Abstract
APOBEC3H (A3H) is the most polymorphic member in the APOBEC3 family. Seven haplotypes (hap I–VII) and four mRNA splicing variants (SV) of A3H have been identified. The various haplotypes differ in anti-HIV activity, which is attributed to differences in protein stability, sub-cellular distribution, and/or RNA binding and virion packaging. Here we report the first comparative biochemical studies of all the A3H variants using highly purified proteins. We show that all haplotypes were stably expressed and could be purified to homogeneity by E. coli expression. Surprisingly, four out of the seven haplotypes showed high cytosine (C) deaminase activity, with hap V displaying extremely high activity that was comparable to the highly active A3A. Furthermore, all four haplotypes that were active in C deamination were also highly active on methylcytosine (mC), with hap II displaying almost equal deamination efficiency on both. The deamination activity of these A3H variants correlates well with their reported anti-HIV activity for the different haplotypes, suggesting that deaminase activity may be an important factor in determining their respective anti-HIV activities. Moreover, mC deamination of A3H displayed a strong preference for the sequence motif of T-mCpG-C/G, which may suggest a potential role in genomic mC modification at the characteristic “CpG” island motif.
Graphical abstract
Introduction
The seven APOBEC3 (A3) family members of DNA cytosine deaminases, which are clustered on human chromosome 22 (reviewed in(1) and references therein), play a defensive role against endogenous retroelements and infectious retroviruses (2–6). A3H is the most divergent member of the A3 family, containing a Z3 Zn-coordination domain that is phylogenetically distinct from the Z1- and Z2-type domains of other A3 proteins (7,8). A3H is also the most polymorphic of the A3 family (7–9); A3H mRNA can undergo alternative splicing to generate four splicing variants (SVs) (Fig. 1A): SV154, SV182, SV183, and SV200 (4,10,11), and seven distinct human A3H haplotypes (hap I–VII) have been identified, which are comprised of various combinations of five single nucleotide polymorphisms (Fig. 1B, 1C) (4,10,11).
Fig. 1. The splicing variants (SVs) and haplotypes of A3H.
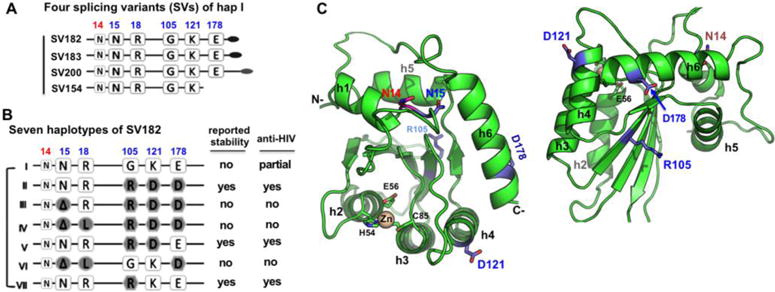
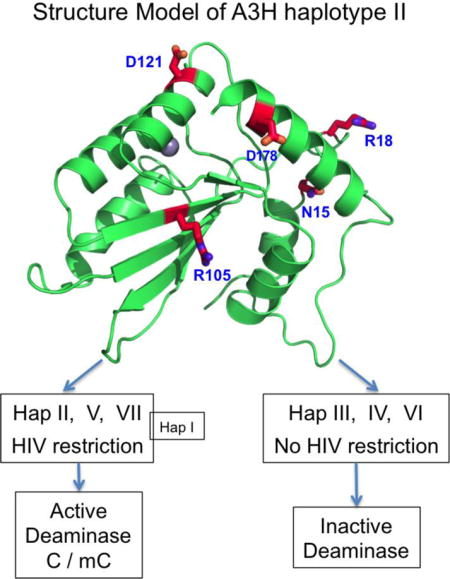
(A) Schematic overview of the four splicing variants based on haplotype I (hap I), with varying C-terminal extension. (B) The sequence variation of the seven haplotypes at the haplotype position 15, 18, 105, 121, and 178 based on SV182. The position 14 (colored in red), a conserved N in all A3H variants, is also shown. The reported stability inside cells and the anti-HIV activity for each haplotypes are listed on the right side based on literature. The weak anti-HIV activity of hap I is reported based on cell culture studies (10,11,17,18,27). (C) Two views of a modeled A3H hap II structure based on A3C (PDB ID 3VOW), which, like several previously published A3H models (13,18,57), shares the same core fold with all the known APOBEC structures. The locations of the haplotype residues (in sticks) are mapped to the modeled A3H structure. N15 at the end of helix 1 (h1) is buried, while its neighboring residue N14 is exposed. The R105 is on δ4, the opposite side of h2, h3 and h4. D121 is at the beginning of h4, and D178 is near the end of h6.
Evidence indicates that the seven A3H haplotypes possess different anti-viral activities. Only hap II, hap V, and hap VII are reported to effectively restrict Vif-deficient HIV-1 (4,10–12), and previous literature suggests that the anti-HIV activity of A3H can be through both deaminase dependent and independent pathways (10,13). Previous studies indicate that the different anti-HIV activities of A3H variants can be attributed to the following factors: variation in protein stability of the haplotypes (4,14–16), different subcellular localizations (17,18), different binding affinities to RNA (18) and relative levels of virion packaging (12). Additionally, A3H has been shown to be found in different oligomeric forms, and it can oligomerize in cells (16,19,20).
The distribution of A3H haplotypes in the human population is correlated with geographical location (4,10). Evidence indicates that the more stable A3H haplotypes can suppress HIV replication effectively enough to delay infection (21), and a higher frequency of the highly active and stable hap II is present in Africa, possibly a result of the long term presence of the HIV viral pathogen endogenous to the region (4,22–24). On the other hand, a majority of people of non-African descent carry A3H alleles for an inactive form of the A3H haplotypes, and variants of Vif extracted from HIV-1 strains infecting these populations are less effective at degrading stable A3H alleles prior to adaptation (4,10,15,21). In fact, it was shown that adaptive changes in viral Vif sequences could be attributed to the absence or presence of the different antiviral A3H haplotypes (15,20,21,24–27). These observations provide strong evidence for a significant role of A3H in immune defense against HIV infection.
