of proteins. For each PRIAM profile containing more than one sequence, a HMM is generated by taking the original set of protein sequences constituting the profile, constructing a multiple alignment using MUSCLE (14) and passing this to the HMMbuild program (part of the HMMER package, available from http://hmmer.wustl.edu).
The SHARKhunt search protocol
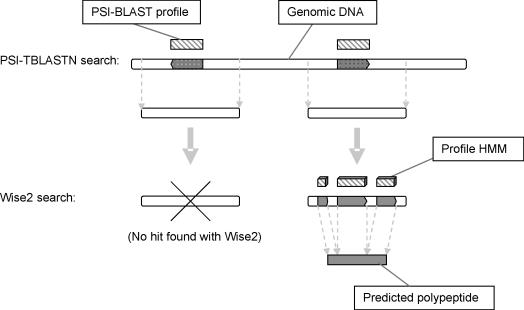
The search method used in SHARKhunt is an automated version of a protocol that we have previously demonstrated to be effective in searching genomic DNA for distant homologues of a set of model sequences (15). The Wise2 package (16) is used to aligning the profile HMMs with genomic DNA and produce predicted polypeptide sequences for the resulting hypothetical gene fragments. Although the Wise2 algorithm is very powerful, it is too slow to be used to search through a whole genome. Hence, we apply a preliminary filtering step, where PSI-TBLASTN (12) is used to search the genome for regions with some similarity to each original PRIAM profile (hits with E-value < 1.0). These regions are then extracted from the genome and passed to Wise2 for further analysis with the appropriate HMM (Figure 1). Any resulting Wise2 hits are assessed by using PSI-BLAST to compare the predicted polypeptide translation with the original PRIAM profile, and the E-values and locations of predicted coding regions are output.
Figure 1.
The SHARKhunt search protocol is a two-stage, profile-based method that aims at detecting all regions of a genome homologous to an enzyme model. A preliminary PSI-TBLASTN search (12) identifies regions with some similarity to a PRIAM profile. These regions are extracted from the genome and passed to the Wise2 program (16) to attempt a homology-based gene structure prediction. If Wise2 detects sufficient similarity between the model HMM and the DNA region, an intron/exon structure will be predicted and the resulting polypeptide sequence output. This sequence is then aligned with the original PRIAM profile to obtain an E-value score for each hit found.
In cases where several different profiles produce hits to the same region of DNA, we take the function of that region to be the same as the best hit (i.e. the lowest E-value). Some enzymes require more than one functional unit in order to operate, or are represented by more than one homologous family of proteins. This has already been taken into account in the generation of the PRIAM profiles and their associated logical AND/OR rules (11). To assert the presence of an enzymatic function in the genome, the hits must agree with the relevant PRIAM rule.
The SHARKhunt package (including the programs MUSCLE, HMMer, PSI-BLAST and Wise2 and the necessary PRIAM profile data) is available to download from the metabolic SearcH And Reconstruction Kit (metaSHARK) website (http://bioinformatics.leeds.ac.uk/shark/). The package comprises two executable scripts:
SHARKhunt invokes a Java program to run the profile searches. The output files created by this program include a FASTA format library of predicted polypeptide sequences, a GFF (gene feature format, see http://www.sanger.ac.uk/Software/formats/GFF/) file containing the locations of the predicted gene structures on the DNA sequence, a list of EC numbers representing the functions detected within the input DNA, together with their PSI-BLAST E-value confidence scores, and an eXtensible Markup Language (XML) file, which may be uploaded to the metaSHARK website (see above URL) for easy browsing and visualization of the results.
SHARKmodel allows users to create their own PSI-BLAST and HMMer profile models from sets of homologous polypeptide sequences for customized searches using the SHARKhunt protocol.
Verification of search method
To confirm that the search protocol is effective in detecting enzyme-encoding genes within eukaryotic DNA, SHARKhunt was run on the genome of the human malaria parasite, Plasmodium falciparum (17). The profile data used for this search were a special ‘jacknife’ dataset, prepared from the original PRIAM models by removing all sequences from Plasmodium spp. and re-making the affected profiles (38 out of 2562 profiles).
The results were compared with the enzymatic functions annotated for this organism in the initial genome publication (17) (data available at the PlasmoDB website: http://www.plasmodb.org/) and the subsequent re-annotation of metabolic enzymes prepared by the PlasmoCyc team (18) (http://plasmocyc.stanford.edu/).
As a comparison with an existing method based on polypeptide sequence input, searches were also run against P.falciparum with the PRIAM software (11), using the same set of jacknife PSI-BLAST profiles and all predicted polypeptides from the original genome annotation.
Analysis of preliminary genome sequences
As a practical example of how metaSHARK may be used to investigate metabolic pathways within unfinished genomes, we applied the software to the available genomic DNA sequences of another apicomplexan parasite, the coccidian Eimeria tenella (http://www.sanger.ac.uk/Projects/E_tenella/).
A full description of the E.tenella genome project has been given elsewhere (19). Briefly, more than 855 000 reads with an average read length of 525 bp have yielded ∼45 Mb of unique sequence for a coverage of ∼7.5×. The data are presently assembled into 8718 contigs and the G+C content of the genome is ∼53%. Little annotation has yet been undertaken on this genome, and ab initio genefinding is hampered by the current lack of well-characterized gene structures in Eimeria spp. As such, it presents us with a good example of an organism for which the metaSHARK system may be of use.
