Abstract
Triplication of whole autosomes or large autosomal segments is detrimental to the development of a mammalian embryo. The trisomy of human chromosome (Chr) 21, known as Down's syndrome, is regularly associated with mental retardation and a variable set of other developmental anomalies. Several mouse models of Down's syndrome, triplicating 33–104 genes of Chr16, were designed in an attempt to analyze the contribution of specific orthologous genes to particular developmental features. However, a recent study challenged the concept of dosage-sensitive genes as a primary cause of an abnormal phenotype. To distinguish between the specific effects of dosage-sensitive genes and nonspecific effects of a large number of arbitrary genes, we revisited the mouse Ts43H/Ph segmental trisomy. It encompasses >310 known genes triplicated within the proximal 30 megabases (Mb) of Chr17. We refined the distal border of the trisomic segment to the interval bounded by bacterial artificial chromosomes RP23-277B13 (location 29.0 Mb) and Cbs gene (location 30.2 Mb). The Ts43H mice, viable on a mixed genetic background, exhibited spatial learning deficits analogous to those observed in Ts65Dn mice with unrelated trisomy. Quantitative analysis of the brain expression of 20 genes inside the trisomic interval and 12 genes lying outside on Chr17 revealed 1.2-fold average increase of mRNA steady-state levels of triplicated genes and 0.9-fold average down-regulation of genes beyond the border of trisomy. We propose that systemic comparisons of unrelated segmental trisomies, such as Ts65Dn and Ts43H, will elucidate the pathways leading from the triplicated sequences to the complex developmental traits.
Keywords: dosage-sensitive genes, Down's syndrome, Morris water maze, mouse segmental trisomy, quantitative RT-PCR
In human, mouse, and rat genomes, 2–5% of each sequence consists of recent segmental duplications of up to 200 kbp in size (1–3). These tolerated duplications are fixed in populations and represent a potential raw material for evolution of species (4, 5). In contrast, the majority of segmental duplications that occur de novo are strongly selected against because of the associated genetic disorders (6). Extra copies of large autosomal segments and autosomal trisomies are mostly incompatible with human and mouse embryonic development. Only trisomies of human chromosomes (Chr) 21, 18, and 13 survive postnatally as semilethal conditions accompanied by severe developmental anomalies known as Down's syndrome (DS) and Edwards and Patau syndromes (7). The most frequent and thoroughly studied trisomy of Chr21 (DS) is associated with ≈80 clinical phenotypes, a variable subset of which is present in each DS individual. The high phenotypic variability is a characteristic feature of aneuploidy syndromes, but its nature remains unclear. The detailed analysis of rare cases of segmental trisomy 21 led to attempts to define the DS critical region of Chr21 responsible for most DS clinical phenotypes (8, 9). However, studies in human patients as well as in mouse models showed that such straightforward mapping is a difficult if not impossible task (10, 11). Thus, despite the vast amount of detailed clinical, biochemical, and genetic data pertaining to DS and recent availability of the complete, annotated sequence of human Chr21, the mechanism by which the three copies of Chr21 impair development is still largely unknown.
Three mouse models have been introduced specifically to study DS. Ts65Dn is a mouse segmental trisomy of distal 17 megabases (Mb) of Chr16 syntenic to a large portion of human Chr21, which triplicates 104 orthologs of genes of human Chr21 and recapitulates several phenotypic features of DS patients (12). The other two partially trisomic mouse models include Ts1Cje and Ms1Ts65 with smaller subsets of human Chr21 orthologs (13, 14). Recently, a mouse model with one or three copies of the Chr16 interval corresponding to DS critical region was generated by chromosome engineering (11).
To distinguish between the developmental instability hypothesis and specific effects of dosage-sensitive genes, the existing mouse models may be compared with a segmental trisomy of similar size but with unrelated sets of triplicated genes. Here, we evaluated the Ts43H/Ph mouse segmental trisomy unrelated to previous models. We identified the distal boundary of the trisomic segment and showed that Ts43H/Ph triplicates >310 known genes in the proximal 30 Mbp of Chr17. Because cognitive impairment belongs to the few clinical features common to all individuals with DS and is replicated in the Ts65Dn mouse model, we studied the spatial learning of Ts43H/Ph mice in a Morris water maze. Subsequently, we quantified the brain transcription of 20 triplicated genes and 12 adjacent genes present in two copies on the same chromosome.
Methods
Mice. The mouse inbred strains T43H/Ph, t121/Ph, and PWD/Ph were maintained in standard conditions with controlled light and temperature at the Specific Pathogen-Free facility of the Institute of Molecular Genetics, Academy of Sciences of the Czech Republic (Prague). T(16;17)43H belongs to a series of mouse autosomal translocations causing male sterility and aneuploidy (15). The T43H translocation originated by sperm X-irradiation of a (C3Hx101)F1 male, is highly asymmetrical and involves Chr16 and Chr17 (16, 17). The PWD/Ph inbred strain originated from wild mice of Mus musculus musculus (sub)species (18). The T43/t121/PWD trisomic mice (Ts43H) were prepared by crossing +T43/t121+ females with PWD/Ph males and selecting for the offspring with three allelic forms of the Igf2r gene. The mice were used in experiments at 12 months of age. Principles of laboratory animal care (National Institutes of Health guidelines) as well as specific Czech Laws No. 246/1992 and 162/93 Sb (Protection of Animals Against Cruelty) were observed. The Institute of Molecular Genetics of the Academy of Sciences of the Czech Republic is accredited for work with laboratory animals (accreditation no. 1020/454/A/00).