So far, three APOBEC members, A3A, A3B and AID, have also been shown to deaminate methylated C (mC), with A3A showing the strongest activity (28–31). While the cellular functions of mC deamination for A3A and A3B have not been fully explained, the mC deamination activity associated with AID has been proposed as an alternative demethylation pathway for regulating methylation patterns in mouse germ cells (32), and for cell reprogramming in inducing pluripotent stem cells (33,34). To date, no other APOBEC protein outside of these three APOBEC members (A3A, A3B and AID) have been shown to have significant levels of mC deamination activity.
Despite mounting studies on the different variants of A3H, there is currently no complete systematic study comparing the deamination and DNA substrate binding activities of all the naturally occurring A3H variants. It is also unknown whether A3H can deaminate mC. Here we have described the expression and purification of all seven A3H haplotypes and four SVs in an E. coli expression system. We have performed systematic comprehensive biochemical studies of these variants and show that four haplotypes of A3H (I, II, V, VII) displayed high activity on both C and mC substrates, even though significant differences in overall activity levels exist among these variants. Surprisingly, mC deamination activity of hap II, the highest among all haplotypes, is comparable to normal cytosine deamination by the same variant. This side-by-side biochemical study of the haplotypes of A3H reveals that deaminase activity correlates well with their differential anti-HIV activity shown by previous cellular studies. Furthermore, the highly active mC deamination ability of A3H on motifs containing -mCpG- suggests the possibility of modifying mC in CpG islands of the genome in multiple cell types that express A3H.
Results
Comparing the deamination activity of the four SVs and seven haplotypes
The four A3H SVs (SV154, SV182, SV183 and SV200) vary in their C-terminal extension (Fig. 1A). We cloned and purified recombinant proteins of the four SVs of A3H haplotype I (hap I) from E. coli, and SDS-PAGE analysis of the purified A3H variants showed the isolated A3H proteins contained little to no detectable contaminating proteins (Supplemental Fig. 1A, 1B). The purified A3H proteins were assayed for deaminase activity using a FAM labeled 30 base oligonucleotide containing the known preferred sequence motif (5′-TCA-3′). The result showed that the shortest SV154 had no activity, but SV182, SV183 and SV200 all showed strong deaminase activity (Fig. 2A). Interestingly, the deaminase activity of SV200 was about 1.5 fold lower than SV182/SV183, suggesting that the additional C-terminal residues of SV200 somehow had an inhibitory effect on deaminase activity.
Figure 2. Deaminase activity of A3H splicing variants (SV) and haplotypes.
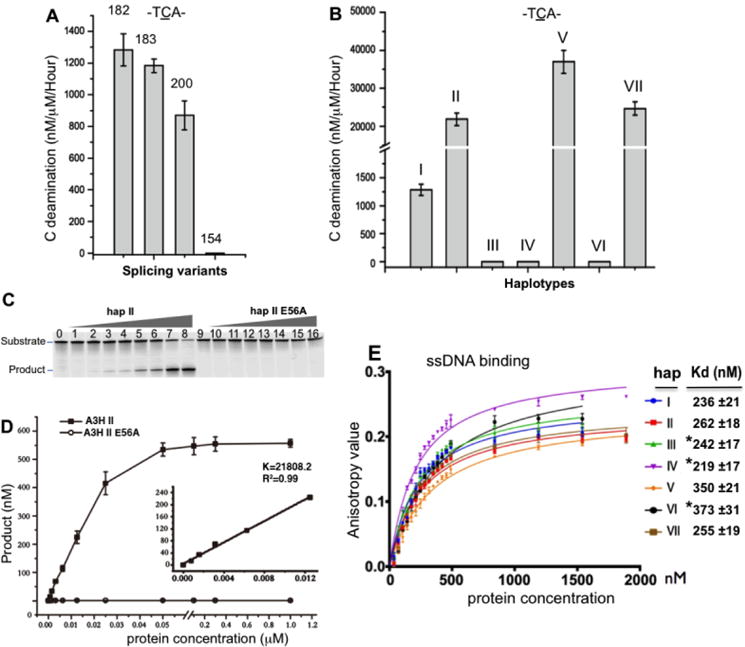
(A) The comparison of the deaminase activity on C (TCA motif) of the four SVs from A3H hap I. (B) The comparison of deaminase activity on C (TCA motif) of the seven haplotypes (hap I–VII) from A3H SV182, with hap V having the highest activity. (C, D) Representative gel result of deamination assay of hap II of A3H SV182 and its inactive mutant E56A (panel-C), and the dose response curve of its deaminase activity (in panel-D). Inset: the slope was calculated by linear regression using data from liner range. (E) The result of a ssDNA (30 nt) binding assay, showing no significant differences in ssDNA binding affinity for all seven haplotypes. The apparent Kd values for hap I–VII are listed on the right side. The error bars in panel-E represent the Standard Error of the Mean (SEM) in three independent experiments (see methods). * indicates hap III, IV, and VI that are inactive for deamination and for anti-HIV activity in panel-E.
We also examined the deaminase activity of all seven haplotypes (hap I-VII) using splicing variant SV182 of A3H (Fig. 1B). SDS-PAGE analysis revealed that the purified proteins for A3H hap I-VII consisted of essentially one major band free of any major contaminating proteins (Supplemental Fig. 1A), indicating that all seven haplotypes were stably expressed and soluble. The subsequent deamination assay revealed that four out of the seven haplotypes, specifically hap I, II, V and VII, were highly active in terms of C deamination (Fig. 2B). Among them, hap V had the highest deaminase activity, with a measured value of 36,986 nM/μM/hour, which approximated that of A3A – generally regarded as the most active in the family (28,31,35–37). Hap II and VII were about 1.7 times less active, and hap I was about 17 times less active than hap V. We also could not detect any deaminase activity for hap III, IV and VI, even at protein concentrations as high as 10 μM. A construct with the catalytically inactive mutation at E56A of hap II was used as a negative control for this deaminase assay (Fig. 2C). One common feature of the three inactive haplotypes is that they all contained the N15 deletion (ΔN15, Fig. 1B, 1C), indicating that this N15 deletion is sufficient to abolish deaminase activity, as noted previously by Zhen at al. (27).
