Abstract
Worldwide HIV-1 vaccine efforts are guided by the principle that HIV-specific T cell responses may provide protection from infection or delay overt disease. However, no clear correlates of T cell-mediated immune protection have been identified. Here, we examine in a HLA-B27+ HIV seronegative vaccinee persistent HIV-specific vaccine-induced anti-Gag CD4+ and CD8+ T cell responses. Although these responses exhibited those characteristics (multifunctionality, appropriate memory phenotype, and targeting of epitopes associated with long-term nonprogression) predicted to correlate with protection from infection, the subject became HIV infected. After HIV infection, the vaccine-induced CD8+ T cells expanded, but both CD4+ and CD8+ T cell responses acquired the functional and phenotypic patterns characteristic of chronic HIV infection. The virus quickly escaped the vaccine-induced T cell response, and the subject progressed more rapidly than expected for someone expressing the HLA-B27 allele. These data suggest that control of HIV by vaccine-elicited HIV-specific T cell responses may be difficult, even when the T cell response has those characteristics predicted to provide optimal protection.
Keywords: acute infection, correlate of protection, multiparameter flow cytometry
Several phase I and II clinical trials investigating HIV vaccine candidates conducted worldwide are based on the prevailing notion that both HIV-specific CD4+ and CD8+ T cell responses may be required to confer protection from disease progression (1). However, the correlates of immune protection remain unclear and controversial.
The properties likely to be required in a protective HIV-specific T cell response include high frequency, polyclonal, multifunctional CD4+ and CD8+ T cells of broad specificity, capable of proliferation and differentiation from central memory to effector memory phenotype that ideally recognize conserved HIV epitopes (1). Further, T cells have a myriad of functions, yet those that are critical for controlling HIV replication, preventing disease progression, or providing vaccine-induced protection are unknown. We know that CD8+ T cells place substantial pressure on HIV with repercussions for virus fitness (2), and that CD4+ T cell responses may be pivotal for the generation of long-lasting, fully functional CD8+ T cell responses (3, 4). Although selective T cell memory phenotypes are likely to play a decisive role in long-term protection from challenge against chronic viral infections in animals (5), these features are still unresolved for chronic virus infection in humans.
Here, we present the sentinel analysis of vaccine-induced multifunctional anti-HIV Gag-specific CD4+ and CD8+ T cell responses in a HLA-B*2705+ HIV-vaccine recipient, their changes after infection, and how they compare with those detectable in two nonvaccinated HLA-B*2705+ chronically HIV-infected individuals. The results underscore our lack of understanding regarding the immune correlates of protection both in HIV disease and vaccine models.
Materials and Methods
Subjects and Study Protocols. Peripheral blood mononuclear cells (PBMC) were obtained from a HLA-B*2705 positive volunteer (202-T07) enrolled in the National Institutes of Health-sponsored AIDS Vaccine Evaluation Group protocol 202 that evaluated the immunogenicity of a recombinant canarypox vector vCP205 (expressing gp120MN, the transmembrane portion of gp41LAI 688–712, GagLAI, and proteaseLAI; Pasteur-Merieux Connaught Laboratories, North York, ON, Canada) (6). This individual received four vector injections over a 6-month period. For comparative purposes, we studied anti-Gag T cell responses in two chronically HIV-1-infected HLA-B*2705-positive subjects who were enrolled at the Adult HIV Clinic of the University of Alabama at Birmingham. PBMC samples were obtained from all of the subjects under signed informed consent approved by all of the institutions. For subject 202-T07, postinfection time points are referred to as “acute” and “chronic,” where acute is the initial clinic visit and chronic is 28 months after the acute time point. The MHC class I haplotypes of the subjects in this study are as follows: 202-T07: HLA-A2, B27/44, and Cw2/5; 2714A: HLA-A2/30, B27/42, and Cw2/17; 3963I: HLA-A1/2, B8/27, and Cw4/6.
Peptides. The 15- and 20-mer overlapping peptides of the HIV-1HXB2 subtype B gene products were provided through the National Institutes of Health AIDS Research and Reference Reagent Programs. The 9-mer overlapping peptides and optimal epitopic peptides were purchased at >90% HPLC purity from SynPep, Dublin, CA. For the purpose of epitope mapping, a peptide matrix analysis was used as described in ref. 7.
Antibodies Source. The source for each antibody used in this study is reported in Supporting Text, which is published as supporting information on the PNAS web site.
CTL Studies. PBMC were separated from 50 ml of acid citrate dextrose anti-coagulated blood, and CD8+ CTL responses were evaluated by using autologous 51Cr-labeled target cells pulsed with defined peptides, infected with the appropriate recombinant vaccinia constructs, or with autologous virus isolates as reported in refs. 7 and 8. To generate KK10-specific CD8+ T cell lines, PBMC were stimulated and cultured according to the method in ref. 7.
IFN-γ ELISpot Assay. A modified ELISpot assay was used to detect peptide-specific release of IFN-γ by cryopreserved PBMC as described in ref. 7.
T Cell Receptor B (TCRB) Gene Expression Analysis. B27/KK10 tetramer+CD8+ T cells were sorted to >99% purity and the CDR3 region of the TCRB gene was sequenced according to procedures in ref. 9 (see Supporting Text for details).