Genotyping. Genomic DNA was prepared as described in ref. 19. A 1-kb region in the Igf2r intronic DMR2 was sequenced and analyzed by lasergene software (DNASTAR, Madison, WI). SNPs from T43H/T43H, t121/t121, and PWD/Ph inbred strains were identified and used to design the allele-specific single nucleotide primer extension assay (SNuPE) primers, S1, S2, and S3, respectively. SNuPE was performed as described in ref. 20 with modifications (21). For SNuPE-based genotyping, a 272-bp fragment from region 2, containing the polymorphic sites, was amplified with primers F1 and R1 and purified on agarose gel. Ten nanograms of the purified PCR product were used as a SNuPE template with 0.5 units of Taq DNA polymerase (MBI Fermentas, St. Leon-Rot, Germany), 1 mM MgCl2, and 1 μCi of (1 Ci = 37 GBq) dTTP (NEN) in a final volume of 10 μl. The primer sequences are available on request.
Whole-Genome Mouse Bacterial Artificial Chromosome (BAC) Microarray Comparative Genome Hybridization (CGH). The mouse whole-genome BAC array used in this study contained 2,803 unique BAC clones from mouse genomic libraries spaced at 1-Mb intervals (22). Array CGH was performed essentially as described in ref. 22. Briefly, control DNAs (T43H/Ph, PWD/Ph, and t121/Ph) and DNA from trisomic mice (Ts43H) (1 μg) were labeled with Cy3-dCTP or Cy5-dCTP using random-primer labeling (BioPrime DNA Labeling Kit, Invitrogen). Then, un-incorporated nucleotides were removed by using Microspin G50 columns (Amersham Pharmacia), and labeled DNAs were combined, mixed with 135 μg of mouse Cot-1 DNA (Invitrogen) and yeast tRNA (600 μg), and precipitated. The samples were resuspended in hybridization buffer [50% formamide/10% dextran sulfate/0.1% Tween 20, 2× SSC (0.3 M sodium chloride/0.03 M sodium citrate)/10 mM Tris·HCl, pH 7.4], denatured at 70°C for 10 min, and incubated at 37°C for 60 min. The slides were prehybridized under coverslips in 30 μl of hybridization buffer containing denatured herring sperm DNA (880 μg) (Sigma) and mouse Cot-1 DNA (135 μg) at 37°C for 60 min in a humid chamber. After removal of the prehybridization solution, the prehybridized labeled genomic DNA was added, and the slide with a coverslip was placed in a slide mailer containing Whatman 3MM paper saturated with 2× SSC and 20% formamide, sealed with parafilm, and incubated at 37°C for 48 h. Slides were washed in PBS/0.05% Tween 20 for 10 min at room temperature, in 50% formamide/2× SSC for 30 min at 42°C, and in PBS/0.05% Tween 20 for 10 min at room temperature and then dried by centrifugation at 150 × g for 5 min.
Image Scanning and Data Processing. Hybridized microarrays were scanned by using a ScanArray 5000XL scanner (Packard BioScience) and processed by using quantarray software (Packard BioScience) according to the manufacturer's instructions. We performed all CGH experiments in a fluorochrome-reversed pair of two-color hybridizations. For all data points, we obtained quadruple measurements derived from duplicate spots on arrays from both hybridizations.
Behavioral Testing. The Morris water maze was adapted for mice. Its size was 80 cm in diameter and 30 cm in depth, surrounded by a 40-cm-high Perspex wall and filled with 25°C water. The animals were trained to find an invisible circular escape platform (11 cm in diameter) hidden 0.5 cm below the surface of 25-cm-deep water. The maze was located in an experimental room rich in environmental cues. If the mice failed to find the hidden platform in <1 min, they were guided to the platform by the experimenter. In both successful and unsuccessful searches, the mice were given 20 s to rest on the platform and to familiarize themselves with the position of the goal. The intervals between individual trials were ≈25 min; the animals were trained in seven daily sessions, with four trials per session. The trajectories were monitored with a computerized tracking system (23), and escape latencies and path lengths were measured and evaluated. On days 5–7, the escape latencies had stabilized, and their average was considered as an asymptotic value. On day 4, at the fifth trial, the escape platform was removed, and the annuli crossings (number of crossings over the position of the goal) were calculated and compared with the number of annuli crossings over similar locations in other quadrants. In the reversal test performed on day 8, the escape platform was placed in the opposite quadrant of the pool, and the mice were given six trials to find it. Finally, a Visible Shifted Platform experiment was conducted on day 9, in which the escape platform was raised 0.5 cm above the water surface and labeled by a colored strip on the circumference and a black-and-white cue hanging above it. Twelve trisomic and 13 wild-type (WT) littermate animals were given six trials each to find the goal, the position of which changed from trial to trial.