In order to test whether each of these haplotypes had similar binding affinities for the ssDNA substrate, we performed a DNA binding assay using the same 30 nt ssDNA substrate used in the deaminase assay. The data revealed that all active and inactive haplotypes had similar binding affinities, with apparent Kd values within the range of 219–373 nM (Fig. 2E), suggesting that the difference in deaminase activity between the seven haplotypes was not caused by different binding affinities for ssDNA substrates. Therefore, the deaminase activity differences are likely caused by other features that are associated with the amino acid differences at the five different haplotype positions (Fig. 1B).
Evaluation of contribution of different haplotype residues to deaminase activity
Among the haplotype mutation positions (15, 18, 105, 121, and 178) of A3H, position 15 is N (N15, or ΔN15 in the three non-active haplotypes) as is the immediately previous position (N14) (Fig. 1A–C, Fig. 3A). By aligning the sequences of all human APOBEC domains, it is clear that N15 is absolutely conserved in both the inactive and active human APOBECs. However, residue 14 normally diverges into two categories: either an N/K14 for all active APOBECs (Fig. 3A), or an R/E/Y14 for all inactive APOBECs (Fig. 3A, Supplemental Fig. 2). We wanted to investigate how mutations at N14 and N15 would impact the deaminase activity of A3H. Results showed that in A3H hap II, N14 can be replaced with K14 without losing the wild-type level of activity (Fig. 3B). However, a mutation of N14 to R, E, or Y resulted in much lower activity (Fig. 3B), confirming that the type of residue is indeed important at position 14. The N14R mutant showing low activity is consistent with similar observations for A3A: the gene for an A3A equivalent in dogs and pandas has an R at the 14th position on loop-1, while A3A from tamarin has a T at the 14th position; the activity of these A3A orthologs are about half of those from human or horse, which have an R at the 14th position on loop-1 (38). These results are also consistent with previous findings that loop-1 of A3A is important for its high deaminase activity (31,36,37). As for N15, since all three N15 deletion haplotypes (Fig. 1B) have no detectible A3H deaminase activity (Fig. 2B), we tested the effect on deamination activity of a mutation to alanine at that site. The result indicated that N15A had no detectible deaminase activity (Fig. 3B). A structural model of A3H showed that N15 is between helix-1 and loop-1 and interacts with helix-6 to provide stability for loop-1 around the active center (Fig. 1C). Thus, an N15 deletion or N15A mutation probably alters the loop-1 conformation around the active center in a subtle way that doesn’t cause obvious loss of substrate binding affinity, but is sufficient to disturb the orientation of the target C/mC bound at the pocket enough to abolish deamination.
Figure 3.
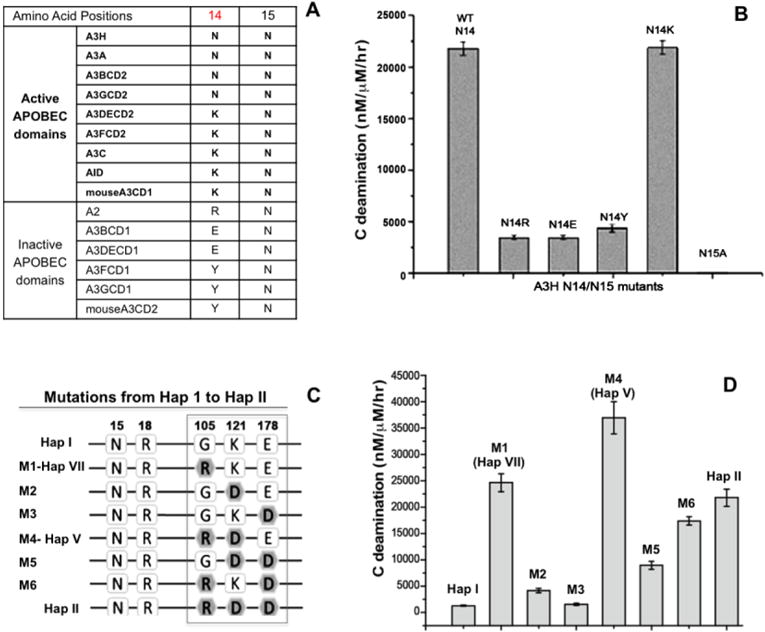
Mutational study to evaluate the effects of amino acids from the haplotypes on deaminase activity. (A) The sequence alignment at the residue number 14 and 15 of A3H with the active APOBEC domains and the inactive APOBEC domains. (B) The comparison of the deaminase activities of A3H mutants with substitutions at N14 and N15. The result indicates that position 14 with N14/K14 shows activity, but having the other three residues was about 10 times less active. For position 15, N15A mutation resulted in a complete loss of activity, similar to the ΔN15 mutant in hap III, IV, and VI. (C) Schematic overview showing the sequences at the haplotype residue position 105, 121 and 178 for hap I (first sequence) and hap II (last sequence), and the different combinations of mutations evolving from hap I to hap II. (D) The comparison of the deaminase activities of the different combination mutants are shown in panel-C. Error bars represent s.d. from the mean of three independent replicates.
We also examined the residues at the three remaining positions that vary between the haplotypes, residues 105, 121, and 178 (Fig. 1B), with various amino acid combinations differentiating the four catalytically active hap I, II, V, and VII (Fig. 2B). A3H hap I and II differ the most at these three positions: with G105K121E178 for hap I, and R105D121D178 for hap II (Fig. 3C, boxed). Other haplotypes have varying combinations of these six residues. Because none of these three locations are around the active center (Fig. 1C), or expected to directly affect catalysis, and given that the purified proteins of these four haplotypes are all soluble and stable, it is puzzling why these amino acid variations lead to different deamination efficiencies in our assay.
In order to evaluate the contribution of each of the three haplotype residues on deaminase activity, we generated six step-wise mutations (M1-M6, Fig. 3C) between hap I (at the top) to hap II (at the bottom). Among them, mutants M1–M3 contained a single point mutation, and M4–M6 contained combined double mutations. The mutant M1 clone was equivalent to hap VII, and M4 was equivalent to hap V (Fig. 3C). Compared with A3H hap I, M2 and M3 showed a minor increase in deaminase activity (Fig. 3D), whereas the M1 (G105R) mutant showed a striking increase. This activity of M1 is comparable to that of hap II and hap VII (Fig. 2B, Fig. 3D), suggesting that, among the three haplotype positions, an arginine at 105 has the most impact on overall deamination activity. Among the combined double mutants, M4 (M1+M2, or G105R+K121D) had a further 54% increase in activity than the M1 (G105R) single mutation (Fig. 3D), suggesting that position D121 co-operates positively with R105 to enhance the deaminase activity. However, M6 (M1+M3, or G105R+E178D) had a ~40% reduction in activity compared to the M1 (G105R) mutation (Fig. 3D), suggesting that position D178 co-operates negatively with R105 to attenuate the activity. The negative impact of D178 (vs E178) is also consistent with the activity levels of hap II (with D178) and hap V (with E178), which differ only at this position (Fig. 3D). These additional mutational studies verified that the amino acid types at these three haplotype positions are indeed sufficient to alter the deaminase activity, but without obvious difference of measurable binding affinity for ssDNA substrates.