For comparison with SHARKhunt, we ran the PRIAM software on all E.tenella open reading frames (ORFs) of 50 amino acids or longer. In addition, the best available ab initio gene predictions for this species were obtained using the genefinding program TwinScan2 (20), trained on a set of 350 human-annotated genes from the related coccidian, Toxoplasma gondii (Aaron J. Mackey, personal communication). The polypeptide translations of these gene predictions were used in a PRIAM analysis, in parallel with the genomic ORF data. The PRIAM software was also run on all ORFs of at least 50 amino acids found within the clustered (98% similarity) EST+ORESTES data available for E.tenella (downloadable from the Sanger Institute website, see http://www.sanger.ac.uk/Projects/E_tenella/). These clusters were obtained by using publicly available ESTs from the NCBI and open reading frame-expressed sequence tags (ORESTES) cDNA reads generated by Alda M. Madeira and Arthur Gruber at the University of Sao Paulo, Brazil (unpublished data). ORESTES reads are cDNA fragments synthesized by a low stringency RT–PCR process using arbitrary 18–25mer primers (21).
Effect of variation in genome coverage on performance
To investigate how the number of enzymatic functions predicted by SHARKhunt is affected by variations in the coverage of the input genome sequence, we re-ran the E.tenella searches using an earlier genome assembly based on an ∼4.3× genome coverage (from December 2002), and also on an assembly constructed using the program Phrap (http://www.phrap.org/) from sets of reads representing ∼0.5× coverage (all data obtained from the Sanger Institute website).
RESULTS AND DISCUSSION
Verification of search method
Owing to its high A+T content and phylogenetic distance from any well-studied model species, it is particularly difficult to establish a gold standard for the full set of enzymes encoded by P.falciparum. However, there has been a substantial recent effort by Yeh et al. (18) working on the PlasmoCyc project to catalogue the enzymatic reactions believed to operate in this parasite, and to provide an extensive re-annotation of the genome sequence to identify as many of the enzymes implicated as possible. We chose to take as our standard the set of all EC-labelled reactions within the PlasmoCyc database for which a PRIAM profile was available (henceforth referred to as PlasmoCyc-RXN). Eliminating enzymes not contained in PRIAM gives a fair test of the capabilities of both PRIAM and SHARKhunt, since neither system is able to detect an enzyme for which it has no model. It should be noted that this standard set of EC numbers includes both the enzymes annotated by PlasmoCyc (the set PlasmoCyc-ANN) and all enzymatic functions that are assumed to be present based on our current knowledge of Plasmodium biology, but that have not yet been identified within the genome. This choice of standard has been made so that predictions that are in agreement with known biology are treated as true positive even if they do not yet have gene annotations in PlasmoCyc. Although this avoids unfairly penalizing the search methods tested, it also means that a large number of false-negative predictions are likely to be made, and hence that the sensitivity scores quoted will appear lower than is usual. Nevertheless, the choice of standard does allow a good comparison between the search methods used and with the original genome annotation.
The numbers of true and false positives predicted by SHARKhunt and PRIAM, working from the P.falciparum genomic DNA sequence and annotated polypeptide sequences, respectively, are shown in Table 1 for the three E-value cut-offs 10−30, 10−20 and 10−10 (see Table legend for definitions of true and false-positive predictions). These E-value scores are calculated in the same way for both programs, by using the PSI-BLAST software to measure similarity between a polypeptide and a PRIAM profile, and hence they offer a directly comparable measure of performance. In addition, we have included the corresponding numbers of true and false-positive predictions derived from the original P.falciparum manual annotation (17) (discarding EC numbers not found in PRIAM—we label this set PlasmoDB-ANN), as compared with PlasmoCyc-RXN. The SHARKhunt program performs almost as well in this comparison as the PRIAM automated enzyme annotation software, which is working with the set of annotated polypeptides taken from PlasmoDB. This indicates that the presence of introns in many P.falciparum genes does not adversely affect the SHARKhunt predictions made for this genome. Both programs make approximately the same numbers of predictions at the three E-value cut-offs taken, although the specificity is slightly higher for PRIAM than SHARKhunt at more relaxed cut-offs (2% higher for E-value < 10−10). At an E-value cut-off of 10−10, both programs are able to detect significant numbers of true positives that were missed in the original human annotation, achieving ∼54% sensitivity as compared with ∼46% for PlasmoDB-ANN.
Table 1.
Performance of SHARKhunt and PRIAM for P.falciparum, at varying E-value cut-offs
| E-value cut-off | Total predictions | TP | FP | FN | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|
| SHARKhunt (on genomic DNA) | 10−30 | 235 | 198 | 37 | 265 | 42.8 | 84.3 |
| 10−20 | 273 | 223 | 50 | 240 | 48.2 | 81.7 | |
| 10−10 | 333 | 247 | 86 | 216 | 53.3 | 74.2 | |
| PRIAM (on annotated polypeptides) | 10−30 | 229 | 193 | 36 | 270 | 41.7 | 84.3 |
| 10−20 | 269 | 222 | 47 | 241 | 47.9 | 82.5 | |
| 10−10 | 328 | 250 | 78 | 213 | 54.0 | 76.2 | |
| PlasmoDB-ANN (original human annotation) | n/a | 232 | 214 | 18 | 249 | 46.2 | 92.2 |
| PlasmoCyc-ANN (human reannotation) | n/a | 304 | 304 | n/a | 159 | 65.7 | n/a |
Note: PlasmoDB-ANN, the set of EC numbers assigned in the original P.falciparum genome annotation (17) for which PRIAM (11) profiles were available; PlasmoCyc-RXN, the set of all EC-labelled reactions within the PlasmoCyc database (18) for which a PRIAM profile was available; PlasmoCyc-ANN, the subset of PlasmoCyc-RXN for which enzymes have been annotated by PlasmoCyc curators; TP, number of true positives (predictions that were also contained in PlasmoCyc-RXN); FP, number of false positives (predictions that were not in PlasmoCyc-RXN); and FN, number of false negatives (PlasmoCyc-RXN functions that were not detected).