Quantitative Expression Analysis. Twenty genes situated proximally from the Ts43H breakpoint within the trisomic segment of Chr17 and 12 genes distal to this region were selected for quantitative real-time RT-PCR analysis from brain-specific EST libraries. The primers were designed by using the lightcycler probe design software (Roche) to span at least one intron to avoid false signal due to DNA contamination. Total RNA (3 μg) was reverse-transcribed with random hexamers in a 25-μl final volume. A 1 μl-sample of cDNA, diluted 1/4, was used as a template in real-time RT-PCR with the FastStart DNA Master SYBR Green I kit (Roche). The same aliquots of reverse-transcription reactions without added reverse transcriptase were used as negative controls. Three housekeeping genes, β-actin, Gapdh, and Hprt were tested as standards for normalization. Because β-actin and Hprt proved to have an almost identical expression profile as Gapdh, the latter was chosen for normalization. The data were analyzed by using lightcycler software (Version 3.5.3, Roche). Three Ts43H female adult brains were compared with three WT euploid female siblings, and each quantitation was performed in duplicate on the independent cDNA samples, creating six values for Ts43H mice and the same number for their control siblings.
Statistical Analysis. Statistical evaluations were performed by using statistica software (StatSoft, Tulsa, OK). The comparison of the means was performed by using ANOVA or unpaired two-sample t test as indicated in Results.
Results
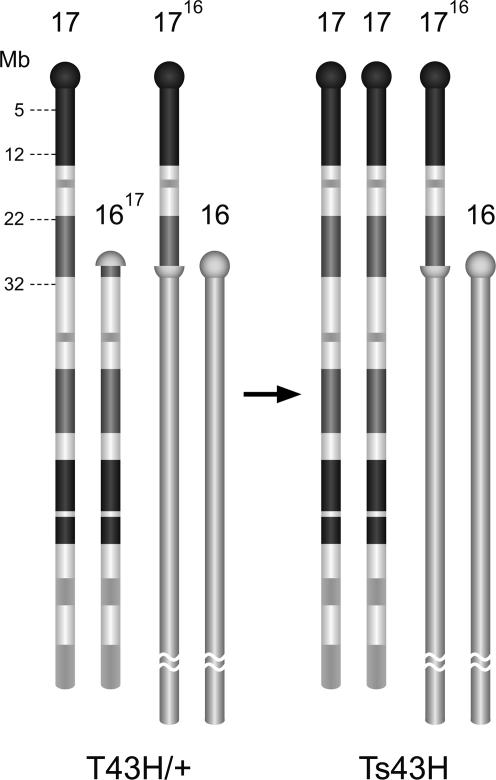
Generation of Viable Ts43H Mice with Segmental Trisomy of Chr17. Mice with Ts43H partial trisomy were identified among the progeny of crosses of female T43H/+ reciprocal translocation heterozygotes with WT males. Because of adjacent-2 disjunction from the T43/+ translocation quadrivalent, the Ts43H trisomics possess two intact Chr17s and one modified Chr16 with 30 Mb of the proximal part of Chr17 on the top of its centromeric heterochromatin (Fig. 1). The associated segmental monosomy of Chr16 deletes the most proximal tip of centromeric heterochromatin. In earlier cytogenetic studies, the Ts43H trisomics were recognized by the presence of a marker 1716 chromosome with an interstitial band of centromeric heterochromatin 16 and by the simultaneous absence of the other translocation product, Chr1617, which was the smallest marker chromosome in the complement (Fig. 1 and ref. 24). Here, we used the allele-specific SNuPE to genotype SNPs of the Igf2r gene as described in ref. 21. The +T43/t121+ females were crossed with PWD males, and the offspring with Ts43H trisomy were recognized by the simultaneous presence of three allelic forms, T43H, t121, and PWD, of the Igf2r gene. At weaning, 20.7% of the progeny were T43/t121/PWD segmental trisomics (data not shown).
Fig. 1.
The scheme of chromosomes involved in T(16;17)43H translocation and Ts43H segmental trisomy. One translocation breakpoint is located within centromeric heterochromatin of Chr16, and the other is in the B3 band of Chr17. The segmental (partial) trisomy of proximal 30 Mb of Chr17 is associated with monosomy for the tip of centromeric heterochromatin of Chr16.
