Abstract
Liver cancer is one of the leading causes of cancer-associated mortalities worldwide, partly due to the absence of effective therapeutic targets and diagnostic biomarkers. Therefore, novel molecular targets are critical to develop new therapeutic approaches for liver cancer. In the present study, ceramide synthase-4 (CERS4) was investigated as a novel molecular target for liver cancer. High expression of CERS4 in liver cancer tissues was detected by reverse transcription polymerase chain reaction and western blot analysis. Subsequently, CERS4 was silenced by lentivirus-mediated RNA interfere, and the proliferation rates of liver cancer cells were significantly suppressed (P<0.001). In addition, the weight and volume of the tumors were reduced subsequent to silencing of CERS4 in liver cancer cells, revealed by an in vivo study using Balb/c nude mice. In addition, the nuclear factor (NF)-κB signaling pathway was affected following knockdown of CERS4 in liver cancer cells. The present results proposed that CERS4 is an important regulator of liver cancer cell proliferation and indicated that CERS4 may be a potential anticancer therapeutic target and a promising diagnostic biomarker for human liver cancer.
Keywords: ceramide synthase-4, liver cancer, cell proliferation
Introduction
Hepatocellular carcinoma (HCC), the fifth most common cancer worldwide, is also the third leading cause of cancer-associated mortalities (1,2). It accounts for ~695,900 mortalities per year, half of which occur in China, as a result of the high chronic hepatitis B virus infection incidence (3). At present, surgical resection remains the primary approach for treating HCC. However, <20% of patients receive timely radical surgical resection mainly due to advanced cancer, which includes high degrees of malignancy, early metastasis and the lack of effective therapies (4,5). Therefore, it is desirable to reveal the molecular mechanisms of carcinogenesis, to identify novel prognostic markers that may detect HCC earlier or facilitate the development of effective therapeutic strategies, including antibody or immune-based treatments for advanced cancer.
Numerous transcription factors or signaling pathways, including P53, Wnt signaling pathway key members [including glycogen synthase kinase-3β (Gsk-3β) and β-catenin] and nuclear factor (NF)-κB signaling, contribute to the proliferation of cancer cells (6–8). Particularly, NF-κB signaling, including NF-κB transcription factor, inhibitor of κ-light polypeptide gene enhancer in B-cells kinase γ (Ikbkg) and TRAF family member associated NF-κB activator (Tank), is involved in several aspects of tumorigenesis, including the prevention of apoptosis, an increase in the metastatic potential of tumor cells and cancer cell survival and proliferation (9,10). Although these observations demonstrated that NF-κB signaling had an important role in liver cancer tumorigenesis and cells proliferation, its upstream regulators remain to be elucidated.
In comparison with non-transformed cells, carcinoma cells exhibit increased metabolic autonomy in taking up and metabolizing nutrients, which support cell growth and proliferation (11). In the plasma membrane, sphingolipids, together with cholesterol, form lipid microdomains, which are hypothesized to function as structural scaffolds and platforms for signal transduction. The biosynthesis of this plasma membrane is essential for cell growth and proliferation (12,13). Sphingolipids, particularly ceramides, have also been implicated in regulating physiological activity, including apoptosis induced by stress stimuli, such as radiation and chemotherapeutic drugs (14).
Ceramide synthase-4 (CERS4), one of the six mammalian CERSs, catalyzes an amide bond between asphingoid base and a fatty acyl-coenzyme A (15,16). Mutations in this gene are involved in human diseases, including the disease of sphingolipidoses, which are characterized by the accumulation of specific sphingolipid subtypes (14). It was shown that ceramide synthase is involved in tumorigenesis (17,18); however, its roles in HCC have not been studied. The present study revealed that CERS4 was highly expressed in HCC cells, and the functions and mechanisms of CERS4 in HCC were investigated.
Materials and methods
Cell line and human samples
The human 293T cell line and liver cancer HepG2 and Huh7 cell lines were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Human samples were obtained from the People's Hospital of Zhengzhou (Zhengzhou, China) by surgery between March 2014 and May 2014, according to procedures approved by the Ethics Committee at the Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China). Informed consent was obtained from all of the three patients that participated in the present study.