A3H haplotypes are highly active in assays for mC deamination
Previous work has not shown whether A3H can deaminate mC. We examined deaminase activity on the same 30 nt oligonucleotide substrate except with the target C on the trinucleotide motif TCA switched to mC. We first tested for mC deamination using the four SVs of hap I. The result clearly showed that the three SVs showing C deamination activity were all similarly highly active in terms of mC deamination (Fig. 4A). We then examined the mC deamination activity for all seven haplotypes of A3H SV182. The results showed that all four haplotypes that were active on C also efficiently deaminated mC (Fig. 4B). Interestingly, unlike the C deamination activity where hap V is the most active, the one with the highest mC deamination activity is hap II, with about four times more activity on mC deamination than hap V.
Figure 4.
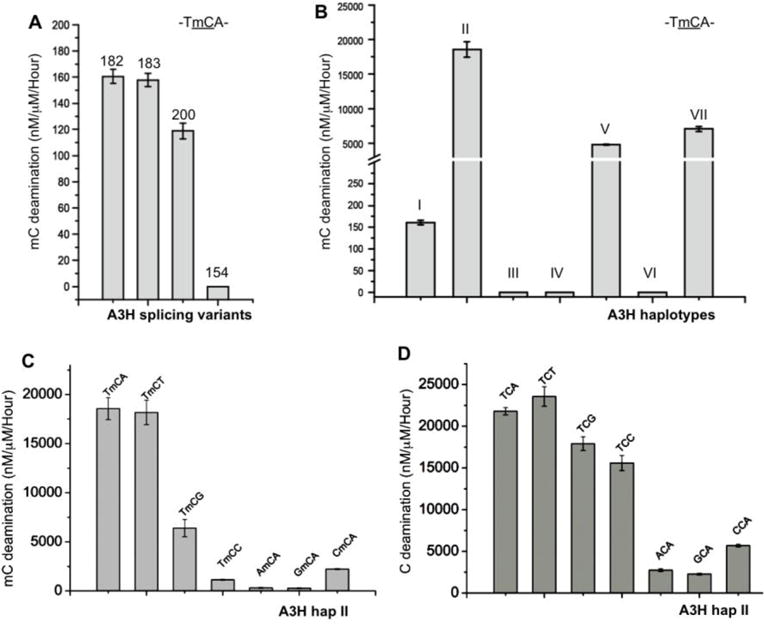
Highly active mC deaminase activity of A3H. (A) The deaminase activity on mC (TmCA motif) by the four different A3H SVs of hap I. Error bars represent s.d. from the mean of three independent replicates. (B) The deaminase activity on mC (TmCA motif) by the seven A3H haplotypes I–VII, with hap II having the highest activity. (C, D) The tri-nucleotide motif preference for mC (panel-C) and C (panel-D) deaminase activity by A3H hap II. Error bars represent s.d. from the mean of three independent replicates.
In order to evaluate the relative deamination activity on mC vs C, we calculated the selectivity factor for mC deamination as described in Fu et al.(31), by dividing the mC activity by the C activity and multiplying by 100. Previously, as a comparative benchmark, we showed that A3A has a selectivity factor of 12.7 for mC deamination, which is 6-fold higher than both A3B and the reported activity for AID (29,31). Here, we show that different A3H variants had different selectivity factors for mC deamination (Table 1), with most having a mC selectivity factor value around 12–15, on par with the highly active A3A (31). However, hap II had a remarkable mC selectivity factor of 85, indicating almost equal efficiency for both mC and C deamination. Hap VII also showed a mC selectivity factor of 28 (Table 1). These results clearly showed that A3H hap II had a higher mC selectivity factor than any other APOBEC characterized to date.
Table 1.
The deaminase activity and mC selectivity factor of A3H variants
| Deaminase Activity | hap I | Splicing variant 182 (SV182) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SV 200 | SV 183 | SV 182 | SV 154 | hap I | hap II | hap III | hap IV | hap V | hap VI | hap VII | |
| Act. on Ca (nM product /μM A3H/hr) | 870 ±92 | 1183 ±42 | 1283 ±102 | 0 | 1424 ±105 | 21809 ±429 | 0 | 0 | 36986 ±3046 | 0 | 24657 ±1713 |
| Act. on mCa (nM product /μM A3H/hr) | 119±6 | 158±5 | 160±5 | 0 | 215 ±17 | 18559 ±1121 | 0 | 0 | 4807 ±76 | 0 | 7050 ±336 |
| mC selectivity factorb | 137 | 13.4 | 12.5 | 0 | 15.1 | 85.1 | 0 | 0 | 13.1 | 0 | 28 6 |
Note:
Deaminase activity (nM product /μM enzyme/hr) for the C and mC deamination was calculated from the initial linear range of dose response assays for each protein constructs using the 30 nt ssDNA substrate containing motif TCA. SD was estimated from data collected in three independent experiments.
The mC selectivity factor was calculated [mC/C activity × 100], where the deaminase activity for the corresponding C and mC for each A3H variant in this table was used for the calculation.
A trinucleotide sequence of TCA is the preferred motif for normal C deamination by A3H (7,11,13,14,22). We examined the substrate motif preference for mC deamination using A3H hap II on a 30 nt ssDNA containing the following motifs: TmCA, TmCT, TmCG, TmCC, AmCA, GmCA and CmCA (Supplemental Table S1). For comparison, these same motifs with a corresponding normal C as the target were also included in this assay. The results showed that A3H had the highest activity on TmCA/T (Fig. 4C), which is followed by TmCG, with the least preferred sequence motifs being AmCA and GmCA. The tri-nucleotide motif preference for normal C deamination is similar but not identical to that of mC deamination (Fig. 4D). Generally speaking, normal C deamination appears to be less discriminating for the third residue of the tri-nucleotide motifs.