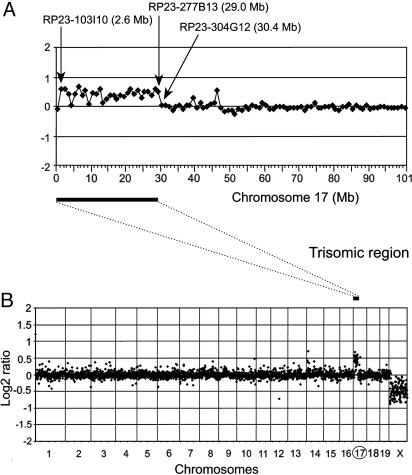
T43H Breakpoint Defined by BAC-Array CGH. The segmental trisomy Ts43H/Ph encompasses the proximal part of Chr17. Genetically, the distal border of the trisomic interval is defined by the position of the T43H translocation breakpoint, tightly linked to the H2-K gene (no recombinant among 218 N2 animals) (24). Because reciprocal translocations can potentially suppress re-combination in the vicinity of the translocation breakpoint, we performed physical mapping of the T43H breakpoint by microarray-based CGH (22). The global CGH analysis was performed to compare the DNA content between Ts43H trisomic individuals and, respectively, T43H/Ph, PWD/Ph, and t121/Ph euploid (disomic) parental strains. All six analyses showed the same profile as presented in Fig. 2. We compared male and female DNAs as an internal control of each experiment (Fig. 2). The mean log2 ratios of the ChrX (male vs. female hybridization) ranged between –0.42 and –0.47 compared with an ideal value of –1 for a 1:2 ratio. The underestimation of the ChrX ratio was reported previously in human-array CGH experiments and is thought to result from incomplete suppression of repetitive sequences in the X-chromosomal BACs or from cross-hybridization of autosomal or Y-chromosomal sequences with strong homology to ChrX (25). Analysis of the ratio profile confirmed the presence of a trisomic region in the Ts43H mouse covering the proximal 30 Mb of Chr17. The mean log2 ratios of the trisomic region ranged between 0.3 and 0.4 compared with an ideal value of 0.58 for a 3:2 ratio. The distal end of the Ts43H trisomic region was localized to the interval bounded by BACs RP23-277B13 (linear location 29.0 Mb) and RP23-304G12 (linear location 30.4 Mb). The CGH mapping was further specified by subsequent genomic SNP analysis of the Cbs gene (location 30.2 Mb) coding for cystathionine-β-synthase. Sequencing of PCR products, by using primers Cbs7 (5′-GGCCATGACCCTGCTTAG-3′) and Cbs8 (5′-ATGCCTTAGATGTGGTGATGG-3′), revealed a sequence polymorphism in the region 30212936 –30212984 bp on Chr17 (www.ensembl.org/Mus_musculus). Sequence analysis of genomic DNA showed that the trisomic individuals do not carry the T43H allele of the Cbs gene and are therefore disomic for this locus. Thus, the distal end of the Ts43H trisomic region must be upstream of the Cbs gene.
Fig. 2.
Array CGH analysis of Chr17 segmental trisomy, showing hybridization between DNA from Ts43H (male) and t121 (female). (B) CGH profile of all autosomes and the X-chromosome, showing the copy number gain for Chr17 and copy number loss for ChrX. (A) CGH profile of the Chr17. The trisomic region is in between RP23–103l10 (linear location 2.6 Mb) and RP23–277B13 (linear location 29.0Mb). The translocation T(16;17)43H breakpoint is situated in the interval between BACs RP23-277B13 and RP23-304G12.
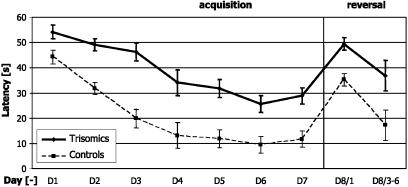
Deficit of Spatial Learning of Ts43H Trisomics in the Morris Water Maze Test. Escape latencies were used as a critical indicator of spatial learning, i.e., the ability to acquire, store, and recall spatial information. The performance of 12-month-old mice significantly improved over 7 training days in both groups, but WT littermates of the Ts43H mice showed significantly faster progress in solving the task (Fig. 3). Their escape latencies decreased significantly (P < 0.01, unpaired two-sample t test) from the value of 43 ± 3 s (mean ± SEM) on day 1 to the asymptotic level of 11 ± 3 s on days 5–7. Their trisomic siblings improved their performance, too, from 54 ± 3 on day 1 to the asymptotic level 29 ± 3 s that was significantly higher (P < 0.05, unpaired two-sample t test) than that of controls. Two-way ANOVA with repeated measures on days showed significant main effects of groups F(1, 23) = 30.72 (P < 0.01), of days F(6, 138) = 42.40 (P < 0.01), and no significant interaction F(6, 138) = 1.94 (P = 0.56). Post hoc tests indicated that trisomic animals had significantly longer escape latencies than control mice on days 1–7 despite considerable variability between individual animals (Fig. 4A).
Fig. 3.
Learning curves of trisomic (solid line) and WT (dashed line) animals. Average latencies (±SEM) show that trisomics performed significantly worse than controls from day 1 on and that their performance did not improve as much as in controls during further training. The trisomic animals were initially ≈10% slower than controls, but their escape latencies were >2-fold longer on day 6. The results of the reversal experiment are shown on day 8. Escape latencies equal f (group, day).