Intracellular Cytokine Staining and Degranulation Assay. PBMC were thawed and rested overnight at 37°C, in 5% CO2 atmosphere, in RPMI medium 1640 containing 10% FCS (R10). The next day, the cells were adjusted to 1 × 106 per ml. In four-color flow, cells were stimulated and stained according to methods in ref. 7. In nine-color flow, cells were stimulated and stained as described in ref. 10. We used the following mAbs: αCD107a-Alexa 680, αCD4-Cascade Blue, αCD14/CD19-Cy5PE, αCD3-Cy7APC, αCD8-TRPE, α-IFN-γ-FITC, αTNFα -Cy7PE, and αIL-2-APC. In all experiments, a negative control (αCD28/49d), and a positive control (staphylococcal enterotoxin B, 10 μg/ml, Sigma-Aldrich) were included.
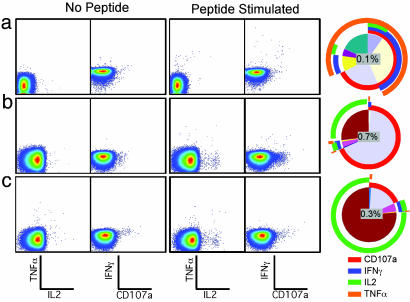
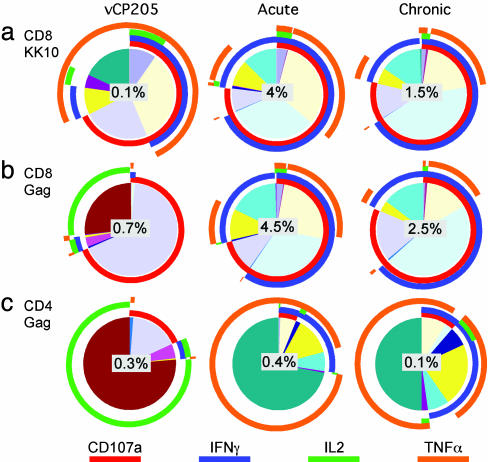
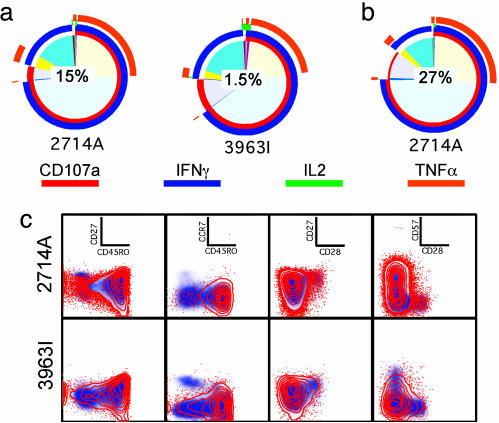
The results are reported as pie charts that depict the contribution of various responding T cell populations (background adjusted) to the total peptide-specific response (see Figs. 2, 4, and 5). Each portion of the pie chart represents a single population of CD8+ or CD4+ T cells expressing the marker depicted by the concentric colored arcs (red, CD107a; blue, IFN-γ; green, IL-2; orange, TNF-α) surrounding the pie chart. The color table used to generate the pie charts within all figures is matched, allowing direct comparison of the relative size of each responding population between each pie chart.
Fig. 2.
Fine characterization of vaccine-induced T lymphocyte functional responses by using nine-color flow cytometry. The two left dot plots of each row represent the unstimulated control, and the two right dot plots show cells stimulated with KK10 peptide (a) or CD8+ (b) or CD4+ (c) overlapping Gag peptides. The staining combination for each column of dot plots is shown at the bottom of the figure. The pie charts (see Materials and Methods) depict the contribution of various responding CD8+ (a and b) or CD4+ (c) T cell populations (background adjusted).
Fig. 4.
Shift in T cell functional profile in 202-T07 after infection. The pie charts (see Materials and Methods) depict the background-adjusted T cell functional response to the KK10 epitope (CD8) (a), HIV Gag (CD8) (b), and HIV Gag (CD4) (c). The value in the center of each pie chart represents the total response to the respective stimuli. Left shows vaccine-induced responses, Center shows responses during the acute phase, and Right shows chronic phase responses.
Fig. 5.
HLA B27-restricted KK10 responses in HIV-infected controls. The pie charts depict the CD8+ T cell response to KK10 (a) in 2714A and 3963I and HIV Gag (b) in 2714A. The value in the center of each pie chart represents the total response to the respective stimuli. (c) Infection-induced KK10-specific CD8+ T cells have a predominant effector/effector memory phenotype. Red contour overlays depict the B27 KK10 tetramer+ CD8+ T cells, whereas the blue density plot depicts total CD8+ T cells.