Cell viability assay
The cell proliferation rate was evaluated by MTT assay. Briefly, HepG2 and Huh7 cells were seeded onto 96-well plates (1×103 cells/well), and cell proliferation was documented at 12, 24, 48, 72 and 96 h. The number of viable cells was assessed by measuring the absorbance at 490 nm using a microplate reader (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Colony forming assay
Various groups of HepG2 cells were maintained in RPMI-1640 complete media (10% fetal bovine serum and 1% penicillin/streptomycin). The HepG2 cells were then plated on 6-well plates (500 cells/well), followed by incubation at 37°C overnight. The media was changed every 2 days and the cells were cultured for ~2 weeks to form colonies. After 14 days, each well was washed with 1 ml PBS, and 1 ml of crystal violet solution (1% crystal violet and 10% ethanol; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was added to each well, followed by incubation for 10 min at room temperature. The excess crystal violet was washed out with PBS and the colonies were counted using NIH Image J software (NIH, Bethesda, MD, USA).
Cell cycle assay
Cell cycle analysis of HepG2 cancer cells was performed according to the manufacturer's protocol (BD Cycle Test Plus DNA Reagent kit; BD Biosciences, Franklin Lakes, CA, USA). Different groups of HepG2 cells were seeded on T25 flasks and incubated at 37°C for 48 h. Following incubation, the cells of each group were harvested. Methanol (90%) was then added to the harvested cells for re-suspending and fixing for 30 min. After 30 min, the cells were centrifuged at 400 × g for 5 min at room temperature and washed twice with PBS. The pellets were then re-suspended in propidium iodide and incubated at 37°C for 1 h. A fluorescence-activated cell sorting machine (FACS Calibur flow cytometer; BD Biosciences) was utilized to analyze the data.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR was performed to detect mRNA expression levels of CERS4, P53, Gsk-3β, β-catenin1, Ikbkg and Tank, according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). Total cellular RNA was extracted from HepG2 or Huh7 cells from different groups by lysing cells with TRIzol® reagent (Gibco; Thermo Fisher Scientific, Inc.), followed by centrifugation at 10,000 × g for 15 min at 4°C with chloroform (in the ratio of 5:1). The supernatant was centrifuged with isopropanol (in the ratio of 1:1) at 8,000 × g for 10 min at 4°C. The RNA pellet was washed with 75% ethanol and solubilized with DNase and RNase free water. The RNA was quantified by measuring absorbance at 260 nm using NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.). Single stranded cDNA was prepared using the Prime-Script RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The mRNA expression of the target gene was determined by SYBR-Green assays. The SYBR-Green qPCR kit was purchased from Roche (Roche Diagnostics GmbH, Mannheim, Germany). qPCR was performed using an Applied BioSystems 7300 sequence detection system (Thermo Fisher Scientific, Inc.). All experiments were performed in triplicate. The relative gene expression levels were calculated using the 2−ΔΔCq analysis tool (19). Primers were as follows: CERS4 forward, 5′-TCGGTCCTGTACCACGAGTC-3′ and reverse, 5′-GCCTGATTAGCAGTGAGAGGTAG-3′; β-catenin forward, 5′-CATCTACACAGTTTGATGCTGCT-3′ and reverse, 5′-GCAGTTTTGTCAGTTCAGGGA-3′; Gsk-3β forward, 5′-AGACGCTCCCTGTGATTTATGT-3′ and reverse, 5′-CCGATGGCAGATTCCAAAGG-3′; P53 forward, 5′-GAGGTTGGCTCTGACTGTACC-3′ and reverse, 5′-TCCGTCCCAGTAGATTACCAC-3′; Ikbkg forward, 5′-CGGCAGAGCAACCAGATTCT-3′ and reverse, 5′-CCTGGCATTCCTTAGTGGCAG-3′; and Tank forward, 5′-AGCAGAGAATACGTGAACAACAG-3′ and reverse, 5′-CAGAAGCAATGTCTACCTTTGGT-3′. The GAPDH internal control primers were GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western blot analysis