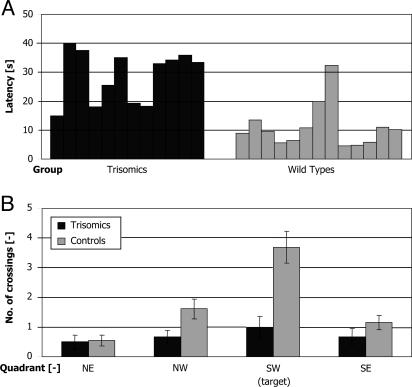
Fig. 4.
Performance of trisomic and WT mice in the Morris water maze. (A) Asymptotic escape latencies of individual trisomic (black) and WT (gray) animals. Asymptotic escape latencies were taken as average of escape latencies on the last three consecutive days on which the performance did not significantly differ (days 5–7). Note the increased variability of the performance of trisomic animals. (B) Annuli crossings of trisomic (black) and WT (gray) animals on day 4 during 30-s probe trials. Control animals passed more frequently across the target annulus than across the symmetrical ones. Trisomic animals visited each quadrant with similar frequency as controls, but they did not pass more frequently across the target annulus.
Probe trials. In probe trials, the animals were allowed to swim for 1 min in an empty pool, and the time spent in each quadrant and the number of annuli crossings were evaluated. In this way, the accidental finding of the target platform was excluded. The quadrant search indicated that both groups preferred the west part of the pool, but only the WT mice spent significantly more time in the goal quadrant than the trisomic mice (t23 = 3.3, P < 0.05). Annuli crossings (Fig. 4B) revealed a more apparent between-group difference. Control animals crossed the target position in the correct quadrant significantly more often than trisomic animals on day 4 both in 30-s (t23 = 4.1, P < 0.01) and 60-s (t23 = 3.9, P < 0.01) searches. Control animals also crossed the target position in the correct quadrant significantly more often than in places corresponding to a similar position in other quadrants.
Reversal experiment. In the reversal experiment, the escape platform was placed into the opposite quadrant of the pool. This arrangement is sensitive to the type of search strategy. Whereas animals with a cognitive spatial map performed badly in the first reversal trial and improved their performance rapidly in following trials, the performance of impaired animals remained unchanged. The mean escape latencies on day 7, the first reversal trial on day 8, and the average of reversal trials 3–6 on day 8 were evaluated and compared by using two-way ANOVA with repeated measures on trials. The escape latencies of the first reversal trials were significantly longer than the mean escape latencies of day 7 in both groups, but only control animals improved their performance significantly in following reversal trials; their first reversal trial latency was significantly different from the mean of reversal trials 3–6 on day 8 [F(2, 36) = 8.55, P < 0.01]. The trisomic animals did not improve significantly under the same conditions.
Visible platform experiment. Search of a visible platform allows comparison of sensorimotor abilities of both groups of animals. Means of trials 3–6 performed on day 9 were compared by using one-way ANOVA. No significant difference was found (t23 = 1.9, P > 0.05) between trisomic and control animals in this task. The relatively long latency of escape to the visible platform is probably due to the fact that it was a novel task and that the goal position was changing from trial to trial.
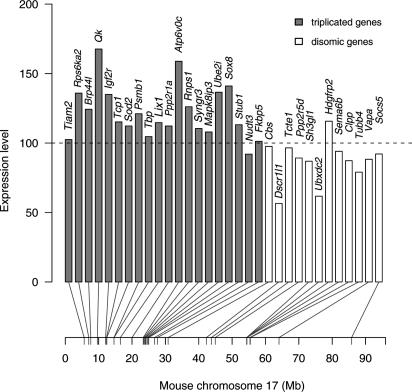
Brain Expression of Triplicated Genes. To evaluate the brain transcription of genes within the triplicated region of Chr17, 20 genes in the interval 0–30 Mb were selected, based mostly on their abundant/specific expression in brain (Fig. 5). A set of 12 genes distal to this interval served as disomic controls. The whole-brain expression levels of selected genes in trisomic animals were compared with the values obtained from their euploid control littermates. The average level of transcription of triplicated genes in Ts43H mice was increased 1.2-fold compared with control euploid siblings. Overall, the level of expression of triplicated genes was variable, with only two genes, Qk and Atp6v0c, reaching or exceeding the expected (based on dosage) 150% level. The expression levels of three genes, Nudt3 (26.2 Mb), Fkbp5 (27.0 Mb), and disomic Cbs (30.2 Mb), located most closely to the translocation breakpoint, did not differ from controls. Two genes, Dscr1l1 and Ubxdc2, in the disomic part of Chr17 were considerably down-regulated in trisomic brains. The average level of expression of 12 disomic Chr17 genes was down-regulated 0.9-fold compared with euploid siblings.
Fig. 5.
Relative expression levels of 32 genes on Chr17 in brains of Ts43H mice. Genes are arranged according to their position on Chr17 with the centromere on the left. The expression level 100 corresponds to the values obtained in control euploid littermates. The expression levels of 20 triplicated genes (gray columns) were variable with 1.2-fold change compared to euploid littermates. The average expression of 12 disomic genes was 0.9-fold down-regulated with regard to euploid controls.