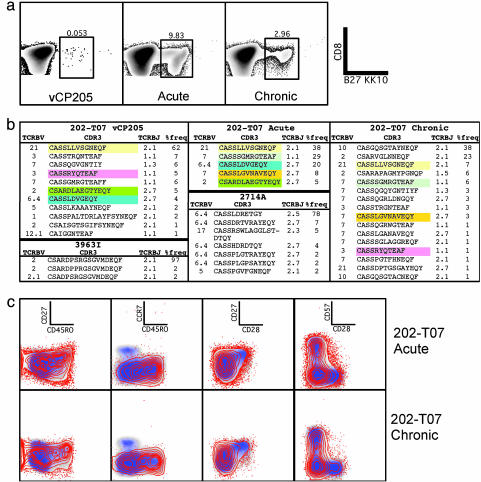
Memory Phenotyping. PBMC were rested overnight and stained with phycoerythrin (PE)-labeled B27/KK10 tetramers for 15 min, followed by mAbs αCD45RO-TRPE, αCD28-APC, and αCD57-Cascade Blue in the following combinations: 1: αCD3-Cy7APC, αCD8-Cy5.5APC, αCD27-Cy5PE, αCD45RA-Cy5.5PE, αCCR7-Cy7PE, and αCD14/19-FITC; and 2: αCD3-Cy5PE, αCD8-Cy7PE, αCD27-FITC, αCCR7-Alexa 680. Although only the particular combinations of mAbs that gave the best discrimination of the various memory phenotypes of the total CD8+ T cells and the tetramer+ population were selected for presentation, all stains could be used to confirm the memory phenotype of the cells being examined. For Figs. 1c, 3c, and 5c, the panels represent, from left to right, the following combinations of markers: CD27/CD45RO, CCR7/CD45RO, CD27/CD28, and CD57/CD28.
Fig. 1.
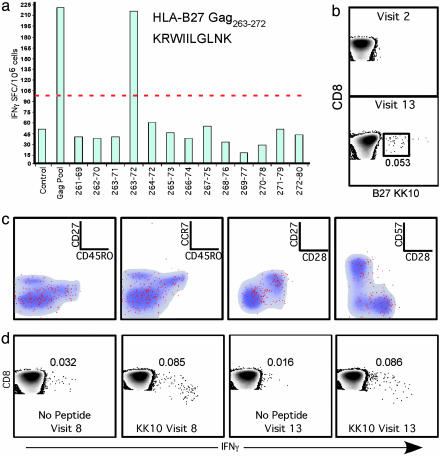
Initial characterization of the vaccine-induced CD8+ T cell response. (a) IFN-γ ELISpot was performed to map the vaccine-induced response in HIV Gag. Either Gag Pool or individual peptides overlapping by a single amino acid spanning the region of HIV Gag p24261–280 were used on PBMC obtained at visit 8. (b) The KK10-specific CD8+ T cells could be detected at visits 8 (data not shown) and 13, but not before the first immunization (visit 2), by using direct staining with B27/KK10 tetramer. The number shown on the plot represents the percentage of B27/KK10+ CD8+ T cells. (c) Memory phenotype of the KK10-specific CD8+ T cells, shown in red, are overlaid onto the total CD8+ T cell memory phenotype (gray/blue density plot). (d) The KK10-specific CD8+ T cells produce IFN-γ after peptide stimulation. The values on the plots represent the percentage frequency of IFN-γ-producing CD8+ T cells.
Fig. 3.
Comparison of 202-T07 KK10-specific CD8+ T cell responses pre- and postinfection. (a) Expansion of KK10-specific CD8+ T cells after HIV infection. CD8 expression is shown on the y axis; B27/KK10 tetramer binding on the x axis. Values shown on the density plots depict the percentage of KK10-specific CD8+ T cells. (b) Vaccine-induced KK10-specific clonotypes are recalled after infection. The table depicts the TCRB variable family, CDR3 sequence, TCRB joining (TCRBJ) family, and percentage frequency of each clonotype obtained from 202-T07 [before infection (202-T07 vCP205), during the acute period (202-T07 acute), and through the chronic infection period (202-T07 chronic)] and the two chronically HIV-infected control subjects, 2714A and 3963I. Matching CDR3 clonotype sequences between different time periods are highlighted in identical colors. (c) Vaccine-induced KK10-specific CD8+ T cells shift to a predominant effector/effector memory phenotype after infection. Red dots/contour overlays depict the B27 KK10 tetramer+ CD8+ T cells. The blue/gray density plot depicts total CD8+ T cells.
Flow Cytometer. Four-color analysis was performed on a FACS-Calibur. Nine- and 10-color analysis was performed on either a FACS Digital Vantage (Becton Dickinson) equipped for 12-color analysis or an LSR II (Becton Dickinson) equipped for 18-color analysis. Between 100,000 and 500,000 events were collected for each sample. Data were analyzed by using flowjo software (Tree Star, San Carlos, CA).
Viral Sequencing. Viral RNAs were isolated from 280-μl patient plasma with the Qiagen (Valencia, CA) viral mini kit. We sequenced plasma virus from an acute visit and 12, 24, and 32 months (chronic) postdiagnosis samples for patient 202-T07. One sample obtained at time of study from 3963I and 2714A subjects was also analyzed. The first strand of DNA was synthesized by using either random hexamer primers or a gene specific primer cgagA 5′-TGATAAAACCTCCAATTCCCCCTAT-3′ according to manufacturer's protocol. Two rounds of PCR were carried out to obtain sufficient quantities of amplification products for further subcloning (PBS1A 5′-TTTGCCTGTACTGGGTCTCTCTGGTT-3′ and cgagA in the first round; PBS1C 5′-GCTTAAGCCTCAATAAAGCTTGCCTT-3′ and cgagB 5′-AATACTGTATCATCTGCTCCTGTATC-3′ in the second round). About 20 clones from each sample were sequenced on a PRISM 3100 Genetic Analyzer (Applied Biosystems).