The standard western blot analysis procedure was utilized to detect the protein expression levels of CERS4, NF-κB and GAPDH in HepG2 or Huh7 cells. The cells were washed twice with ice-cold PBS and lysed using a protein sample buffer kit (Beyotime Institute of Biotechnology, Shanghai, China), according to the manufacturer's protocol. Total cell lysates were then centrifuged (10,000 × g 15 min; 4°C), and the supernatants were used for additional processing. The bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) was used to determine the protein concentration. Total protein (20 µg) was separated by SDS-PAGE and electro-blotted to a polyvinylidene fluoride membrane (Merck Millipore). The membranes were blocked with 5% bovine serum albumin for 2 h at room temperature, and incubated overnight at 4°C with different primary antibodies: Anti-CERS4 antibody (dilution, 1:2,000; cat. no. ab118379; Abcam, Cambridge, UK); anti-NF-κB (dilution, 1:2,000; cat. no. ab32360; Abcam); and anti-GAPDH antibody (dilution, 1:5,000; cat. no. ab181602; Abcam). Following incubation with primary antibodies, the horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibodies (dilution, 1:5,000; cat. no. SC-2004; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were utilized and incubated for 1 h at room temperature. Protein bands were visualized using an electro chemiluminescence assay kit (Beyotime Institute of Biotechnology) and a luminescent image analyzer (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA).
Animal studies
Male Balb/c nude mice (5 weeks old) were purchased from the SLAC Laboratory Animal Company (Shanghai, China). The total number of mice used is the present study was 21 and the average weight of these mice was ~15 g. All mice were maintained in the specific-pathogen-free conditions at 22±2°C and 40–70% relative humidity throughout the experiments according to animal welfare regulations and protocols approved by the Institutional Animal Care and Use Committee of Sichuan University (Sichuan, China). Different groups of HepG2 liver cancer cells (1×107 cells/mouse) were injected subcutaneously into the right forelimb axillaries of Balb/c nude mice to generate tumors in mice. The mice were sacrificed by decapitation following 4 weeks and the weight and volume of the tumors from each mouse were evaluated.
Statistical analysis
All the results represented three or more independent experiments, with the data expressed as the mean ± standard deviation. Differences between the control and treatment groups were analyzed using Student's t-test with SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
CERS4 is highly expressed in liver cancer tissues
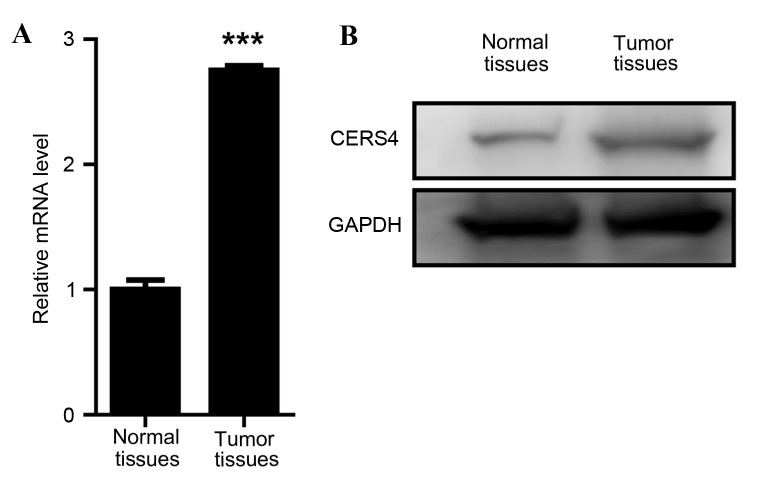
To investigate the function of CERS4 in liver cancer, the expression levels of CERS4 in liver cancer tissues and paired normal liver tissues were evaluated. Firstly, a RT-qPCR assay was applied to determine the mRNA levels of CERS4. The data revealed that the mRNA relative level of CERS4 in liver cancer tissue was ~3 times that of the paired normal liver tissue, indicating a high expression level of CERS4 in liver cancer tissue (Fig. 1A). Western blot analysis was also performed to evaluate the protein expression level of CERS4, and this reconfirmed that the expression level of CERS4 in liver cancer tissue was high compared with paired normal liver tissue (Fig. 1B). The data demonstrated that CERS4 was highly expressed in liver cancer, indicating that CERS4 may serve an important role in the regulation of liver cancer cells.