Discussion
Despite the vast amount of detailed clinical, biochemical, and genetic data pertaining to DS and the availability of the complete, annotated sequence of human Chr21 (26, 27), the molecular mechanisms of adverse effects of a supernumerary Chr21 on human development remain elusive. Two working hypotheses have been proposed. According to the “dosage-sensitive genes” hypothesis, a subset of genes on a triplicated chromosome is directly responsible for particular pathological traits associated with trisomy. Consequently, the DS critical region was defined as a minimal interval of Chr21 that carries the dosage-sensitive genes necessary and sufficient for typical features of DS individuals (8, 9, 28). The observations of cardiac pathology, malformation of cerebellum with reduced granular cell density, the deficit of cognitive functions, or characteristic craniofacial dysmorphology in DS individuals as well as in mouse models were in accord with the hypothesis. However, the attempts to delineate DS critical region more precisely and to map the individual loci responsible for particular complex traits mostly failed (9, 11). According to an alternative hypothesis, the overexpression of a large number of triplicated genes causes disruption of genetic homeostasis that impairs gene regulation of developmental processes (29–31). We suggest that comparison of Ts43H segmental trisomy introduced in this study with nonoverlapping segmental trisomies of Chr16 could help distinguish between the complex phenotype anomalies representing a putative genomic response to aneuploidy and those features that can be directly ascribed to particular triplicated genes or their combination (32).
The BAC array CGH and SNP analysis of the Cbs gene localized the T43H breakpoint in between the 29.0 and 30.2 Mb interval on Chr17, which delimited the distal end of the Ts43 trisomic segment. The refined mapping allowed us to determine the minimum (318) and the maximum (330) number of triplicated genes. Adjacent and overlapping the T43H breakpoint is a region of synteny to human Chr21 encompassing 21 gene orthologs. Of them, nine genes (D17Ertd488e, Abcg1, Tff3, Tff2, Tff1, Tmprss3, Ubash3a, Tsga2, and Slc37a1) are located within the minimum estimate trisomy interval, whereas the other six genes (Cn9a, Q8CB29, Wdr4, 1500032D16Rik, Q9D2L2, and Pknox) are located in the gray zone between 29.0 and 30.2 Mb, and their triplication status is thus uncertain. The remaining six genes (Cbs, U2af1, Cryaa, Snfllk, Hsf2bp, and 2600005C20Rik) are in the disomic part of Chr17, distal to the Ts43H breakpoint. The Ts43H trisomy is larger and carries more known genes than Ts65Dn, which spans 23.3 Mb with 133 genes. The Ts43H displays even a higher number of triplicated genes than human trisomy 21, which triplicates 269 known genes in 46.9 Mb (ensembl, Mouse Genome Assembly, National Center for Biotechnology Information Build 35, and www.ensembl.org/Homo_sapiens/mapview?chr=21).
Segmental trisomy of Chr16 in the Ts65Dn mouse has been widely used as a model in studies of neurological disorders associated with DS (33). Place navigation learning in the Morris water maze was found to be severely impaired in the Ts65Dn mice (12, 34), but their deficit also extended to the cued version of the task. The thigmotaxic behavior forcing them to swim around the circumference of the pool (35, 36) could account for their failure to find the escape platform inside the pool. This result was not the case in the present experiments with the Ts34H mice that were readily entering the inner parts of the pool. The Ts65Dn mice are hyperactive (12, 37), whereas the Ts34H mice had the same exploratory activity in the open field as their WT littermates. Nevertheless, the spatial learning deficit of Ts65Dn and Ts43H mice is comparable.
A poorly understood feature of DS and murine segmental trisomies is the high incidence of associated prenatal mortality (38, 39). The majority of pregnancies with human trisomy 21 end as spontaneous abortions. The Ts43H/Ph segmental trisomy was recovered in 11% of adult progeny of Ts43H/+ male heterozygotes, whereas the same cross yielded the expected 50% of trisomic 18-day embryos (39) The viability of Ts43H trisomy was markedly dependent on the genetic background, falling to zero on the C57BL/10 background (J. Čapková, S.G., and J.F., unpublished data).
Recently, a series of works reported global gene expression in various tissues of Ts65Dn and Ts1Cje trisomics as well as in brain samples and cell lines of DS fetuses (40–45). All works confirmed elevated average expression of triplicated genes, albeit within a wide range of 1.1- to 1.5-fold. Conflicting results were reported on the variation within the group of triplicated genes and on the effect of trisomy on the gene expression in the rest of the genome. Our observation of 1.2-fold average overexpression of triplicated genes in Ts43H differed from the expression studies of adult cerebellum in Ts65Dn and newborn brains in Ts1Cje, which showed an ≈1.5-fold change. However, even these studies differed in the effect of trisomy on dys-regulation of disomic genes, which was not detected in the Ts1Cje study (40) but affected Ts65Dn trisomics (44, 45) and DS (42) individuals. The expression of three genes (Tiam2, Nudt3, and Fkbp5) of 20 analyzed triplicated genes and the Igf2r imprinted gene reported previously (21) was compensated, showing the expression ratio close to 1.0. It will be interesting to elaborate on a possible position effect of interrupted centromeric heterochromatin of Chr16 on the adjacent genes (Nudt3 and Fkbp5).