Supporting Information. For further information, see Figs. 6–9, which are published as supporting information on the PNAS web site.
Results
Study Participant Clinical Course. HIV-negative subject 202-T07 was immunized with a recombinant canarypox vector vCP205. His final vaccination occurred 168 days postfirst immunization (DPFI), and he completed his final study visit on day 364 (visit 13). Eight hundred DPFI (acute visit), the subject returned to the clinic complaining of flu-like symptoms, and he admitted having unprotected anal intercourse with an undisclosed HIV-infected homosexual partner. Testing revealed a positive HIV ELISA antibody test, an indeterminate Western blot, and a plasma viral load of 234,695 copies per ml indicative of recent HIV infection. He began taking antiretroviral therapy that was discontinued on 1,240 DPFI after he became only partially compliant. Although he remained free of symptoms attributable to HIV off therapy, his CD4 count continued to decline and viral replication was not effectively controlled (Supporting Text and Fig. 6 include detailed clinical course).
Characterization of vCP205-Induced Immune Responses. No Env-specific antibody responses were detectable during the vaccine trial (6). Two weeks after the last immunization (visit 8), cytotoxic T cell activity against HIV Gag was detected by 51Cr release assay after antigen-specific in vitro expansion (28% specific lysis; data not shown). This response was mapped by IFN-γ ELISpot to the HLA B*2705-restricted epitope Gag263–272, KRWIILGLNK (KK10; Fig. 1a). The KK10 response was detectable by using HLA-B27 KK10 (B27/KK10) tetramers (Fig. 1b) at both visits 8 (data not shown) and 13 but not before the first vaccination (visit 2). Phenotypically, the B27/KK10 tetramer+ CD8+ T cells comprised central and effector/effector memory cells defined as CD27+CD28+ CCR7+CD45RO+CD57– and CD27–CD28–CCR7–CD45RO– CD57±, respectively, six months after the final vaccination (visit 13) (Fig. 1c). The KK10-specific CD8+ T cells were functional as determined by intracellular cytokine staining for IFN-γ (Fig. 1d), at both visits 8 and 13.
A more detailed analysis of the KK10-specific CD8+ T cells revealed a heterogenous functional profile (Fig. 2a; each sector of the pie chart represents a subset expressing the marker depicted by the concentric colored arcs). The total response to this epitope was 0.1% of all CD8+ T cells; the dominant effector function was TNF-α production (≈75% of responding cells). About two-thirds of responding CD8+ T cells degranulated (CD107+) (10). IL-2 was produced by ≈10% of responding cells. IFN-γ production only accounted for ≈50% of the total response. Therefore, we reexamined the T cell response to full-length Gag by using overlapping peptides with multiparameter flow cytometric analysis, and found a much higher frequency of CD8+ T cell response (0.7%), dominated by cells that only degranulated (Fig. 2b). Additionally, we noted an IL-2-producing population that constituted >25% of the Gag response. The proportion of IFN-γ-producing CD8+ T cells responding to the overlapping Gag peptides (≈0.05%) was equivalent to the proportion of KK10-specific CD8+ T cells. We also observed a substantial CD4+ T cell response to Gag (0.3%, Fig. 2c), dominated by cells that produced IL-2 alone. Only a small number of IL-2+CD4+ T cells produced IFN-γ (0.02%). No T cell responses to HIV Env or Nef (encoded and not contained in the vaccine vector, respectively) were detected. Because insufficient PBMC samples remained to map the epitopes recognized by both the Gag-specific CD4+ and CD8+ T cell response, it can be assumed that subject 202-T07 recognized at least one CD4 epitope and two CD8 epitopes (based on the larger response frequency) after the final immunization dose.
Recall of Vaccine-Induced KK10-Specific CD8+ T Cell Response After Infection. After infection, the B27/KK10 tetramer+CD8+ T cell population had expanded from 0.05% (visit 13; 364 DPFI) to 9.8% of total CD8+ T cells (acute visit, 800 DPFI), but decreased to 2.96% at 1,700 DPFI (chronic visit) (Fig. 3a). The sequence of the TCRB CDR3 region of the B27/KK10 tetramer+CD8+ T cells from T cell lines obtained from visit 13 and cells isolated directly ex vivo from the acute and chronic visits indicate that the dominant vaccine-induced clonotype that expanded in vitro was also the dominant clonotype observed during acute infection (Fig. 3b). Two subdominant vaccine-induced clonotypes were present during acute infection, one of which had become substantially more prevalent. Only two vaccine-induced clonotypes were maintained through to chronic infection, at which time they were both subdominant. The other clonotypes present at the chronic visit appear to have arisen de novo after infection. None of the clonotypes observed in 202-T07 were similar to the KK10-specific CD8+ T cell clonotypes identified in two HIV-infected subjects with HLA-B27-restricted KK10-specific CD8+ T cell responses (Fig. 3b).
