The publishers would like to apologize for an error in one of the labels in Figure 1B. The correct complete figure and its legend are given on the next page.
Figure 1.
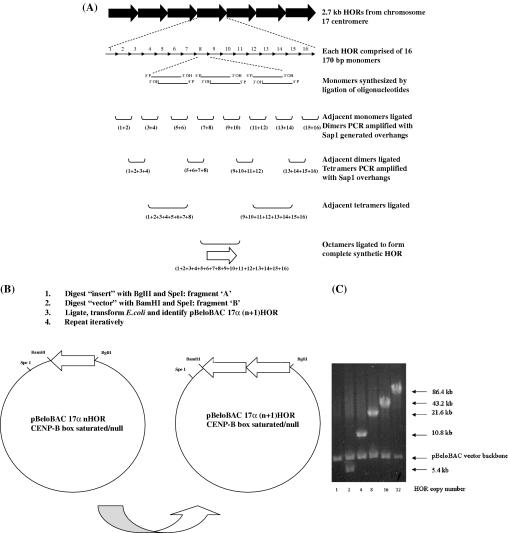
(A) Outline of iterative scheme for synthesis of mutated versions of D17Z1 alpha-satellite arrays. Each of the 16 individual monomers comprising a single 2.7 kb higher-order repeat (HOR) was synthesized as 2–3 oligonucleotide pairs (60–100 bp each), which were directly ligated together and gel purified. Adjacent HORs were subsequently ligated to form dimers as shown, and PCR modified to introduce SapI recognition sites at both ends as appropriate. Digestion with SapI allows seamless ligation of adjacent dimers to create tetramers without introduction of extraneous non-alpha-satellite sequences. Two additional rounds of serial ligation resulted in formation of a complete synthetic HOR, which was subcloned into the BAC vector pBeloBAC. (B) Outline of scheme for directional multimerization of mutated HORs. A synthetic, 86 kb alpha-satellite array consisting of 32 tandem copies of the HOR was created as follows. pBAC17α1 was digested with BglII and SpeI and the alpha-satellite containing fragment (fragment ‘A’) was isolated and gel purified. The same construct was separately digested with BamHI and SpeI, and the larger fragment (fragment ‘B’) was isolated and gel purified. Ligation of fragment ‘A’ to fragment ‘B’ is directional, resulting in head-to-tail multimerization of adjacent repeats. The resulting pBAC17α(n + 1) construct was then isolated following transformation of the ligation reaction into E.coli. This process was repeated iteratively to create the final pBAC17α32 arrays. (C) Pulsed-field gel electrophoresis (PFGE) analysis of intermediates in the construction of 17α32 HOR/BeloBAC constructs. Each intermediate was digested with NotI, which drops out the entire subcloned alpha-satellite array from the pBeloBAC vector backbone. Each lane shows the completed intermediate en route to 32 copies of the mutated HOR. The number beneath each lane corresponds to the number of copies of the HOR in each construct, and the arrows to the right indicate the size of the alpha-satellite insert and the vector backbone. The insert in lane labeled 1 is 2.7 kb and therefore too small to be resolved by PFGE.