Figure 1.
CERS4 expression levels in colorectal cancer. (A) Reverse transcription-quantitative polymerase chain reaction assay (n=3). (B) Western blot analysis. Values are presented as the mean ± standard error of mean. ***P<0.001. CERS4, ceramide synthase-4.
CERS4 is effectively silenced by lentivirus-mediated RNA interfere (RNAi)
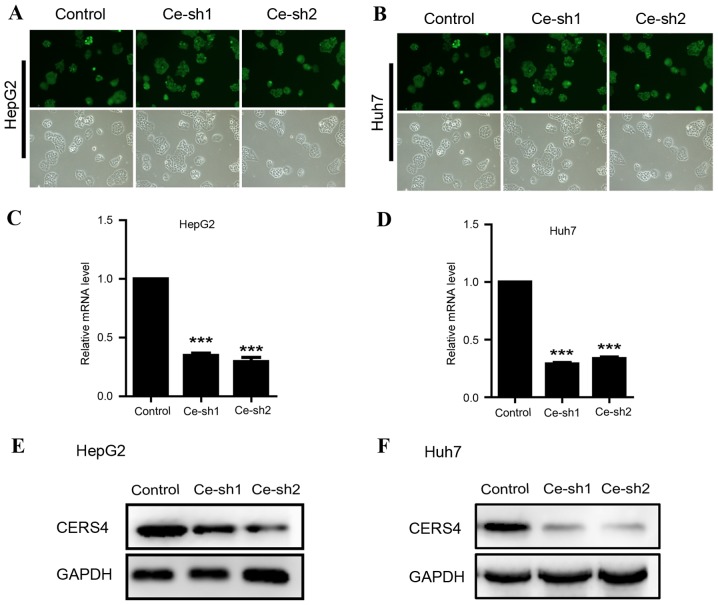
Due to the high expression of CERS4 in liver cancer tissue, lentivirus-mediated RNAi technology was applied to knockdown the CERS4 expression in HepG2 and Huh7 liver cancer cell lines, in order to reveal the function of CERS4 in liver cancer cells. CERS4 short hairpin RNA (shRNA) targets were cloned into lentivirus vectors and the lentivirus was packaged to infect HepG2 and Huh7 cells. The infecting efficiency was >90%, as assessed with green fluorescent protein, indicating successful lentivirus infection in liver cancer cells (Fig. 2A and B). Quantification analysis by RT-qPCR revealed that lentivirus-mediated RNAi evidently reduced (P<0.001) CERS4 mRNA expression levels by 70% in liver cancer HepG2 cells (Fig. 2C). Similarly, CERS4 mRNA levels were markedly decreased (P<0.001) following lentivirus-mediated RNAi in liver cancer Huh7 cells (Fig. 2D). The protein levels of CERS4 were also decreased in HepG2 and Huh7 liver cancer cells infected by CERS4 shRNA lentivirus (Fig. 2E and F). In conclusion, the results demonstrated that the expression of CERS4 was effectively silenced by CERS4 shRNA lentivirus in HepG2 and Huh7 liver cancer cells.
Figure 2.
Silencing of CERS4 expression by lentivirus-mediated RNAi. Representative images of (A) HepG2 and (B) Huh7 cells infected by recombinant lentivirus. Reverse transcription-quantitative polymerase chain reaction assay to detect the knockdown efficiency of CERS4 in (C) HepG2 and (D) Huh7 cells (n=3). The knockdown efficiency of CERS4 determined by western blot analysis in (E) HepG2 and (F) Huh7 cells. The values are presented as the mean ± standard error of the mean. ***P<0.001. CERS4, ceramide synthase-4.