This test demonstrates that our software can detect enzyme-encoding genes with an accuracy equivalent to that of PRIAM, but with the advantage that it may be applied to any genome, regardless of whether a reliable set of predicted polypeptides is available. PRIAM runs many times faster than SHARKhunt (just over 1 h on a 1.8 GHz Linux node for this genome, compared with over 100 h for SHARKhunt on the same machine), but requires a good annotation of gene structures to produce reliable results.
Analysis of preliminary genome sequences
The coccidian parasite E.tenella is known to have large numbers of introns in its genes. This can make homology searches difficult in the absence of an annotated genome: protein versus DNA searches, such as TBLASTN (12), may fail owing to sizeable breaks in coding DNA, and protein versus protein searches [e.g. BLASTP (12)] applied to ORFs may also give disappointing results, since a gene with several introns may be split over many short ORFs. These factors make the E.tenella genome an interesting test case for SHARKhunt.
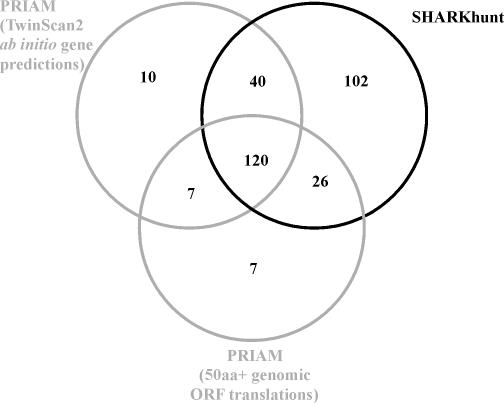
Since no annotation is available for this genome, we do not have a standard against which to compare the search results and so cannot obtain reliable numbers of true- and false-positive predictions. We can, however, compare the predictions made by PRIAM from the sets of genomic ORFs and ab initio gene predictions with the SHARKhunt results from the analysis of the genomic DNA sequences, under the assumption that the false-positive rate will remain relatively similar to that measured in the P.falciparum analysis for both programs. The enzymatic functions asserted by each of these three methods at an E-value cut-off of 10−20 are shown in the form of a Venn diagram in Figure 2. PRIAM predicted 177 functions from the ab initio genes and 160 from the ORF data, whereas metaSHARK predicted a total of 288 functions, covering the majority (89%) of PRIAM predictions and including an additional 102 that were not detected by PRIAM in either polypeptide dataset derived from the genomic DNA. It is notable that the ab initio gene predictions made using T.gondii training data were only slightly more informative for enzymatic function than the set of ORFs of length 50 amino acids or more, demonstrating that genefinding software trained on a species related to the target genome may not always produce a comprehensive set of predictions.
Figure 2.
Comparison of asserted enzymatic functions in E.tenella obtained by running PRIAM on the ab initio gene predictions generated by TwinScan2 (20) trained on T.gondii gene models and on all genomic ORFs over 50 amino acids, and by running SHARKhunt on the full set of genomic DNA contigs. An E-value cut-off of 10−20 was used for each method. Note that the SHARKhunt analysis predicts 102 functions that were missed in both PRIAM analyses.
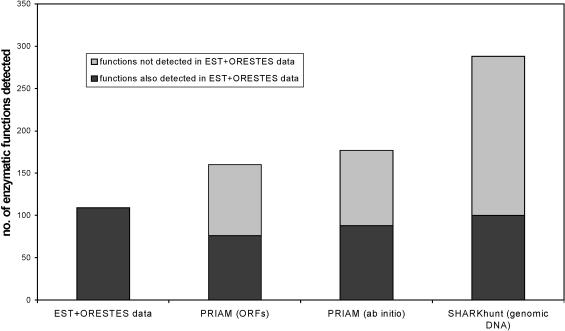
Figure 3 compares the predictions made by PRIAM from the EST+ORESTES ORFs with those made by PRIAM and SHARKhunt based on the genomic data. All results here were generated using an E-value cut-off of 10−20. The relatively low number of functional predictions made from the EST+ORESTES ORFs (109 functions) is explained by the fact that enzymes are generally expressed at very low levels in the cell, so will be detected at a correspondingly low rate by EST or ORESTES sequencing. However, the fact that almost all (92%) of the functions predicted from these expression data are also present in the SHARKhunt results for genomic DNA shows that our software is successful in detecting E.tenella genes, despite the large number of introns present. SHARKhunt detects considerably more of these EST+ORESTES-derived functions within the genome than PRIAM does (81 and 70% for the ab initio and ORF data, respectively).
Figure 3.
Bar chart showing numbers of predicted enzymatic functions in E.tenella obtained by SHARKhunt, by PRIAM run on our ab initio gene predictions and by PRIAM run on genomic ORFs of at least 50 amino acids. The bar to the left shows the number of functions predicted by PRIAM run on ORFs of at least 50 amino acids from the E.tenella EST+ORESTES data. The proportion of these functions also predicted by each of the three genomic methods is shown in black for each method. An E-value cut-off of 10−20 was used for all searches.
Overall, these results demonstrate that the homology-based genefinding method used by SHARKhunt can be a much more effective way to detect enzymatic functions within an unannotated genome with high intron number than existing methods based on the analysis of polypeptide sequences, even when gene models from a related organism are available.