A3H efficiently deaminates mC in the context of CpG
As shown in Fig. 4C, A3H hap II efficiently deaminates mC to T in the sequence motif TmCG, which is over two orders of magnitude higher than for AID and A3B (29–31). Considering that A3H expresses in multiple cell types (7,12,39), this raised an interesting possibility that A3H may deaminate mC in mCpG islands in order to initiate an alternative demethylation pathway of mC (34). To further evaluate the effect of the nucleotide immediately 5′ or 3′ of the mCpG with respect to mC-deamination efficiency by A3H, FAM 30 nt substrates containing N-mCG-N (N being any nucleotide, Supplemental Table S1) were used for a deamination assay with A3H hap II. For comparison, the same deamination assay using substrates containing N-CG-N was performed as well. The result showed that both the residue immediately before and after the mCpG impacted the mC deamination efficiency. The preferred nucleotide upstream of the mCpG motif was T, followed by C, with G and A being roughly the least preferred (Fig. 5A). Surprisingly, the downstream residue of the T-mCpG motif also showed a significant impact, with G or C being the most preferred, followed by A, with T being the least preferred (Fig. 5A). The normal C deamination by hap II showed a very similar trend, with instead a slight preference for A residues downstream of CpG, followed by G and C (Fig. 4B).
Figure 5.
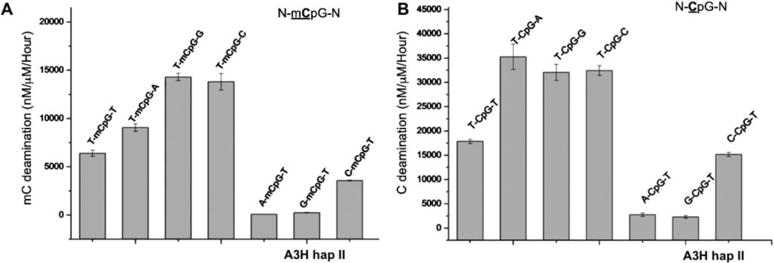
Examination of nucleotide sequence effect for deaminase using DNA substrate containing N-CpG-N or N-mCpG-N (the underlined C and mC are deamination target residue, and N being any nucleotide). (A) The deaminase activity of A3H hap II on the N-mC pG-N substrates, showing that T-mCpG-C/G are the best substrate. Error bars represent s.d. from the mean of three independent replicates. (B) The deaminase activity of A3H hap II on the N-CpG-N containing substrates, showing that T-CpG-C/G/A are the best substrates than others.
When the various substrate motifs were tested with A3H hap I, the deaminase activities were about 8–25 times lower than those of hap II when comparing the same sequence motif (Fig. 5, Fig, Supplemental Fig. 3). Additionally, while both hap I and hap II use T as the preferred 5′ upstream nucleotide in the N-CpG-N or N-mCpG-N motifs, they differ in the preferred downstream nucleotide. For example, a downstream A (T-mCpG-A) motif gave the third highest activity for hap II (Fig. 5A), but it yielded the highest activity for hap I (Supplemental Fig. 3A). For the non-methylated substrate, T-CpG-T showed much less deaminase activity than T-CpG-A/G/C for hap II (Fig. 5B), whereas T-CpG-T showed equal activity as with T-CpG-A/G/C for hap I (Supplemental Fig. 3B). Taken together, the results showed that A3H hap I and II both deaminate mC efficiently in DNA containing any T-mCpG-N sequence, with hap I having its most preferred motif of T-mCpG-A, and hap II having its most preferred motif of T-mCpG-G/C. A3H hap II approximately displayed an order of magnitude higher activity than hap I.
Analysis of the oligomeric status of A3H versus deamination activity
All previous A3H activity data gathered as part of this analysis utilized MBP-A3H fusion proteins that were eluted directly from the affinity column, which was essentially free of contaminating proteins via SDS-PAGE. In order to examine the oligomeric status of the purified MBP-A3H proteins, we selectively analyzed the purified protein of two representative A3H haplotypes: hap II as an example of one showing high activity, and hap III as an inactive comparison. A combination of size exclusion chromatography (SEC) and multi-angle light scattering (MALS) was the primary method of analysis. SEC revealed that the active hap II and the inactive hap III have similar oligomeric forms in vitro, with a large oligomeric form (larger than 880 kDa) and a small dimer/monomer form (around 100 kDa) (Fig. 6A-B). Subsequent MALS experiments on the samples isolated from the two peaks showed that the larger peak had a molecular weight (Mwt) of 982 kDa (Fig. 6C), which corresponds to the Mwt of an oligomer form of more than 14 subunits, whereas the smaller peak had a Mwt of 106 kDa (Fig. 6D), which is somewhere between a monomeric and a dimeric form. Interestingly, a side-by-side deaminase assay of the two oligomer forms of hap II revealed that the larger oligomeric form is over two times more active than the dimer/monomer species in both C- and mC deamination (Fig. 6E), which suggests that A3H can oligomerize into multiple copy oligomer forms and that oligomerization may enhance the deaminase activity.
Figure 6.