CERS4 knockdown inhibits the proliferation of liver cancer cells
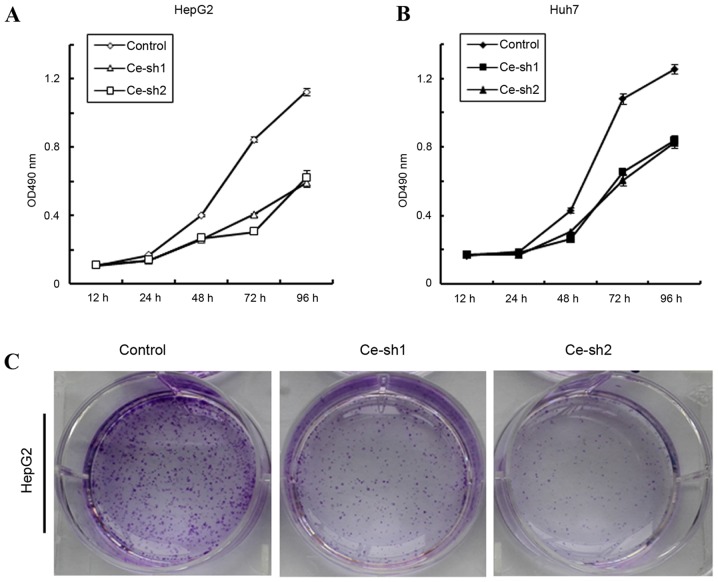
To investigate whether CERS4 affected the proliferation of liver cancer cells, MTT assay was performed. The proliferation of HepG2 and Huh7 liver cancer cells was documented at 12, 24, 48, 72 and 96 h. According to the results, the proliferation rates of CERS4 silenced HepG2 liver cancer cells were markedly reduced (P<0.001) compared with that of the scramble control group (Fig. 3A). For another established liver cancer cell line, Huh7, the proliferation rates also decreased markedly (P<0.001) following knockdown of CESR4 (Fig. 3B). Furthermore, the colony formation assay was also performed to evaluate the effect of CERS4 knockdown on the colony formation ability of HepG2 liver cancer cells. Compared with the scramble control group, the number of cell colonies by crystal violet staining in the CERS4 silenced groups was reduced (Fig. 3C). Therefore, the present results indicated that CERS4 performs an important role in the regulation of liver cancer cells proliferation.
Figure 3.
Effects of CERS4 knockdown on the proliferation of liver cancer cells. (A) The proliferation rates of HepG2 cells detected by MTT assay (n=3). (B) The proliferation rates of Huh7 cells detected by MTT assay (n=3). (C) Knockdown of CERS4 inhibited colony formation of HepG2 cells. CERS4, ceramide synthase-4; OD, absorbance.
CERS4 suppression affects the cell cycle of liver cancer cells
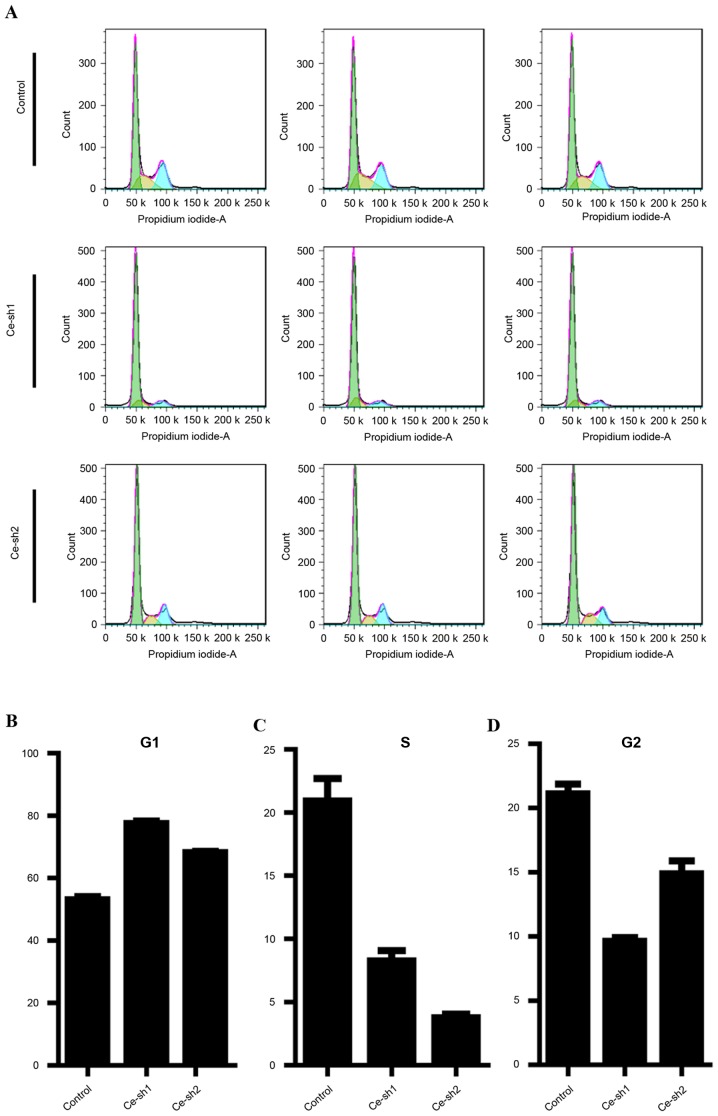
As CERS4 performed an important role in the regulation of liver cancer cells proliferation, flow cytometry was performed to analyze the cell cycle distribution of the HepG2 liver cancer cells following CERS4 shRNA lentivirus infection (Fig. 4A). In the G0/G1 phase of the cell cycle, a higher percentage of cells accumulated following suppression of CERS4 by lentivirus-mediated RNAi compared with the scramble control group (Fig. 