Effect of variation in genome coverage on performance
Since SHARKhunt is expected to be useful for the analysis of preliminary genome sequence data, it is interesting to investigate how performance is affected by low genome coverage. Taking the SHARKhunt results (at an E-value cut-off of 10−20) for the current E.tenella assembly (∼7.5× coverage) as the standard, we calculated the percentage of these functions detected by SHARKhunt using an assembly at ∼4.3× generated in 2002 and using our own Phrap assembly of reads totalling ∼0.5× coverage. These results are shown in Table 2.
Table 2.
Comparison of SHARKhunt performance for E.tenella at varying levels of genome coverage
| Genome coverage | No. of functions predicted at an E-value cut-off of 10−20 | Percentage of functions predicted using ∼7.5× assembly |
|---|---|---|
| ∼7.5× | 288 | 100 |
| ∼4.3× | 274 | 95 |
| ∼0.5× | 117 | 41 |
The ∼7.5× and ∼4.3× assemblies were obtained directly from the Sanger Institute Eimeria project website (http://www.sanger.ac.uk/Projects/E_tenella/). The ∼0.5× assembly was generated with Phrap (http://www.phrap.org/) using a subset of the raw reads obtained from the same site.
It is notable that the reduction from ∼7.5× to ∼4.3× coverage has very little effect on the predictions made by SHARKhunt. With a further reduction to ∼0.5× coverage, the program still detects over 40% of the enzymes found using the ∼7.5× assembly. This implies that even very small genome sequencing projects should have sufficient data to give a general picture of the metabolic capabilities of an organism, and to support wet-lab experiments to search for missing genes in known pathways.
Network visualization with SHARKview
In order to assist researchers in interpreting the results of a genome search, we have constructed an online database of known metabolic processes, visualized graphically as a network upon which the SHARKhunt results may be projected.
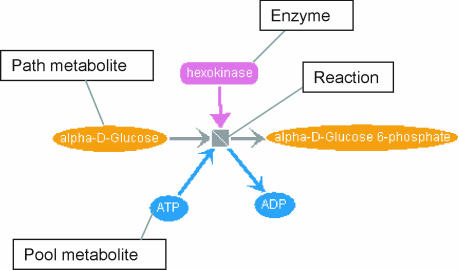
A specialized database (SHARKdb) was developed using the pure Java object-oriented database management system, ozone (http://www.ozone-db.org), which is freely available open source software. Our overall metabolic network was imported to SHARKdb from the KEGG/LIGAND database (6) (release 26), with corrected reaction equations (including reversibility information and definitions of path and pool metabolites, see legend to Figure 4 for definitions) derived from the data of Ma and Zeng (22).
Figure 4.
A SHARKview Petri net representation of the reaction alpha- d-glucose + ATP → alpha-d-glucose-6-phosphate + ADP, catalysed by the enzyme hexokinase. The orange ovals represent ‘path’ metabolites, considered to lie on metabolic pathways as source, intermediate or sink compounds. Blue ovals represent ‘pool’ metabolites, such as water or ATP, which act as co-factors for metabolic pathways but are not generally considered as intermediates. Reactions are shown as square nodes with open-headed arrows, indicating the consumption and production of metabolites. Enzymes are shown as rounded rectangles, connected to the reactions they catalyse with full-headed arrows. Petri net representations for biological pathways are reviewed in (24).
The generic metabolic network from KEGG is stored in the database in the form of a Petri net (23). This is a mathematical graph structure that is gaining popularity in the field of biological network analysis (24). It is especially well suited to the representation of metabolic networks, since reactions and the enzymes that catalyse them are treated as separate objects, allowing a single enzyme to catalyse several different reactions (e.g. with alternative substrates), or a single reaction to be catalysed by several different enzymes. Figure 4 shows a Petri net representation of a simple enzymatic reaction. In addition to its intuitive graphical representation, the Petri net structure will make computational analysis of the metabolic networks inferred from SHARKhunt results much more convenient in future studies, e.g. in the computation of elementary flux modes representing metabolic pathways (4).
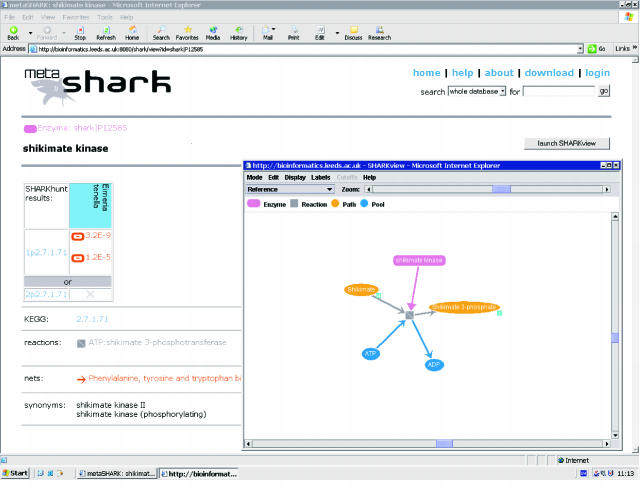
The metaSHARK package includes a novel network visualization tool called SHARKview (Figure 5). This has been developed from the TouchGraph dynamic graph layout library (http://www.touchgraph.com) and runs as an applet in the user's web browser. The Petri net structure of the metabolic network is clearly presented, with circular or elliptical nodes (‘places’ in Petri net terminology) representing compounds, and square nodes (‘transitions’) representing reactions. The arrows (‘arcs’) linking compounds and reactions show the consumption and production of metabolites during the reaction. Enzymes are depicted as rounded rectangles, connected to reactions by a different type of arc called a ‘test arc’. The test arc indicates that, unlike a compound, an enzyme is not consumed by the reactions in which it participates. Most reactions are reversible and can have their displayed direction changed by the user, but those identified as irreversible by Ma and Zeng (22) cannot be changed. An irreversible reaction is shown with a diagonal white stripe across the reaction node.
