Abstract
MicroRNAs (miRNAs/miRs) regulate the expression of target genes and are considered to be associated with human cancer. The aim of the present study was to screen novel miRNA biomarkers in esophageal cancer (EC). The miRNA expression profile GSE41268 was extracted from Gene Expression Omnibus database, and differentially expressed miRNAs between whole saliva samples from patients with EC and healthy controls were identified using the Linear Models for Microarray Data package. Then, the targets of these miRNAs were screened using the miRecords database and used to construct the regulatory network. Gene ontology and pathway enrichment analyses were performed for the target genes of differentially expressed miRNAs to predict their potential functions. A total of 18 differentially expressed miRNAs were identified in saliva samples from patients with EC, and 43 validated target genes corresponding to 7 upregulated miRNAs were identified. Then, 6 miRNAs (miR-144, miR-451, miR-98, miR-10b, miR-486-5p and miR-363) and their target genes were used to construct a regulatory network. Within the network, miR-144 may target Notch homolog 1, fibrinogen α chain and fibrinogen β chain; miR-451 may regulate murine thymoma viral oncogene homolog 1, matrix metalloproteinase (MMP)9 and MMP2; miR-98 may directly target E2F transcription factor (E2F) 1, E2F2 and v-myc avian myelocytomatosis viral oncogene homolog (MYC); miR-10b may modulate peroxisome proliferator-activated receptor α and Kruppel-like factor 4; miR-485-5p and miR-363 may regulate TNF receptor superfamily member 5 and cyclin-dependent kinase inhibitor 1A. In addition, E2F1, E2F2 and MYC were associated with the cell cycle, which was the most significantly enriched function and pathway in EC. The results of the present study suggested that miR-144, miR-451, miR-98, miR-10b and miR-363 may be involved in EC by regulating their target genes, and may be used as biomarkers for EC.
Keywords: esophageal cancer, microRNA, network, target gene, differentially expressed
Introduction
Esophageal cancer (EC), also known as oesophageal cancer, is one of the most aggressive tumors of the gastrointestinal tract, and ranks the sixth most common cause of cancer mortality (1). According to the statistical data of National Central Cancer Registry of China, an estimated 287,632 patients were diagnosed with and 208,473 patients succumbed to EC in 2010 in China (2). Although the EC-associated mortality rate has decreased over the past 30 years, it remains a major cancer burden in high risk areas (3). Thus, the early diagnosis and treatment of EC are vital.
Previous studies have indicated that tumor-specific microRNAs (miRNAs/miRs) are considered to be potential biomarkers for the early diagnosis and treatment of EC (4–6). miRNAs, a class of endogenous short non-protein-coding RNA molecules, bind to their target sites within the mRNA molecules via base-pairing with complementary sequences at a post-transcriptional level, and thus certain miRNAs are considered to be oncogenes or tumor suppressor genes (7–9). Previous studies have suggested that miRNAs are primarily expressed in EC tissue and plasma. For example, miR-145, miR-133a and miR-133b were demonstrated to function as tumor suppressors by targeting FSCN1 in esophageal squamous cell carcinoma (ESCC) (10). miR-375 may inhibit tumor growth and metastasis in ESCC by repressing insulin-like growth factor 1 receptor (11). miR-92a has been revealed to promote tumor metastasis of ESCC via epithelial cadherin (12). In addition, miR-21, miR-184 and miR-221 in the plasma were demonstrated to be correlated with the recurrence of ESCC, and serve as oncogenes in ESCC (13). However, miRNA expression in the saliva of patients with EC remains largely unknown.
Due to the extensive blood supply to the salivary glands, saliva is a promising bodily fluid for use in the early detection of diseases including cancer, infectious and cardiovascular diseases (14,15). Previous studies have also suggested that saliva molecules may be used to detect human organic and systemic diseases (16,17). In addition, Xie et al (18) analyzed the expression levels of miRNAs in saliva samples and revealed that miR-10b, miR-144 and miR-451 in whole saliva were significantly upregulated in patients with EC. These miRNAs demonstrate promise as biomarkers for the detection of EC due to the convenience and noninvasive nature of saliva collection (18). However, the target genes of theses miRNAs were not mentioned and the precise functions of these miRNAs remain to be fully understood. Using the same data as Xie et al (18), the present study aimed to identify the validated target genes of the miRNAs that were differentially expressed between whole saliva samples from patients with EC and healthy controls, and to analyze the potential functions of the targets by gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
Materials and methods
Microarray data and data preprocessing
The miRNA expression profile (GSE41268) was extracted from the Gene Expression Omnibus (GEO) database (19) (http://www.ncbi.nlm.nih.gov/geo/) based on the Agilent-021827 Human miRNA Microarray platform (V3; miRBase release 12.0 miRNA ID version; Agilent Technologies, Inc., Santa Clara, CA, USA). This dataset was deposited by Xie et al (18). The microarray contained 10 chips of whole saliva samples from 7 patients with EC and 3 healthy controls. The pathology of the patients with EC was squamous cell carcinoma: 1 patient was stage I, 1 patient was stage II, 3 patients were stage III and 2 patients were stage IV. The probe-level data in CEL files were converted into the miRNA expression profiles. For miRNAs corresponding to multiple probes that exhibited a plurality of expression values, the miRNA expression values of those probes were averaged. Log2 transformation was performed on probes that mapped with the miRNA names labeled in the annotation platform to normalize the data from a skewed distribution to the approximate normal distribution (20).