4B). Correspondingly, the percentage of cells in the S phase was decreased following lentivirus infection (Fig. 4C). Similarly, in the G2/M phase of cell cycle, the percentage of cells was also reduced following silencing of CERS4, indicating a G0/G1 phase arrest subsequent to depletion of CERS4 (Fig. 4D). The results revealed that knockdown of CERS4 suppressed the growth of liver cancer cells, possibly via induction of cell cycle arrest.
Figure 4.
Effects of CERS4 knockdown on cell cycle progression of liver cancer cells. (A) Flow cytometric histograms of HepG2 cells under each cell cycle phase. Statistical analysis of (B) G0/G1, (C) S and (D) G2/M phases of the cell cycle in HepG2 cells following knockdown of CERS4. CERS4, ceramide synthase-4.
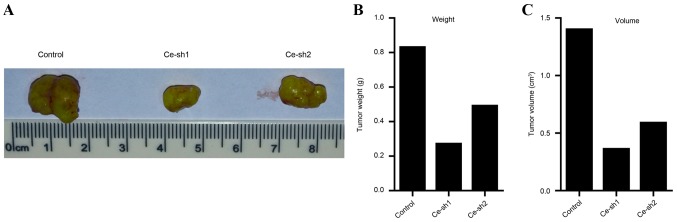
Silencing of CERS4 suppresses the development of liver cancer in vivo
The present results demonstrated that the proliferation rate was inhibited subsequent to the expression of CERS4 being silenced by lentivirus-mediated RNAi technology in vitro. Therefore, an in vivo study using tumor-bearing nude mice models was then performed to determine whether silence of CERS4 suppresses the development of liver cancer in vivo. Tumors were generated by injecting different groups of HepG2 liver cancer cells, including the scramble control group, Ce-sh1 group and Ce-sh2 group into subcutaneous tissues of Balb/c nude mice. The mice were sacrificed by cervical dislocation and the solid tumors were removed and arranged (Fig. 5A). Quantification analysis of the weight of tumors suggested that the weights of the Ce-sh1 and Ce-sh2 groups were decreased compared with the scramble control group (Fig. 5B). Additionally, the volume of tumors was also measured and the results demonstrated that the volume of tumors was reduced following silencing of CERS4 (Fig. 5C). The present results revealed that CERS4 depletion may suppress the development of liver cancer in vivo.
Figure 5.