Figure 5.
The metaSHARK web interface (http://bioinformatics.leeds.ac.uk/shark/) includes an applet, SHARKview, for the navigation and visualization of metabolic networks. Links to KEGG (6) and PRIAM (11) provide further information about the network and sequence data used.
The SHARKview applet permits users to browse the database, starting from any compound, reaction, enzyme or pre-defined network (derived from the KEGG pathway diagrams). A blue tag at the corner of a node indicates that it has more neighbours in the database than are currently loaded by the browser. These neighbours may be retrieved on demand in order to build up user-defined sub-networks and pathways, a function not possible with the static KEGG pathway diagrams.
SHARKview initially displays only the generic network structure stored in the database, but the results of a SHARKhunt search of a particular genome can also be uploaded and superimposed on the metabolic network. To represent the strength of the evidence for each enzyme, nodes are coloured according to E-value, with cut-offs at 10−20 and 10−10. This colour scheme helps the user to pick out the encoded pathways, and also makes it easier to spot potential ‘missing enzymes’, where most of a pathway has strong evidence, but one or more of the enzymes have not been detected by the software or have only tentative assignments. The evidence for each SHARKhunt prediction may be viewed in more depth on its own webpage, which provides direct links to the NCBI BLAST server for further sequence analysis. Uploaded data are stored in the user's private workspace, which is protected by a password. This workspace may also be used to store customized SHARKview network diagrams for later retrieval.
As well as examining a single metabolic annotation, SHARKview can also visualize comparisons between the SHARKhunt results for two different genomes. The user selects the two genomes to be compared and an E-value cut-off above which an enzyme assertion is ignored. Colour coding then indicates the enzymes found in each genome and those common to both genomes. Careful use of this feature should make it easy to detect differences in the metabolic capabilities of related species, or to highlight potential drug targets where a pathogen has an essential pathway that the host lacks.
To enable the metaSHARK system to be used with genome annotation data from other sources (either from a manual annotation or from a program, such as PRIAM), or alternatively with data derived from microarray experiments, we have also made it possible to upload a simple list of EC numbers and visualize them with the SHARKview applet in the same way as the SHARKhunt XML results. Users may choose to supply extra information for each EC number, such as the corresponding gene name or probe ID, and using a simple syntax these may even be displayed as web links, turning metaSHARK into a dynamic interface to other online databases. An example of such an application can be found on the website, where we provide links to the available PlasmoDB and PlasmoCyc annotations for P.falciparum enzymes.
Case study: coenzyme A biosynthesis in P.falciparum and E.tenella
Coenzyme A is an enzyme cofactor that is essential to all organisms, taking part in many metabolic pathways, including fatty acid synthesis and degradation, pyruvate oxidation and glyceride synthesis. The biosynthetic pathway for coenzyme A is shown in Figure 6. The full pathway (from 3-methyl-2-oxobutanoate) is present in bacteria, plants and fungi, but animals lack the initial steps and are dependent on food and intestinal bacteria for a supply of pantothenate, a B-group vitamin and coenzyme A precursor (25). We will use coenzyme A biosynthesis as an example to show how metaSHARK can be used to examine the evidence for the presence of a particular metabolic pathway in a sequenced organism.
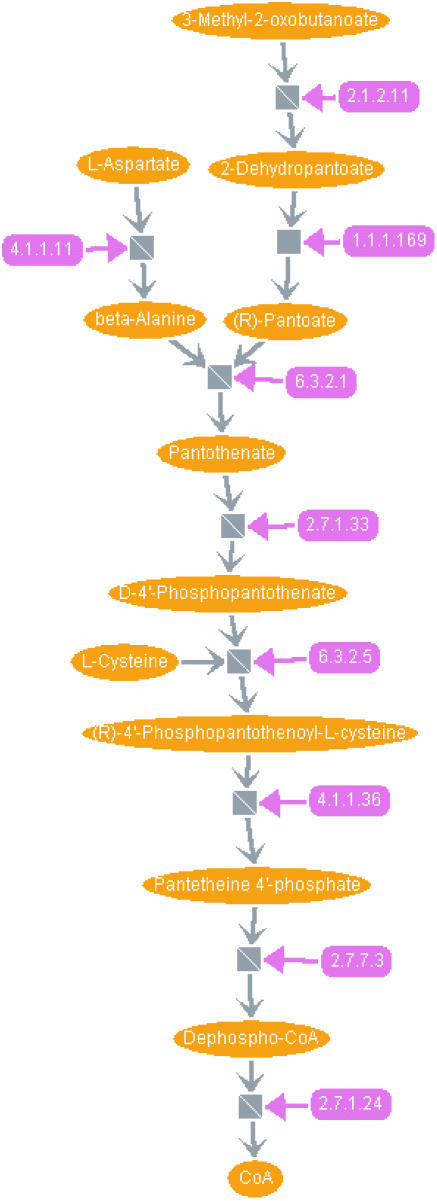
Figure 6.