Further analysis of the global gene expression pattern at different stages of development in several tissues is needed to be able to answer questions evoked by the observed 0.9-fold down-regulation of 12 disomic genes in the diploid part of Chr17. The other obvious goal will be to analyze the phenome of different trisomies involving Chr16 and Chr17 and to compare it with transcriptome changes.
Acknowledgments
We thank Dr. M. F. Lyon for helpful comments on the manuscript and Drs. P. Divina, R. Ivanek, R. Storchova, and Z. Trachtulec for discussions. This work was supported by Ministry of Education, Youth, and Sports of the Czech Republic Grant No. 1M0512-1M638805002, Grant Agency of the Academy of Sciences Grants AVOZ50111922 and AVOZ50110509, and Grant Agency of the Czech Republic Grant 309/03/0715. J.F. was supported as an International Scholar of the Howard Hughes Medical Institute by Grant 55000306.
Author contributions: J.B. and J.F. designed research; T.V., M.O., S.G., R.B., N.C., and J.F. performed research; T.V., M.O., S.G., P.S., R.B., N.C., A.B., J.B., and J.F. analyzed data; N.C. and A.B. contributed new reagents/analytic tools; and J.B. and J.F. wrote the paper.
Abbreviations: BAC, bacterial artificial chromosome; CGH, comparative genome hybridization; Chr, chromosome; DS, Down's syndrome; Mb, megabases; SNuPE, single nucleotide primer extension assay.
References
- 1.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. A., Church, D. M., Ventura, M., Rocchi, M. & Eichler, E. E. (2004) Genome Res. 14, 789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuzun, E., Bailey, J. A. & Eichler, E. E. (2004) Genome Res. 14, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
- 5.Lynch, M. (2002) Science 297, 945–947. [DOI] [PubMed] [Google Scholar]
- 6.Emanuel, B. S. & Shaikh, T. H. (2001) Nat. Rev. Genet. 2, 791–800. [DOI] [PubMed] [Google Scholar]
- 7.Pearson, P. L. (2001) in Encyclopedia of Genetics, eds. Brenner, S. & Miller, J. H. (Academic, London), Vol. 4, pp. 2056–2058. [Google Scholar]
- 8.Delabar, J. M., Theophile, D., Rahmani, Z., Chettouh, Z., Blouin, J. L., Prieur, M., Noel, B. & Sinet, P. M. (1993) Eur. J. Hum. Genet. 1, 114–124. [DOI] [PubMed] [Google Scholar]
- 9.Korenberg, J. R., Chen, X. N., Schipper, R., Sun, Z., Gonsky, R., Gerwehr, S., Carpenter, N., Daumer, C., Dignan, P., Disteche, C., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 4997–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves, R. H., Baxter, L. L. & Richtsmeier, J. T. (2001) Trends Genet. 17, 83–88. [DOI] [PubMed] [Google Scholar]
- 11.Olson, L. E., Richtsmeier, J. T., Leszl, J. & Reeves, R. H. (2004) Science 306, 687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves, R. H., Irving, N. G., Moran, T. H., Wohn, A., Kitt, C., Sisodia, S. S., Schmidt, C., Bronson, R. T. & Davisson, M. T. (1995) Nat. Genet. 11, 177–184. [DOI] [PubMed] [Google Scholar]
- 13.Sago, H., Carlson, E. J., Smith, D. J., Kilbridge, J., Rubin, E. M., Mobley, W. C., Epstein, C. J. & Huang, T. T. (1998) Proc. Natl. Acad. Sci. USA 95, 6256–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dierssen, M., Fillat, C., Crnic, L., Arbones, M., Florez, J. & Estivill, X. (2001) Physiol. Behav. 73, 859–871. [DOI] [PubMed] [Google Scholar]
- 15.Lyon, M. & Meredith, R. (1966) Cytogenetics 5, 335–354. [DOI] [PubMed] [Google Scholar]
- 16.Searle, A. G., Ford, C. E., Evans, E. P., Beechey, C. V., Burtenshaw, M. D., Clegg, H. M. & Papworth, D. G. (1974) Mutat. Res. 22, 157–174. [DOI] [PubMed] [Google Scholar]
- 17.Beechey, C. & Evans, E. (1996) in Genetic Variants and Strains of the Laboratory Mouse, eds. Lyon, M., Rastan, S. & Brown, S. (Oxford Univ. Press, Oxford), Vol. 2, pp. 1469–1470. [Google Scholar]
- 18.Gregorova, S. & Forejt, J. (2000) Folia Biol. (Prague) 46, 31–41. [DOI] [PubMed] [Google Scholar]
- 19.Forejt, J. & Gregorova, S. (1992) Cell 70, 443–450. [DOI] [PubMed] [Google Scholar]