Ceramide synthase-4 knockdown suppresses tumor growth in xenograft mouse models. (A) Images of tumors removed from the mice of each groups. (B) Weight (g) and (C) volume (cm3) of tumors separated from the mice of different groups.
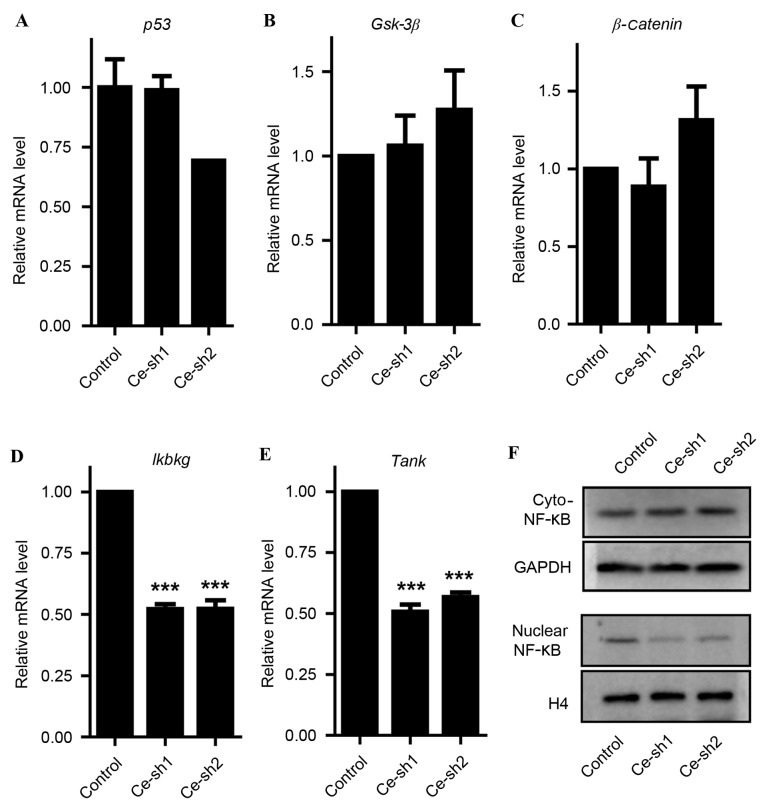
CERS4 regulates the NF-κB signaling pathway
In an effort to reveal the essential molecular mechanism involved in the inhibition of liver cancer cells proliferation in vitro, and attenuation of tumor development in vivo induced by CERS4 silencing, a number of known key genes that perform important roles in regulating cancer cell proliferation, including P53, Gsk-3β, β-catenin, Ikbkg and Tank were assessed. Through RT-qPCR analysis, no significant effects of CERS4 knockdown on P53 and the key genes of the Wnt signaling pathway (Gsk-3β and β-catenin1) were observed (Fig. 6A-C). However, the mRNA levels of Ikbkg and Tank involved in the NF-κB signaling pathway decreased dramatically (P<0.001) subsequent to silencing of CERS4 (Fig. 6D and E). Western blot analysis reconfirmed that CERS4 knockdown gained the protein levels of NF-κB in the cytoplasm, but reduced NF-κB in the cell nucleus (Fig. 6F). These results indicated that silencing of CERS4 had an essential effect on the NF-κB signaling pathway in liver cancer cells.
Figure 6.
Effects of ceramide synthase-4 knockdown on the NF-κB signaling pathway. The mRNA levels of (A) p53, (B) Gsk-3β, (C) β-catenin1, (D) Ikbkg and (E) Tank detected reverse transcription-quantitative polymerase chain reaction (n=3). (F) The protein levels of NF-κB in the cytoplasm and cell nucleus detected by western blot analysis. The values are presented as the mean ± standard error of mean.***P<0.001. Gsk-3β, glycogen synthase kinase-3β; Ikbkg, inhibitor of κ-light polypeptide gene enhancer in B-cells kinase γ; NF-κB, nuclear factor-κB.