After infection, de novo HIV-specific CD8+ T cell responses were detected against various viral epitopes in Gag, Pol, Env, Nef, Vif, and Vpr (Fig. 9), but the KK10-specific response was dominant. One KK10-specific CD8+ T cell line was expanded in vitro from PBMC isolated at visit 13, and two were obtained from PBMC isolated at the acute visit. These cell lines were all composed primarily of the dominant vaccine-induced clonotype and lysed autologous CD4+ lymphoblastoid cell targets either coated with KK10 peptide or infected with the autologous virus isolated from the acute visit (data not shown). These findings indicate that the dominant vaccine-induced clonotype maintained proliferative capacity and cytotoxic ability against autologous viruses after infection. None of these CD8+ T cell lines exhibited noncytolytic suppression of viral replication by using methods described in ref. 11 (Fig. 7).
After infection, the B27/KK10 tetramer+CD8+ cells in subject 202-T07 rapidly switched to a predominantly effector (CD27–CD28–CCR7–CD45RO–CD57+)/effector memory (CD27–CD28–CCR7–CD45RO+CD57±) phenotype (Fig. 3c). A small proportion of the KK10-specific cells retained a central memory (CD27+CD28+CCR7+CD45RO+CD57–) phenotype. Similar phenotypic characteristics were observed in the KK10-specific CD8+ T cells in the control subjects (see Fig. 5c).
Shift in T Cell Functional Profile After Infection. Before infection, the vCP205-induced KK10-specific CD8+ T cells in subject 202-T07 produced predominantly TNF-α upon stimulation. After infection, the KK10-specific CD8+ T cells shifted to an IFN-γ-dominated response (Fig. 4a). Whereas ≈75% of the responding cells degranulated upon stimulation, <50% produced TNF-α. The contribution of IL-2-producing KK10-specific CD8+ T cells dropped to <5%. The KK10-specific CD8+ T cells present during the chronic phase of infection exhibited the same functional profile. This profile was also shared with the HLA B27-restricted KK10 responses in two HIV+ control subjects (Fig. 5a) despite differences in disease courses between these subjects. 2714A is a nonprogressor who displays a high-frequency (≈25%) B27/KK10 tetramer+CD8+ T cell response and is able to control viral load. 3963I is a progressor and has required therapeutic intervention because of declining CD4 counts. The only discernible functional difference between these subjects was that, after in vitro stimulation (data not shown), only 40% and 20% of the tetramer+CD8+ T cells were IFN-γ+CD107+ in subjects 202-T07 and 3963I, respectively, compared with 80% in 2714A. The percentage of responding B27/KK10 tetramer+CD8+ cells in subject 202-T07 did not change between the acute and chronic phase, despite an ≈5-fold lower viral load at the latter time point.
Although CD8+ T cells that only degranulated or produced IL-2 dominated the vaccine-induced response to Gag, IFN-γ production dominated after infection similar to the profile exhibited against the KK10 epitope (Fig. 4b). This functional profile remained consistent into the chronic infection phase and was similar to the HIV Gag-specific CD8+ T cell response profile in 2714A (Fig. 5b).
The most dramatic functional shift observed in subject 202-T07 was in the CD4+ T cell response to HIV Gag (Fig. 4c). Before infection, this response was dominated by IL-2. After infection, the response switched to a TNF-α-dominated response, with a near-complete loss of IL-2 production. The CD4+ T cell response to HIV Gag in 2714A was similarly dominated by TNF-α, and IL-2 was nearly absent; however, a large proportion of the responding CD4+ T cells also produced IFN-γ (data not shown), similar to the chronic Gag-specific CD4+ T cell response in subject 202-T07 (Fig. 4c).
Early Viral Escape at the HLA B27 Restricted KK10 Epitope in Subject 202-T07. To determine whether the immunodominant KK10-specific CD8+ T cell response caused viral evolution, we sequenced the Gag region containing this epitope from autologous plasma-derived virus (Fig. 8) and integrated proviral DNA from PBMC (data not shown).
In the acute infection period, there was no evidence of escape at the KK10 epitope. However, by 32 months after diagnosis, mutation R264G at the anchor residue position 2 had occurred to diminish peptide binding to HLA B27 (12). At this time, KK10-specific CD8+ T cells were still present, albeit with a substantially altered clonal hierarchy (Fig. 3b). We determined that this mutation occurred between the second and third year of infection (data not shown). Escape at this epitope was not found in subject 2714A (19 of 19 clones sequenced; data not shown). Some sequence differences were noted in subject 3963I (KRWIIIGLHK, 23 of 23 clones sequenced; data not shown) that do not affect the binding to HLA-B27 (SYFPEITHI, www.syfpeithi.de).
Discussion
Development of an effective vaccine to suppress the worldwide AIDS epidemic is a major and pressing international goal. Here, we have demonstrated that vaccine-induced T cell responses in an HIV-1 vaccine recipient underwent functional and phenotypic changes and failed to prevent infection and disease progression after homosexual transmission. The vaccine-induced response was composed of both helper and cytotoxic T cell components predicted to be necessary for vaccine-induced protection: killing ability, cytokine production, proliferative capacity, polyclonality, and memory phenotype (1). Moreover, the CD8+ T cell response was directed in part against a highly immunodominant and constrained epitope correlated with long-term nonprogression and infrequent and invariably late-stage viral escape (12–14). Therefore, we could have expected a more favorable outcome after exposure to HIV-1.