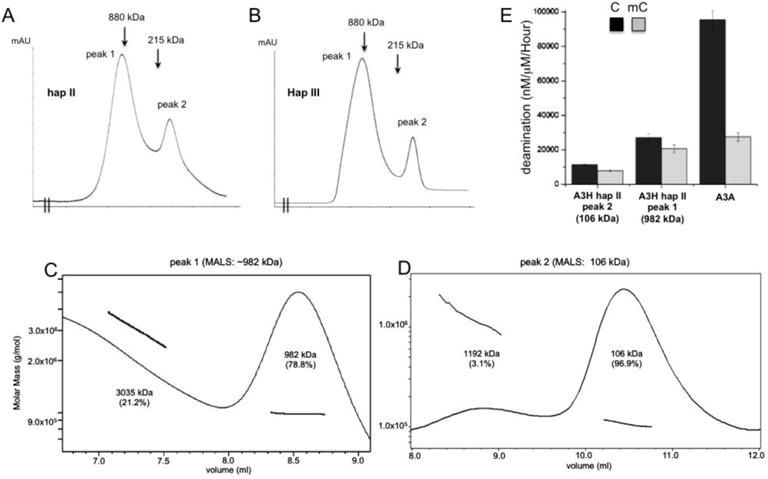
Analysis of the oligomeric status of MBP-A3H hap II and hap III. (A, B) The elusion profiles of A3H hap II (panel A) and hap III (panel B) on Superose 6 size exclusion chromatography. Both haplotypes showed very similar profile, with two major elution peaks. The positions of the molecular markers 880kD (ferritin dimer) and 215kD (catalase) are indicated in the elusion profiles. The calculated molecular weight of monomeric MBP-A3H fusion is 63 kDa. (C, D) The multi-angle light scattering (MALS) analysis of the MBP-A3H hap II protein from peak 1 (panel C) and peak 2 (panel D) shown in panel A. Peak 1 has a major species of ~982 kDa, corresponding to 14–15 monomers, but the broad peak suggests that it should not be a single oligomer form, and rather should contain multiple species of oligomer forms. Peak 2 has a major species of ~106 kDa, which is slightly smaller than a dimer form, but larger than a monomer form. Presence of the 106 kDa species suggests that dimers and monomers may co-exist and equilibrate, i.e. monomers and dimers may associate and dissociate quickly to give an averaged behavior. (E) The side-by-side deaminase activity comparison between the oligomeric (peak 1 in panel A), dimeric/monomeric (peak 2 in panel A) of MBP-A3H hap II, and A3A. The oligomeric form has about 2 times higher deaminase activity than the dimer/monomer form. Error bars represent s.d. from the mean of three independent replicates.
Discussion
Here we described the biochemical characterization of all known SVs and haplotypes of A3H using recombinant proteins purified from E. coli. We showed that all variants of A3H can be stably expressed as soluble proteins and can be purified in large quantity. Three of the four SVs of A3H (SV200, SV183, SV182) are active in deaminating both C and mC. Even though hap I, II, V, and VII all show significant deamination activity on both C and mC, there is a difference between how each one favors the two substrates. It was found that on C substrates hap V showed the most activity, followed by VII, II and I in that order, with hap I having about over 10 fold lower activity than other three active haplotypes (Fig. 2B). On the other hand, for mC deamination, the order was found to instead be hap II, VII, V, and I, with hap I showing about 30–120 fold lower activity the other three haplotypes (Fig. 4B). The only other APOBEC previously known to have as high of a deaminase activity as A3H hap II, V, or VII is A3A. However, the relative activity of mC deamination over C deamination reached ~85% for A3H hap II (a mC selectivity factor of 85), which is far greater than the ~10–12% reported for A3A on the same preferred substrate motif TCA/TmCA (28,31), making A3H the sole APOBEC member to date that possesses a uniquely high selectivity factor for mC deamination. We also showed that ssDNA substrate binding affinity for all seven haplotypes were similar to each other, ruling out DNA binding affinity as the major reason for the observed difference in deaminase activity.
Different A3H variants are distributed globally in distinct human populations, possibly driven by specific selection pressures imposed on humans in different geographic areas. A3H hap II, III and IV are found more frequently in those of African descent, whereas hap I is more frequent in Asian and European ethnic groups (4,7,11,18,22,27), which may reflect the natural history of anti-HIV activity and perhaps some other biological functions of different A3H variants. The seven known haplotypes of A3H differ in their antiviral activities (10,22), and such different anti-viral activities are reported to be related to different factors, which include different protein stability for different haplotypes (4,10–13,15), difference in oligomerization, RNA binding and virion packaging (12,13,18,19), and different sub-cellular localization (17).
The fact that all seven haplotypes can be stably expressed and purified away from contaminating proteins, and are stable at high protein concentrations after over-expression in E. coli cells suggests that they have similar solubilities; all were found to be soluble up to 10 mg/ml. Significantly, all seven haplotypes were shown to have comparable ssDNA binding affinity (Fig. 2E). Furthermore, size exclusion chromatography analysis of purified MBP-A3H showed that the active hap II and the inactive hap III have similar oligomeric forms in vitro, with a large Mwt peak containing multiple oligomeric forms and a small Mwt peak that contained a dimer or monomer form (Fig. 6A–D). Interestingly, a side-by-side deaminase assay showed that the large oligomeric form of A3H hap II was over two times more active than the dimer/monomer species in both C- and mC deamination (Fig. 6E). Previous studies showed that A3H oligomerization can occur in cells and is important for nucleic acid binding, virion packaging and anti-HIV activity (13,16,18–20). The result shown here also suggests that oligomerization may enhance the deaminase activity, which provides further support for the reported role of oligomerization in anti-HIV activity.
Even though all seven haplotypes of A3H were stably expressed and purified from E. coli, previous studies indicated that hap I, III, IV, and VI of A3H are instable in mammalian cells, which suggest that mammalian cells may have mechanisms to regulate the rapid turnover of these A3H haplotypes. In this study, we showed that the three A3H haplotypes with anti-HIV activity (II, V, VII) possess high deaminase activity, and the other three A3H haplotypes without anti-HIV activity (III, IV, VI) have no deaminase activity, which suggests the significance of deamination with regards to HIV restriction. The outlier is hap I, as it was shown to have poor anti-HIV activity in a cell culture assay (10,11,17,18,27) despite showing slightly stronger deamination activity than the potent HIV restriction factors A3G and A3F (28,40–42).This implies that factors other than deamination activity are likely playing a role in the anti-HIV activity of A3H hap I. It was previously reported that A3H hap I was mostly distributed in the nucleus (17,18), which may reduce its opportunity to restrict HIV in the cytoplasm. Therefore, while deaminase activity of the different haplotypes of A3H correlates well with their anti-HIV activity, it appears that other factors in the cellular environment (i.e. subcellular localization, stability, or perhaps virion packaging) play a role in determining the levels of anti-HIV activities of A3H variants.