Discussion
CERS4 is an important enzyme that is critical for ceramide synthase (20). In the present study, CERS4 was revealed to facilitate HCC formation. The mRNA and protein expression levels of CERS4 were higher in HCC tissues compared with normal tissues. Additionally, knockdown of CERS4 suppressed liver cancer cells proliferation in vivo and tumor growth in vitro.
Sphingolipids, particularly ceramide, are structural components of biological membranes. The balance of the key enzymes in these syntheses not only contributes to the membrane formation, but also affects numerous cellular processes, including cell growth, proliferation, differentiation and motility (21). The present results demonstrated that CERS4 is involved in HCC cell proliferation, which lent support to the hypothesis. However, the function of CERS4 in cellular processes remains largely unknown and requires further investigation.
The NF-κB signaling pathway is extensively involved in cell proliferation and the development of tumor formation. The present study revealed that CERS4 affected the NF-κB signaling pathway. Following CERS4 knockdown, the mRNA expression levels of Ikbkg and Tank (which were important components of NF-κB signaling) were downregulated, indicating thatCERS4 is a regulator of NF-κB signaling. In addition, protein level detection demonstrated that CERS4 knockdown gained the protein levels of NF-κB in cytoplasm, but reduced NF-κB in cell nucleus. This reconfirmed that CERS4 performs an important role in regulating NF-κB signaling. In addition, the mRNA levels of other important cell proliferation regulators, including P53 and the key genes of Wnt signaling pathway (Gsk-3β and β-catenin), were also detected. However, silencing of CERS4 in liver cancer cells did not affect the expression of P53, Gsk-3β or β-catenin. Although the present study has provided supporting evidence that CERS4 promotes HCC cell proliferation by regulating NF-κB signaling, but not Wnt/β-catenin signaling, other mechanisms may also be involved in the process, and this requires additional investigation.
In summary, the present study unravels the function of CERS4 and illustrates the molecular mechanisms by which CERS4 is involved in HCC cell proliferation. The present finding that CERS4 functions as an important regulator of HCC development maybe a potential marker for liver tumors and may also facilitate the utility of the precision medicine in HCC.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 31300674 and 31371325).
Glossary
Abbreviations
- CERS4
ceramide synthase-4
- RNAi
RNA interfere
- HCC
hepatocellular carcinoma
- DMEM
Dulbecco's modified Eagle's medium
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
References
- 1.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin. 2012;62:283–298. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Huang L, Liu CF, Cao J, Yan JJ, Xu F, Wu MC, Yan YQ. Risk factors and surgical outcomes for spontaneous rupture of BCLC stages A and B hepatocellular carcinoma: A case-control study. World J Gastroenterol. 2014;20:9121–9127. doi: 10.3748/wjg.v20.i27.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 5.Teoh NC. Proliferative drive and liver carcinogenesis: Too much of a good thing? J Gastroenterol Hepatol. 2009;24:1817–1825. doi: 10.1111/j.1440-1746.2009.06121.x. [DOI] [PubMed] [Google Scholar]
- 6.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 7.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 8.Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 9.Greten FR, Karin M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004;206:193–199. doi: 10.1016/j.canlet.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 10.Karin M, Greten FR. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 11.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: Metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 13.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 14.Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: An overview and current perspectives. Biochim Biophys Acta. 2002;1585:114–125. doi: 10.1016/S1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- 15.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 16.Mizutani Y, Mitsutake S, Tsuji K, Kihara A, Igarashi Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. 2009;91:784–790. doi: 10.1016/j.biochi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds CP, Maurer BJ, Kolesnick RN. Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett. 2004;206:169–180. doi: 10.1016/j.canlet.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 18.White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, Voelkel-Johnson C. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 2009;28:1132–1141. doi: 10.1038/onc.2008.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmitggen TD. Analysis of relative gene expression data using real-time quantitiative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhäberle E, Kaufmann M, Ackermann H, Lötsch J, Schmidt H, et al. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis. 2009;30:745–752. doi: 10.1093/carcin/bgp061. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds CP, Maurer BJ, Kolesnick RN. Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett. 2004;206:169–180. doi: 10.1016/j.canlet.2003.08.034. [DOI] [PubMed] [Google Scholar]