The specific functional ability and phenotype required of vaccine-induced T cells to impart protection against HIV infection and disease progression is unknown. In subject 202-T07, the vaccine-induced T cells, in the presence of persistent antigenic stimulation, rapidly acquired functional and phenotypic properties and impairments characteristic of HIV-specific T cells in nonvaccinated HIV-infected individuals. Moreover, as reported for other anti-HIV CD8 responses (15, 16), the functional ability of the B27/KK10 tetramer+CD8+ T cells in 202-T07 was significantly compromised compared with patient 2714A, who controlled viremia more effectively. This persistent impaired functionality of the CD8+ T cell response in 202-T07 occurred despite the presence of an HIV-specific CD4+ response. Even more dramatic was the rapid loss of IL-2 production by the vaccine-induced Gag-specific CD4+ T cells after infection as also observed in 2714A. The loss of IL-2 production after HIV infection could be a consequence of the infection of antigen-specific CD4+ T cells (17, 18), or due to maturational changes in the CD4+ T cell response. Whether the phenotype and functional response pattern detected in this subject is strictly related to the recombinant canarypox vector or will be recapitulated in other vaccine strategies remains to be determined. Additional analysis of clinical (but not immunological) data from other subjects enrolled in recombinant HIV-1 canarypox vaccine trials, who experienced breakthrough infection, suggest that vaccination did not provide any tangible benefits against HIV infection after exposure (19).
Although the frequency and breadth of the vaccine-induced T cell response may have been suboptimal in subject 202-T07, they were similar to those elicited by other candidate HIV vaccines (20, 21) currently being tested in human trials. Importantly, data collected from the simian immunodeficiency virus model (22, 23) and HIV infection (24, 25) suggest that these two parameters are not likely determining factors in protection from infection and progression.
An escape KK10 mutant epitope (R264G) with impaired binding capacity to HLA-B27 was detected within 32 months of infection. This escape, although rare, occurs on average nine years after infectionk and is associated with disease progression in adults and children (12–14). This observation could herald a potential danger intrinsic to vaccine strategies that elicit T cell responses to HIV, as previously suggested in refs. 12 and 26. Therefore, the role of virus escape from vaccine-induced T cell responses versus the ability of those T cell responses to control viral replication in the acute phase will have to be carefully weighed.
As humoral immunity has been shown to correlate with protection from a variety of acute viral illnesses in humans (27) and in the simian immunodeficiency virus model (28, 29), the lack of detectable vaccine-induced HIV-specific neutralizing antibody responses could have likely contributed to the failure of this vaccine to protect from infection. We also could not detect noncytolytic CD8+ suppressive reactivity in KK10-specific CD8+ T cell lines derived from pre- or postinfection time points in 202-T07. The importance of this deficiency remains to be determined in further vaccine trials (11, 30, 31).
At the first study point after infection, HIV-specific antibodies were already present in subject 202-T07, and the subject immediately began antiretroviral therapy. Thus, we could not determine in this individual whether the peak and set point of virus load were affected by the vaccine-induced responses (26, 32–34). However, time to undetectable virus load in 202-T07 was no different from that observed in other individuals initiating antiretroviral therapy during acute infection (35). Viral rebound to 3–6 × 104 copies per ml after cessation of antiretroviral therapy and decline of absolute CD4+ T cell counts to <400 cells per μl at subsequent visits suggest that the vaccine-induced immune response did not contribute substantially to control of virus replication or CD4 cell loss.
Overall rates of transmission per coital act among heterosexual and homosexual groups do not appear to be significantly different (36), suggesting that similar hurdles may exist for the prevention of sexually acquired HIV in both groups. Subject 202-T07 could have been exposed multiple times to HIV during the 4-month period of high-risk behavior before infection. Once again, infection of subject 202-T07 may have been initially delayed but, ultimately, the vaccine-induced response was insufficient to prevent infection.
Although the data presented here are based on a single individual, the findings highlight the critical need to identify correlates of protection and set parameters that can be used as a yardstick to guide effective vaccine development. Until clear correlates are established for protective immunity against HIV-1 infection, it is likely that vaccine strategies that induce T cell responses in the absence of effective antibody responses may be at risk of failing to protect individuals, regardless of the specificity, functionality, frequency, clonality, and breadth of those responses. Thus, even if a vaccine is strongly immunogenic by all measured parameters, proof of principle and phase III trials should be conducted that measure both efficacy and possible immune correlates of protection.
Supplementary Material
Acknowledgments
We thank the HIV-infected patients, the volunteers enrolled in the AIDS Vaccine Evaluation Group/HIV Vaccine Trials Network (HVTN) vaccine trials, and the AIDS Vaccine Evaluation Group/HVTN investigators and clinical personnel who made this study possible. This work was supported by National Institutes of Health Grants 5RO1-AI50483, R01-AI49126, 5U01-AI46725, 3P30-AI28662, and U01-AI41530. D.A.P. is a Medical Research Council (U.K.) Clinician Scientist. Z.T.C. was supported by the Interdisciplinary Research Training Program in AIDS (5T32 AI07392).