Here we demonstrated for the first time that A3H hap I, II, V, and VII are highly active in mC deamination. While the full biological implications of mC deamination by A3H require future investigation, the mC deamination of AID is reported to be associated with the demethylation of mC and epigenetic regulation (32–34). We show that A3H efficiently deaminates the mC in the motif sequence of T-mCpG-C/G, raising the possibility of modifying genomic mC at mCpG islands through an alternative demethylation pathway by A3H (34). Because methylation and demethylation of the C/mC at CpG islands play an important role in epigenetics of gene regulation in cell growth, differentiation, and development, it would be interesting to see whether mC deamination of A3H has such a role in epigenetic regulation. This is especially intriguing given that A3H is expressed in multiple cell types, and has nuclear partitioning for different variants (7,12,17,39). Meanwhile, the unregulated mC deamination could also be involved with abnormal gene expression and cell growth, leading to diseases such as cancer. Indeed, evidence suggests that the activity of A3H, as well as the other members of the family that are capable of deaminating mC (A3A, A3B, and AID) may have a link to cancer formation (43–48), which implicates the important role of mC modification by APOBECs in biological functions as well as diseases.
Materials and methods
Construct of A3H variants
Genes encoding the seven haplotypes of A3H and splicing variants were synthesized as codon optimized sequences for E. coli expression and cloned into a pMAL-c5X Vector (NEB) using BamH I and Hind III restriction sites for expression in E. coli as MBP-fusion proteins. To construct the inactive mutant (E56A) and the mutants with single or combined mutations at different haplotype positions, the quick change site-directed mutagenesis kit (Agilent) was used. It was confirmed that all clones contained the desired coding sequences for the different A3H constructs by DNA sequencing.
Purification of A3H variants
The protein expression of various A3H constructs in the aforementioned pMAL-A3H plasmid in E. coli cells was performed by adding 0.3 mM IPTG to 1 liter of cell culture in LB media when the OD600 reached ~0.5, with subsequent incubation with shaking at 200 rpm at 16 °C for 12 hours. Cells were collected and lysed by extensive sonication in lysis buffer L (20 mM HEPES, pH 7.5, 250 mM NaCl, 1 mM EDTA, 10 mM DTT, 0.1 μg/μl RNase A). The cell lysate was centrifuged at 12,000 rpm for 30 min, and the supernatant was collected. The supernatant was treated with 2.0 M Urea for 3 hours at room temperature, which dissociated the contaminating E. coli GroEL from the MBP-A3H fusion protein (49,50). The Urea treated supernatant was then dialyzed against lysis buffer overnight at 4 °C with two buffer exchanges at 3 hour intervals, in order to get rid of residual urea. The dialyzed solution was centrifuged again, and the supernatant was passed through a column containing 10 ml of amylose resin (NEB). After extensive washing with 20× the column volume of buffer W (20 mM HEPES (pH 7.5), 250 mM NaCl, 10 mM DTT, 0.1 μg/μl RNase A), the A3H proteins were eluted as an MBP fusion with elution buffer (20 mM HEPES (pH 7.5), 250 mM NaCl, 10 mM DTT, 20 mM maltose). The eluted protein was then concentrated to ~10–20 mg/ml in buffer C (20 mM HEPES, 350 mM NaCl, 10 mM DTT). Finally, the concentrated MBP-A3H fusion proteins were confirmed and quantified by Edman N-terminal sequencing and UV spectrometry, followed by visualization via SDS-PAGE (Supplemental Fig. 1). The purified proteins were stored as 20 μl aliquots at −80° C for activity assays.
Deamination assay
The deamination assay of A3H variants was carried out as described by Fu et al with minor modification (31). In brief, A3H protein was treated with 1mM of EDTA in a 50-μl tube on ice for 10 min prior to performing the assay to inhibit possible contaminations. The 30-nt oligonucleotides which contained normal C or methylated C in the context of various sequence motifs were labeled with FAM at the 5′- or 3′- end and used as substrates in this study (Supplemental Table S1). 9 μl of reaction buffer (20 mM HEPES (pH7.5), 100mM NaCl, 600 nM ssDNA substrate, 1 mM CaCl2, 0.1 μg/μl RNaseA, 1 mM DTT) and 1 μl of A3H protein at different concentrations were mixed and incubated for 1 hour at 37 °C. With the normal C substrates, after denaturing at 95 °C for 5min, E. coli uracil DNA glycosylase was added and incubated at 37 °C for 1 hour for the deamination stage. For methylated C deamination, both thymine DNA glycosylase and a complementary oligonucleotide were added and incubated at 42 °C for 10 hour. Site-specific cleavage was then generated by introducing 150 mM of NaOH and incubation continued for 10 min at 95 °C. Eventually, the reaction mixture was separated on a 20% PAGE-urea gel and visualized with a laser scanner (Bio-Rad). The density of substrate and product in each reaction was determined using the ImageJ software (51). The percentage of product was calculated by dividing the density of the deaminated substrate by the total density of the deaminated and unmodified substrates, and multiplying by 100. The amount of product in each reaction was then calculated by multiplying the starting concentration of substrate by this percentage. The specific velocity of A3H variants was defined as nM/ μM/hour (nM product per μM A3H per hour), which was calculated by linear regression using the data from the linear range of A3H concentration. The data represent the results of three independent experiments.
Steady-state rotational-anisotropy binding assay
A3H-ssDNA binding was monitored by changes in steady state fluorescence depolarization (rotational-anisotropy). 130 μl reaction mixtures, containing 50nM 3′ FAM-labeled 30 nt ssDNA substrate in binding buffer (20 mM HEPES pH 7.0, 25 mM NaCl and 1 mM DTT) and with a varying concentration of 0 to 1,890 nM A3H (hap I to VII), were incubated at 25 °C. Rotational anisotropy was measured using a QuantaMaster QM-1 fluorometer (Photon Technology International) with a single emission channel. Samples were excited with vertically polarized light at 490nm and both vertical and horizontal emission was monitored at 520 nm. Based on the anisotropy value of each reaction, a protein concentration-dependent ssDNA binding curve was fitted with the “one site specific binding” model from Prism 6, and the dissociation constant (Kd) of each protein under the experimental condition was calculated with the same software.
Homology modeling of A3H structure
Generation of an A3H structural model was completed using the Rosetta3.5 software suite (52). A general homology model was first generated by the included threading algorithm for comparative modeling (53), using the previously solved A3C crystal structure(3) as the template PDB (PDB ID 3VOW). The input alignment of A3H and A3C was determined through the HHpred interactive alignment software (54), and 9-mer and 3-mer fragment files for A3H were generated with the online ROBETTA fragment server (55). A total of 1000 comparative structures were generated, with the lowest energy structure selected for further refinement. A further refinement of the loop regions was completed with the Rosetta loop modeling application, through the use of kinematic closure with fragments (56). Bounded and harmonic constraints were incorporated in order to maintain the zinc ion coordination interactions throughout the centroid and full-atom refinement stages, respectively. A total of 50 additional structures were generated and the lowest energy structure was determined.