- 20.Szabo, P. & Mann, J. (1995) Genes Dev. 9, 3097–3108. [DOI] [PubMed] [Google Scholar]
- 21.Vacik, T. & Forejt, J. (2003) Genomics 82, 261–268. [DOI] [PubMed] [Google Scholar]
- 22.Chung, Y. J., Jonkers, J., Kitson, H., Fiegler, H., Humphray, S., Scott, C., Hunt, S., Yu, Y., Nishijima, I., Velds, A., et al. (2004) Genome Res. 14, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaminsky, Y. & Krekule, I. (1997) Physiol. Res. 46, 223–231. [PubMed] [Google Scholar]
- 24.Forejt, J., Capkova, J. & Gregorova, S. (1980) Genet. Res. 35, 165–177. [DOI] [PubMed] [Google Scholar]
- 25.Fiegler, H., Carr, P., Douglas, E. J., Burford, D. C., Hunt, S., Scott, C. E., Smith, J., Vetrie, D., Gorman, P., Tomlinson, I. P. & Carter, N. P. (2003) Genes Chromosomes Cancer 36, 361–374. [DOI] [PubMed] [Google Scholar]
- 26.Hattori, M., Fujiyama, A., Taylor, T. D., Watanabe, H., Yada, T., Park, H. S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D. K., et al. (2000) Nature 405, 311–319. [DOI] [PubMed] [Google Scholar]
- 27.Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A. & Deutsch, S. (2004) Nat. Rev. Genet. 5, 725–738. [DOI] [PubMed] [Google Scholar]
- 28.Rahmani, Z., Blouin, J. L., Creau-Goldberg, N., Watkins, P. C., Mattei, J. F., Poissonnier, M., Prieur, M., Chettouh, Z., Nicole, A., Aurias, A., et al. (1989) Proc. Natl. Acad. Sci. USA 86, 5958–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro, B. L. (1997) Hum. Genet. 99, 421–423. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro, B. L. (1983) Am. J. Med. Genet. 14, 241–269. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard, M. A. & Kola, I. (1999) J. Neural Transm. 57, Suppl., 293–303. [PubMed] [Google Scholar]
- 32.Forejt, J., Vacik, T. & Gregorova, S. (2003) Comp. Funct. Genomics 4, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davisson, M. & Costa, A. (1999) in Mouse Models in the Study of Genetic Neurological Disorders, ed. Popko, K. (Kluwer Academic/Plenum, New York), pp. 297–327.
- 34.Holtzman, D. M., Santucci, D., Kilbridge, J., Chua-Couzens, J., Fontana, D. J., Daniels, S. E., Johnson, R. M., Chen, K., Sun, Y., Carlson, E., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 13333–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escorihuela, R. M., Vallina, I. F., Martinez-Cue, C., Baamonde, C., Dierssen, M., Tobena, A., Florez, J. & Fernandez-Teruel, A. (1998) Neurosci. Lett. 247, 171–174. [DOI] [PubMed] [Google Scholar]
- 36.Escorihuela, R. M., Fernandez-Teruel, A., Vallina, I. F., Baamonde, C., Lumbreras, M. A., Dierssen, M., Tobena, A. & Florez, J. (1995) Neurosci. Lett. 199, 143–146. [DOI] [PubMed] [Google Scholar]
- 37.Coussons-Read, M. E. & Crnic, L. S. (1996) Behav. Genet. 26, 7–13. [DOI] [PubMed] [Google Scholar]
- 38.Epstein, C. J., Cox, D. R. & Epstein, L. B. (1985) Ann. N.Y. Acad. Sci. 450, 157–168. [DOI] [PubMed] [Google Scholar]
- 39.Gregorova, S., Baranov, V. & Forejt, J. (1981) Folia Biol. (Prague) 27, 171–177. [PubMed] [Google Scholar]
- 40.Amano, K., Sago, H., Uchikawa, C., Suzuki, T., Kotliarova, S. E., Nukina, N., Epstein, C. J. & Yamakawa, K. (2004) Hum. Mol. Genet. 13, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 41.Lyle, R., Gehrig, C., Neergaard-Henrichsen, C., Deutsch, S. & Antonarakis, S. E. (2004) Genome Res. 14, 1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.FitzPatrick, D. R., Ramsay, J., McGill, N. I., Shade, M., Carothers, A. D. & Hastie, N. D. (2002) Hum. Mol. Genet. 11, 3249–3256. [DOI] [PubMed] [Google Scholar]
- 43.Mao, R., Zielke, C. L., Zielke, H. R. & Pevsner, J. (2003) Genomics 81, 457–467. [DOI] [PubMed] [Google Scholar]
- 44.Saran, N. G., Pletcher, M. T., Natale, J. E., Cheng, Y. & Reeves, R. H. (2003) Hum. Mol. Genet. 12, 2013–2019. [DOI] [PubMed] [Google Scholar]
- 45.Kahlem, P., Sultan, M., Herwig, R., Steinfath, M., Balzereit, D., Eppens, B., Saran, N. G., Pletcher, M. T., South, S. T., Stetten, G., et al. (2004) Genome Res. 14, 1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]