A SHARKview visualization of the coenzyme A biosynthesis pathway. For an explanation of the symbols used, see legend to Figure 4. Enzymes are labelled by their EC numbers: 2.1.2.11, 3-methyl-2-oxobutanoate hydroxymethyltransferase; 1.1.1.169, 2-dehydropantoate 2-reductase; 4.1.1.11, aspartate 1-decarboxylase; 6.3.2.1, pantoate-beta-alanine ligase; 2.7.1.33, pantothenate kinase; 6.3.2.5, phosphopantothenate-cysteine ligase; 4.1.1.36, phosphopantothenoylcysteine decarboxylase; 2.7.7.3, pantetheine-phosphate adenylyltransferase; and 2.7.1.24, dephospho-CoA kinase. Note that pool metabolites are neglected in this figure.
The nutritional requirements of P.falciparum have been studied in some detail during the development of media for continuous in vitro cultivation (26), during which it has become evident that the intraerythrocytic stages are dependent on the host for a number of essential nutrients. Modifications induced in the erythrocyte membrane by the parasite introduce new permeation pathways that allow it to obtain certain micronutrients that are not ordinarily present in the red blood cell (27,28). Pantothenic acid is one such essential nutrient, which is required in the external medium for growth of P.falciparum (26). In initial studies of coenzyme A requirements in Plasmodium lophurae grown in duck erythrocytes, Bennett and Trager (29) suggested that this parasite lacks the necessary enzymes for coenzyme A synthesis and must, therefore, rely on the enzymes already present in the host cell to convert pantothenate to coenzyme A, which it then absorbs from the cytosol. However, recent experiments with P.falciparum in human erythrocytes by Saliba et al. (30) reached markedly different conclusions. Parasitized erythrocytes were observed to rapidly take up pantothenate from the extracellular medium, while uninfected cells exhibited negligible uptake. The parasites were shown to have the ability to absorb pantothenate and phosphorylate it, indicating the presence of a pantothenate kinase within the parasite. This parasite pantothenate kinase activity was more than 10 times greater than that observed in the host cell cytosol, indicating that the P.falciparum genome must contain a gene encoding pantothenate kinase, and presumably also contains genes for the other enzymes necessary for coenzyme A synthesis from pantothenate. In a subsequent study, Saliba and Kirk (31) described a novel H+-coupled pantothenate transporter within the parasite cell membrane. This system for pantothenate uptake is significantly different from the mammalian Na+:pantothenate symporter, and as such may make a suitable target for the development of new anti-malarial drugs. It will, therefore, be important to characterize the parasite's pathway for pantothenate metabolism in more detail.
Table 3 shows the available annotation from PlasmoCyc for coenzyme A biosynthesis, alongside the results from the SHARKhunt search with an E-value cut-off of 10−10. Although PlasmoCyc shows only the final enzyme in the pathway, dephospho-CoA kinase (EC 2.7.1.24), SHARKhunt also found good evidence for pantothenate kinase (EC 2.7.1.33), phosphopantothenate-cysteine ligase (EC 6.3.2.5) and phosphopantothenoylcysteine decarboxylase (EC 4.1.1.36), the first three steps of the pathway from pantothenate to coenzyme A. In a recent study of the evolution of coenzyme A biosynthesis, Genschel (32) also predicted these four enzymatic functions using BLAST searches against the annotated polypeptides from the P.falciparum annotation, and surmised that P.falciparum, like animals, is able to synthesize coenzyme A from exogenous pantothenate. The only enzyme in this pathway that was not convincingly detected by Genschel was phosphopantetheine adenylyltransferase (EC 2.7.7.3), although a candidate gene (PF07_0018) was detected using PSI-BLAST at an E-value of 10−6. The same candidate was also detected by the SHARKhunt search of genomic DNA with an E-value of 3.0 × 10−6.
Table 3.
Results of searches for enzymes involved in coenzyme A biosynthesis within the P.falciparum and E.tenella genomes
| Enzyme | P.falciparum | E.tenella | |||
|---|---|---|---|---|---|
| PlasmoCyc-ANN (human reannotation) | SHARKhunt (E-value < 10−10) | PRIAM (E-value < 10−10) | SHARKhunt (E-value < 10−10) | ||
| Pantothenate biosynthesis | 2.1.2.11 | • | |||
| 1.1.1.169 | • | ||||
| 4.1.1.11 | |||||
| 6.3.2.1 | • | ||||
| Pantothenate metabolism | 2.7.1.33 | • | • | • | |
| 6.3.2.5 | • | • | |||
| 4.1.1.36 | • | • | |||
| 2.7.7.3 | |||||
| 2.7.1.24 | • | • | • | • | |
A black circle indicates that the enzyme was found using the specified method. Enzymes are labelled by EC number, see legend to Figure 6 for full names. For P.falciparum, The PlasmoCyc-ANN human reannotation (18) is compared with the SHARKhunt results at an E-value cut-off of 10−10. No annotation is currently available for E.tenella, so we compare the PRIAM analysis of all ORFs of 50 amino acids or longer with the SHARKhunt results using the raw genomic DNA sequence, both taken at an E-value cut-off of 10−10.
Compared with Plasmodium, relatively little is currently known about the nutritional requirements of Eimeria spp. Biochemical experiments are difficult to carry out, owing to the limited amount of pure material obtainable from either cell culture or parasites grown in live hosts. In studies carried out using vitamin-deficient diets for Eimeria-infected chicks, Prasad (33) demonstrated that neither E.tenella nor E.acervulina required pantothenic acid in the chick diet for sustained infections. In follow-up work, Warren (34) came to the same conclusions, but observed that a chick diet containing calcium pantothenate had the effect of increasing oocyst output in E.tenella infections. However, a negative result using the ‘deficient diet’ technique does not necessarily imply that the omitted nutrient is not required by the parasite, since it might be synthesized by the host or by its gut flora. Later in vitro experiments by Doran and Augustine (35) with E.tenella grown in chicken kidney cells showed that the omission of pantothenate from the growth medium produced 89% as many parasites as the control medium after two generations.