Author contributions: M.R.B., G.T., P.G., and G.F. designed research; M.R.B., B.E., D.A.P., A.B., Z.T.C., V.T., S.M.W., D.R.A., and G.F. performed research; M.R., F.G., and D.C.D. contributed new reagents/analytic tools; M.R.B., B.E., D.A.P., A.B., Z.T.C., V.T., G.T., F.G., D.C.D., K.J.W., R.A.K., P.G., and G.F. analyzed data; and M.R.B., K.J.W., R.A.K., P.G., and G.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DPFI, days postfirst immunization; PBMC, peripheral blood mononuclear cells; PE, phycoerythrin; TCRB, T cell receptor B.
Data deposition: The nucleotide sequences of HIV isolates reported in this paper have been deposited in the GenBank database (accession nos. AY834776–AY834894).
Footnotes
Ammaranond, P., Guerin, J., Anderson, B., Doong, C., Finlayson, R., Kelly, M., McMurchie, M., Price, R., Cooper, D. & Kelleher, A. D. 11th Conference on Retroviruses and Opportunistic Infections, Feb. 8–11, 2004, San Francisco, CA.
References
- 1.Pantaleo, G. & Koup, R. A. (2004) Nat. Med. 10, 806–810. [DOI] [PubMed] [Google Scholar]
- 2.Goulder, P. J. & Watkins, D. I. (2004) Nat. Rev. Immunol. 4, 630–640. [DOI] [PubMed] [Google Scholar]
- 3.Kalams, S. A., Buchbinder, S. P., Rosenberg, E. S., Billingsley, J. M., Colbert, D. S., Jones, N. G., Shea, A. K., Trocha, A. K. & Walker, B. D. (1999) J. Virol. 73, 6715–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., von Herrath, M. G. & Schoenberger, S. P. (2003) Nature 421, 852–856. [DOI] [PubMed] [Google Scholar]
- 5.Wherry, E. J., Blattman, J. N., Murali-Krishna, K., van der Most, R. & Ahmed, R. (2003) J. Virol. 77, 4911–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belshe, R. B., Stevens, C., Gorse, G. J., Buchbinder, S., Weinhold, K., Sheppard, H., Stablein, D., Self, S., McNamara, J., Frey, S., et al. (2001) J. Infect. Dis. 183, 1343–1352. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari, G., Neal, W., Ottinger, J., Jones, A. M., Edwards, B. H., Goepfert, P., Betts, M. R., Koup, R. A., Buchbinder, S., McElrath, M. J., et al. (2004) J. Immunol. 173, 2126–2133. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, G., Humphrey, W., McElrath, M. J., Excler, J.-L., Duliege, A.-M., Clements, M. L., Corey, L. C., Bolognesi, D. P. & Weinhold, K. J. (1997) Proc. Natl. Acad. Sci. USA 94, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douek, D. C., Betts, M. R., Brenchley, J. M., Hill, B. J., Ambrozak, D. R., Ngai, K. L., Karandikar, N. J., Casazza, J. P. & Koup, R. A. (2002) J. Immunol. 168, 3099–3104. [DOI] [PubMed] [Google Scholar]
- 10.Betts, M. R., Brenchley, J. M., Price, D. A., DeRosa, S. C., Douek, D. C., Roederer, M. & Koup, R. A. (2003) J. Immunol. Methods 281, 65–78. [DOI] [PubMed] [Google Scholar]
- 11.Tomaras, G. D., Lacey, S. F., McDanal, C. B., Ferrari, G., Weinhold, K. J. & Greenberg, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 3503–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelleher, A. D., Long, C., Holmes, E. C., Allen, R. L., Wilson, J., Conlon, C., Workman, C., Shaunak, S., Olson, K., Goulder, P., et al. (2001) J. Exp. Med. 193, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulder, P. J., Phillips, R. E., Colbert, R. A., McAdam, S., Ogg, G., Nowak, M. A., Giangrande, P., Luzzi, G., Morgan, B., Edwards, A., et al. (1997) Nat. Med. 3, 212–217. [DOI] [PubMed] [Google Scholar]
- 14.Feeney, M. E., Tang, Y., Roosevelt, K. A., Leslie, A. J., McIntosh, K., Karthas, N., Walker, B. D. & Goulder, P. J. (2004) J. Virol. 78, 8927–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appay, V., Nixon, D. F., Donahoe, S. M., Gillespie, G. M., Dong, T., King, A., Ogg, G. S., Spiegel, H. M., Conlon, C., Spina, C. A., et al. (2000) J. Exp. Med. 192, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champagne, P., Ogg, G. S., King, A. S., Knabenhans, C., Ellefsen, K., Nobile, M., Appay, V., Rizzardi, G. P., Fleury, S., Lipp, M., et al. (2001) Nature 410, 106–111. [DOI] [PubMed] [Google Scholar]