Supplementary Material
Research Highlights.
Purified the proteins of all four splicing variants (SVs) and seven haplotypes (Hap I–VII) of A3H in large quantity from E. coli expression system.
Showed that three of the four SVs and three haplotypes (hap II, V, VII) all have very high deamination activity on C, with hap I showing ~1/10 of the activities of other three haplotypes.
Revealed that, for A3H hap II, the deamination activity on mC is almost as efficient as on C for the DNA substrates containing -mCpG- or -CpG- motif.
Showed that the deaminase activity of different A3H variants correlates well with their reported anti-HIV activity, indicating the importance of deaminase activity for HIV restriction.
Acknowledgments
We thank the core laboratory in the Center of Excellence in NanoBiophysics at University of Southern California for support in the related biophysics characterization of the purified protein. F.I. is a recipient of the Graduate fellowship from the Nakajima Foundation. This work is supported by the NIH grant R01GM087986 to X.S.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome biology. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. Journal of molecular biology. 2014;426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harbor perspectives in medicine. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aydin H, Taylor MW, Lee JE. Structure-Guided Analysis of the Human APOBEC3-HIV Restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooms M, Majdak S, Seibert CW, Harari A, Simon V. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J Virol. 2010;84:7961–7969. doi: 10.1128/JVI.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra M, Singer D, Mano Y, Hritz J, Nam G, Gorelick RJ, Byeon IJ, Gronenborn AM, Iwatani Y, Levin JG. Sequence and structural determinants of human APOBEC3H deaminase and anti-HIV-1 activities. Retrovirology. 2015;12:3. doi: 10.1186/s12977-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem. 2008;283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, Brown WL, Reilly C, Emerman M, Harris RS. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y, Love RP, Ara A, Baig TT, Adolph MB, Chelico L. Natural Polymorphisms and Oligomerization of Human APOBEC3H Contribute to Single-stranded DNA Scanning Ability. J Biol Chem. 2015;290:27188–27203. doi: 10.1074/jbc.M115.666065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MM, Emerman M. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol. 2011;85:8197–8207. doi: 10.1128/JVI.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One. 2012;7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. Journal of molecular biology. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baig TT, Feng Y, Chelico L. Determinants of efficient degradation of APOBEC3 restriction factors by HIV-1 Vif. J Virol. 2014;88:14380–14395. doi: 10.1128/JVI.02484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Sakurai D, Iwatani Y, Ohtani H, Naruse TK, Terunuma H, Sugiura W, Kimura A. APOBEC3H polymorphisms associated with the susceptibility to HIV-1 infection and AIDS progression in Japanese. Immunogenetics. 2015;67:253–257. doi: 10.1007/s00251-015-0829-2. [DOI] [PubMed] [Google Scholar]
- 23.Naruse TK, Sakurai D, Ohtani H, Sharma G, Sharma SK, Vajpayee M, Mehra NK, Kaur G, Kimura A. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. Journal of human genetics. 2015;61:263–265. doi: 10.1038/jhg.2015.136. [DOI] [PubMed] [Google Scholar]
- 24.Zhao K, Du J, Rui Y, Zheng W, Kang J, Hou J, Wang K, Zhang W, Simon VA, Yu XF. Evolutionarily conserved pressure for the existence of distinct G2/M cell cycle arrest and A3H inactivation functions in HIV-1 Vif. Cell Cycle. 2015;14:838–847. doi: 10.1080/15384101.2014.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooms M, Letko M, Binka M, Simon V. The resistance of human APOBEC3H to HIV-1 NL4-3 molecular clone is determined by a single amino acid in Vif. PLoS One. 2013;8:e57744. doi: 10.1371/journal.pone.0057744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J Virol. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nature chemical biology. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Ito F, Zhang G, Fernandez B, Yang H, Chen XS. DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B. The Biochemical journal. 2015;471:25–35. doi: 10.1042/BJ20150382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nature communications. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain. J Biol Chem. 2015;290:28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byeon IJ, Byeon CH, Wu T, Mitra M, Singer D, Levin JG, Gronenborn AM. Nuclear Magnetic Resonance Structure of the APOBEC3B Catalytic Domain: Structural Basis for Substrate Binding and DNA Deaminase Activity. Biochemistry. 2016;55:2944–2959. doi: 10.1021/acs.biochem.6b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caval V, Suspene R, Vartanian JP, Wain-Hobson S. Orthologous mammalian APOBEC3A cytidine deaminases hypermutate nuclear DNA. Molecular biology and evolution. 2014;31:330–340. doi: 10.1093/molbev/mst195. [DOI] [PubMed] [Google Scholar]
- 39.Lackey L, Law EK, Brown WL, Harris RS. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 2013;12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- 41.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Xiao X, Wolfe A, Chen XS. The in vitro Biochemical Characterization of an HIV-1 Restriction Factor APOBEC3F: Importance of Loop 7 on Both CD1 and CD2 for DNA Binding and Deamination. Journal of molecular biology. 2016 doi: 10.1016/j.jmb.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu M, Wang Y, Wang C, Shen W, Liu J, Geng L, Cheng Y, Dai J, Jin G, Ma H, et al. The eQTL-missense polymorphisms of APOBEC3H are associated with lung cancer risk in a Han Chinese population. Scientific reports. 2015;5:14969. doi: 10.1038/srep14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nature genetics. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience. 2015;2:320–333. doi: 10.18632/oncoscience.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuong KJ, Loeb LA. APOBEC3B mutagenesis in cancer. Nature genetics. 2013;45:964–965. doi: 10.1038/ng.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen XS, Casini G, Harrison SC, Garcea RL. Papillomavirus capsid protein expression in Escherichia coli: purification and assembly of HPV11 and HPV16 L1. Journal of molecular biology. 2001;307:173–182. doi: 10.1006/jmbi.2000.4464. [DOI] [PubMed] [Google Scholar]
- 50.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Molecular cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods in enzymology. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nature methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, Baig TT, Love RP, Chelico L. Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Frontiers in microbiology. 2014;5:450. doi: 10.3389/fmicb.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