Table 3 shows the results obtained by PRIAM on all ORFs over 50 amino acids alongside the SHARKhunt results, both at an E-value cut-off of 10−10. The ORF-based searches have detected only pantothenate kinase (EC 2.7.1.33) and dephospho-CoA kinase (EC 2.7.1.24), so would seem to support the conclusion that E.tenella, like P.falciparum, depends on the host for a source of pantothenate. However, the results obtained using SHARKhunt show very strong evidence for the presence of the full coenzyme A biosynthesis pathway from 3-methyl-2-oxobutanoate. Three enzymes upstream of pantothenate (3-methyl-2-oxobutanoate hydroxymethyltransferase, EC 2.1.2.11; 2-dehydropantoate 2-reductase, EC 1.1.1.169; and pantoate-beta-alanine ligase, EC 6.3.2.1) were detected, along with both enzymes found by PRIAM, a phosphopantothenate-cysteine ligase (EC 6.3.2.5) and a phosphopantothenolycysteine decarboxylase (EC 4.1.1.36). Of the missing two enzymes that were not detected at this cut-off, one, phosphopantetheine adenylyltransferase (EC 2.7.7.3) had a convincing candidate hit at E-value 2.7 × 10−5. No convincing candidate hits were found for aspartate 1-decarboxylase (EC 4.1.1.11). Indeed, genes encoding this enzymatic function remain elusive in many archaeal, fungal and bacterial genomes (32).
The experimental evidence outlined above is compatible with the conclusion that E.tenella is able to synthesize coenzyme A from 3-methyl-2-oxobutanoate. The presence of a pantothenate biosynthesis pathway encoded within the genome would not conflict with the experimentally observed increase in the rate of parasite growth on adding calcium pantothenate to the chick diet (34) or cell culture growth medium (35). It is likely that supplementation of pantothenate would increase growth even if the basal synthetic pathway were active. Alternatively, it remains possible that the enzymes for pantothenate synthesis are present in the genome but are expressed at other stages in the life cycle.
In summary, the SHARKhunt search method was successful in detecting the experimentally demonstrated but as yet unannotated pantothenate to coenzyme A pathway encoded in the P.falciparum genome. However, the SHARKhunt results for the E.tenella genome show strong evidence for the presence of genes coding for the entire bacterial/plant/fungal coenzyme A biosynthetic pathway. The same conclusion might not have been reached with a study of E.tenella ORFs using the PRIAM software, which failed to detect any enzymes upstream of pantothenate at an equivalent E-value cut-off.
The evidence from any form of sequence analysis will always require experimental validation to demonstrate that the genes involved are expressed at a particular life stage and truly perform the roles predicted for them. Nevertheless, this case study is an excellent example of how metaSHARK can be used to make concrete predictions of the metabolic pathways encoded by unannotated genomic sequences and hence guide experimental studies.
CONCLUSION
By working only with genomic DNA, metaSHARK offers an improved level of flexibility and accuracy over existing software for automated enzyme annotation. In this work, we have shown the validity of the SHARKhunt automated annotation protocol by analysis of the genome of P.falciparum, then gone on to demonstrate its usefulness by deriving new knowledge of metabolic pathways within the unannotated genome of E.tenella. We have also introduced a powerful and intuitive pathway navigation and visualization tool in the form of the SHARKview applet. We believe that metaSHARK will be a particularly valuable tool for the study of organisms that are difficult to manipulate in the laboratory, and also for the many organisms for which low-coverage genomic DNA sequences are becoming available. The SHARKhunt software is available to download at http://bioinformatics.leeds.ac.uk/shark/.
Acknowledgments
Metabolic analysis on a genome scale necessarily depends upon a great deal of prior effort, and the authors would like to thank all those upon whose work we are building. Special thanks go to Minoru Kanehisa for permission to use the KEGG network data, to Clotilde Claudel-Renard and Sebastien Carrere for the PRIAM profiles and sequence data and to Hongwu Ma and An-Ping Zeng for supplying their revisions of the LIGAND reactions. Thanks also to Aaron J. Mackey and Martin Fraunholz for kindly supplying us with their latest T.gondii gene models and for running the E.tenella TwinScan2 predictions, to Phil Green for providing the Phrap program, to Matthew Berriman for helpful discussions and to the E.tenella genome consortium for granting permission to publish this analysis of the preliminary genome sequence. The authors are grateful to two anonymous referees for their constructive comments and suggestions. The E.tenella genome sequence data were produced at the Wellcome Trust Sanger Institute during a sequencing project funded by the BBSRC. They can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/Eimeria/tenella/genome/. The original cDNA ORESTES reads were produced at the University of Sao Paulo, Brazil, and are available by request at http://www.lbm.fmvz.usp.br/eimeria/. The authors thank the MRC for funding this work. Funding to pay the Open Access publication charges for this article was jointly provided by the University of Leeds and JISC.