- 17.Douek, D. C., Brenchley, J. M., Betts, M. R., Ambrozak, D. R., Hill, B. J., Okamoto, Y., Casazza, J. P., Kuruppu, J., Kunstman, K., Wolinsky, S., et al. (2002) Nature 417, 95–98. [DOI] [PubMed] [Google Scholar]
- 18.Harari, A., Petitpierre, S., Vallelian, F. & Pantaleo, G. (2004) Blood 103, 966–972. [DOI] [PubMed] [Google Scholar]
- 19.Lee, D., Graham, B. S., Chiu, Y. L., Gilbert, P. B., McElrath, M. J., Belshe, R. B., Buchbinder, S. P., Sheppard, H. W., Koblin, B. A., Mayer, K. H., et al. (2004) J. Infect. Dis. 190, 903–907. [DOI] [PubMed] [Google Scholar]
- 20.Shiver, J. W., Fu, T. M., Chen, L., Casimiro, D. R., Davies, M. E., Evans, R. K., Zhang, Z. Q., Simon, A. J., Trigona, W. L., Dubey, S. A., et al. (2002) Nature 415, 331–335. [DOI] [PubMed] [Google Scholar]
- 21.Mwau, M., Cebere, I., Sutton, J., Chikoti, P., Winstone, N., Wee, E. G., Beattie, T., Chen, Y. H., Dorrell, L., McShane, H., et al. (2004) J. Gen. Virol. 85, 911–919. [DOI] [PubMed] [Google Scholar]
- 22.Allen, T. M., Mortara, L., Mothe, B. R., Liebl, M., Jing, P., Calore, B., Piekarczyk, M., Ruddersdorf, R., O`Connor, D. H., Wang, X., et al. (2002) J. Virol. 76, 4108–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mothe, B. R., Horton, H., Carter, D. K., Allen, T. M., Liebl, M. E., Skinner, P., Vogel, T. U., Fuenger, S., Vielhuber, K., Rehrauer, W., et al. (2002) J. Virol. 76, 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betts, M. R., Ambrozak, D. R., Douek, D. C., Bonhoeffer, S., Brenchley, J. M., Casazza, J. P., Koup, R. A. & Picker, L. J. (2001) J. Virol. 75, 11983–11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Addo, M. M., Yu, X. G., Rathod, A., Cohen, D., Eldridge, R. L., Strick, D., Johnston, M. N., Corcoran, C., Wurcel, A. G., Fitzpatrick, C. A., et al. (2003) J. Virol. 77, 2081–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barouch, D. H., Kunstman, J., Kuroda, M. J., Schmitz, J. E., Santra, S., Peyerl, F. W., Krivulka, G. R., Beaudry, K., Lifton, M. A., Gorgone, D. A., et al. (2002) Nature 415, 335–339. [DOI] [PubMed] [Google Scholar]
- 27.Plotkin, S. A. (2001) Pediatr. Infect. Dis. J. 20, 63–75. [DOI] [PubMed] [Google Scholar]
- 28.Baba, T. W., Liska, V., Hofmann-Lehmann, R., Vlasak, J., Xu, W., Ayehunie, S., Cavacini, L. A., Posner, M. R., Katinger, H., Stiegler, G., et al. (2000) Nat. Med. 6, 200–206. [DOI] [PubMed] [Google Scholar]
- 29.Mascola, J. R., Stiegler, G., VanCott, T. C., Katinger, H., Carpenter, C. B., Hanson, C. E., Beary, H., Hayes, D., Frankel, S. S., Birx, D. L., et al. (2000) Nat. Med. 6, 207–210. [DOI] [PubMed] [Google Scholar]
- 30.Walker, C. M. & Levy, J. A. (1989) J. Immunol. 66, 628–630. [PMC free article] [PubMed] [Google Scholar]
- 31.Toso, J. F., Chen, C. H., Mohr, J. R., Piglia, L., Oei, C., Ferrari, G., Greenberg, M. L. & Weinhold, K. J. (1995) J. Infect. Dis. 172, 964–973. [DOI] [PubMed] [Google Scholar]
- 32.Pal, R., Venzon, D., Letvin, N. L., Santra, S., Montefiori, D. C., Miller, N. R., Tryniszewska, E., Lewis, M. G., VanCott, T. C., Hirsch, V., et al. (2002) J. Virol. 76, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amara, R. R., Smith, J. M., Staprans, S. I., Montefiori, D. C., Villinger, F., Altman, J. D., O'Neil, S. P., Kozyr, N. L., Xu, Y., Wyatt, L. S., et al. (2002) J. Virol. 76, 6138–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellors, J. W., Rinaldo, C. R., Jr., Gupta, P., White, R. M., Todd, J. A. & Kingsley, L. A. (1996) Science 272, 1167–1170. [DOI] [PubMed] [Google Scholar]
- 35.Markowitz, M., Vesanen, M., Tenner-Racz, K., Cao, Y., Binley, J. M., Talal, A., Hurley, A., Jin, X., Chaudhry, M. R., Yaman, M., et al. (1999) J. Infect. Dis. 179, 527–537. [DOI] [PubMed] [Google Scholar]
- 36.Royce, R. A., Sena, A., Cates, W., Jr., & Cohen, M. S. (1997) N. Engl. J. Med. 336, 1072–1078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.