REFERENCES
- 1.Selkov E., Maltsev N., Olsen G.J., Overbeek R., Whitman W.B. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–GC26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 2.Bono H., Ogata H., Goto S., Kanehisa M. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998;8:203–210. doi: 10.1101/gr.8.3.203. [DOI] [PubMed] [Google Scholar]
- 3.Jackson L.K., Phillips M.A. Target validation for drug discovery in parasitic organisms. Curr. Top. Med. Chem. 2002;2:425–438. doi: 10.2174/1568026024607445. [DOI] [PubMed] [Google Scholar]
- 4.Schuster S., Fell D.A., Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 2000;18:326–332. doi: 10.1038/73786. [DOI] [PubMed] [Google Scholar]
- 5.Schilling C.H., Letscher D., Palsson B.Ø. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J. Theor. Biol. 2000;203:229–248. doi: 10.1006/jtbi.2000.1073. [DOI] [PubMed] [Google Scholar]
- 6.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krieger C.J., Zhang P., Mueller L.A., Wang A., Paley S., Arnaud M., Pick J., Rhee S.Y., Karp P.D. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C., Huhn G., Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paley S.M., Karp P.D. Evaluation of computational metabolic-pathway predictions for Helicobacter pylori. Bioinformatics. 2002;18:715–724. doi: 10.1093/bioinformatics/18.5.715. [DOI] [PubMed] [Google Scholar]
- 10.Overbeek R., Larsen N., Walunas T., D'Souza M., Pusch G., Selkov E., Jr, Liolios K., Joukov V., Kaznadzey D., Anderson I., et al. The ERGO genome analysis and discovery system. Nucleic Acids Res. 2003;31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claudel-Renard C., Chevalet C., Faraut T., Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConkey G.A., Pinney J.W., Westhead D.R., Plueckhahn K., Fitzpatrick T.B., Macheroux P., Kappes B. Annotating the Plasmodium genome and the enigma of the shikimate pathway. Trends Parasitol. 2004;20:60–65. doi: 10.1016/j.pt.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Birney E., Clamp M., Durbin R. GeneWise and GenomeWise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh I., Hanekamp T., Tsoka S., Karp P.D., Altman R.B. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirley M.W., Ivens A., Gruber A., Madeira A.M., Wan K.L., Dear P.H., Tomley F.M. The Eimeria genome projects: a sequence of events. Trends Parasitol. 2004;20:199–201. doi: 10.1016/j.pt.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17(Suppl. 1):S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 21.Dias Neto E., Correa R.G., Verjovski-Almeida S., Briones M.R., Nagai M.A., da Silva W., Jr, Zago M.A., Bordin S., Costa F.F., Goldman G.H., et al. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA. 2000;97:3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma H., Zeng A.P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- 23.Petersen J.L. Petri Net Theory and the Modeling of Systems. NJ: Prentice-Hall; 1981. [Google Scholar]
- 24.Pinney J.W., Westhead D.R., McConkey G.A. Petri Net representations in systems biology. Biochem. Soc. Trans. 2003;31:1513–1515. doi: 10.1042/bst0311513. [DOI] [PubMed] [Google Scholar]
- 25.Michal G., editor. NY: Wiley; 1999. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology; p. 115. [Google Scholar]
- 26.Divo A.A., Geary T.G., Davis N.L., Jensen J.B. Nutritional requirements of Plasmodium falciparum in culture. I. Exogenously supplied dialyzable components necessary for continuous growth. J. Protozool. 1985;32:59–64. doi: 10.1111/j.1550-7408.1985.tb03013.x. [DOI] [PubMed] [Google Scholar]
- 27.Ginsburg H., Krugliak M., Eidelman O., Cabantchik Z.I. New permeability pathways induced in membranes of Plasmodium falciparum infected erythrocytes. Mol. Biochem. Parasitol. 1983;8:177–190. doi: 10.1016/0166-6851(83)90008-7. [DOI] [PubMed] [Google Scholar]
- 28.Desai S.A., Bezrukov S.M., Zimmerberg J. A voltage-dependent channel involved in nutrient uptake by red blood cells infected with the malaria parasite. Nature. 2000;406:1001–1005. doi: 10.1038/35023000. [DOI] [PubMed] [Google Scholar]
- 29.Bennett T.P., Trager W. Pantothenic acid metabolism during avian malaria infection: pantothenate kinase activity in duck erythrocytes and in Plasmodium lophurae. J. Protozool. 1967;14:214–216. doi: 10.1111/j.1550-7408.1967.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 30.Saliba K.J., Horner H.A., Kirk K. Transport and metabolism of the essential vitamin pantothenic acid in human erythrocytes infected with the malaria parasite Plasmodium falciparum. J. Biol. Chem. 1998;273:10190–10195. doi: 10.1074/jbc.273.17.10190. [DOI] [PubMed] [Google Scholar]
- 31.Saliba K.J., Kirk K. H+-coupled pantothenate transport in the intracellular malaria parasite. J. Biol. Chem. 2001;276:18115–18121. doi: 10.1074/jbc.M010942200. [DOI] [PubMed] [Google Scholar]
- 32.Genschel U. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol. Biol. Evol. 2004;21:1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 33.Prasad H. The role of some vitamin ‘B’ deficient diets in coccidiosis of the domestic fowl. Indian Vet. J. 1963;40:478–489. [Google Scholar]
- 34.Warren E.W. Vitamin requirements of the Coccidia of the chicken. Parasitology. 1968;58:137–148. doi: 10.1017/s0031182000073492. [DOI] [PubMed] [Google Scholar]
- 35.Doran D.J., Augustine P.C. Eimeria tenella: vitamin requirements for development in primary cultures of chicken kidney cells. J. Protozool. 1978;25:544–546. doi: 10.1111/j.1550-7408.1978.tb04183.x. [DOI] [PubMed] [Google Scholar]