Screening of differentially expressed miRNAs
The Linear Models for Microarray Data package (21) was used to normalize the data and identify the differentially expressed miRNAs between patients with EC and healthy controls. miRNAs with the cutoff criteria of log2fold change (FC) >1 and P<0.05 were considered to be significantly differentially expressed. In order to explore whether the miRNAs were sample-specific, the Pheatmap package (22) in R was used to perform hierarchical clustering by comparing the value of each miRNA in 10 samples.
Target genes for differentially expressed miRNAs
miRecords (http://c1.accurascience.com/miRecords/download.php) is an integrated resource for animal miRNA-target interactions (23). The miRecords database currently contains 1,135 records of interactions between 644 miRNAs and 1,901 target genes in 9 animal species. In the present study, the validated target genes were extracted according to the miRecords database.
Interaction network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING) database is a useful tool that provides multiple experimental and predicted data (24,25). In order to study the associations between miRNAs and their targets, the target genes were scanned by the STRING database, and protein-protein interaction (PPI) pairs with the cutoff criterion of a combined score >0.4 were selected. Then, the regulatory network that contained these miRNAs and the PPI pairs were constructed using Cytoscape software (version 3.2.1; http://cytoscape.org/) (26).
Function enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (27) was used to identify the enriched functions in DEGs. KEGG Orthology-Based Annotation System (KOBAS) is a software to identify significantly enriched pathways using hypergeometric tests (28). To explore the functions and pathways of target genes in the development of EC, the DAVID and KOBAS were used to identify the significantly enriched GO and KEGG categories, respectively. P<0.05 was used as a cutoff criterion.
Results
Screening of differentially expressed miRNAs
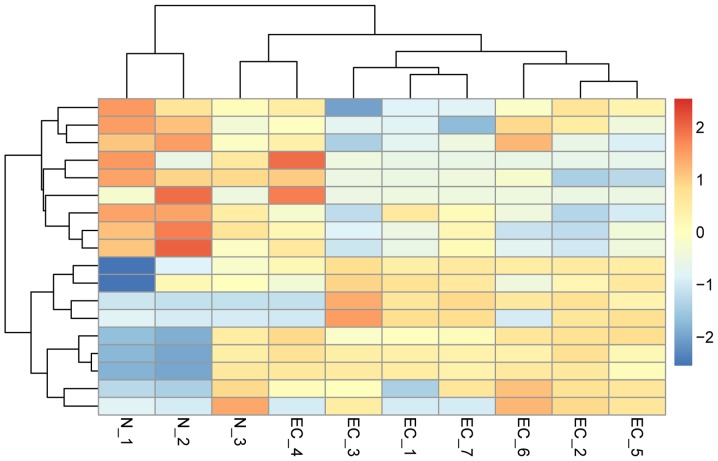
With the cutoff criteria of log2FC >1 and P<0.05, 18 differentially expressed miRNAs were identified in EC samples when compared with the healthy controls (Table I). The top 3 upregulated miRNAs were miR-144, miR-451 and miR-509-3p, while the top 3 downregulated miRNAs were miR-125b-1, miR-129 and miR-636. In addition, to investigate the differences among the 10 samples, a heat map was generated to compare their expression values. The hierarchical clustering analysis revealed a clearly distinct expression of all differentially expressed miRNAs between patients with EC and healthy controls (Fig. 1).
Table I.
Differently expressed miRNAs in esophageal cancer tissue compared with normal tissue.
| miRNA ID | P-value | log2FC | miRNA ID | P-value | log2FC |
|---|---|---|---|---|---|
| hsa-miR-125b-1 | 0.0245 | −4.5437 | hsa-miR-144 | 0.0060 | 6.8525 |
| hsa-miR-129 | 0.0024 | −3.7921 | hsa-miR-451 | 0.0107 | 6.8225 |
| hsa-miR-636 | 0.0422 | −2.1583 | hsa-miR-509-3p | 0.0408 | 5.0251 |
| hsa-miR-1909 | 0.0487 | −2.1353 | hsa-miR-486-5p | 0.0105 | 4.6848 |
| hsa-miR-1274b | 0.0008 | −1.8183 | hsa-miR-10b | 0.0497 | 4.5171 |
| hsa-miR-1274a | 0.0119 | −1.5339 | hsa-miR-363 | 0.0396 | 4.2118 |
| hsa-miR-720 | 0.0453 | −1.4932 | hsa-miR-139-3p | 0.0206 | 3.7638 |
| hsa-miR-371-5p | 0.0206 | −1.3268 | hsa-miR-648 | 0.0454 | 3.7265 |
| hsa-miR-933 | 0.0048 | −1.3112 | hsa-miR-98 | 0.0434 | 3.3743 |
FC, fold change; miRNA/miR, microRNAs.
Figure 1.
Hierarchical clustering heat map of 18 differentially expressed miRNAs. Each column corresponds to a single microarray, whereas each row indicates the expression profile of a single gene. Red and blue represent high and low miRNA expression, respectively. N-1, 2 and 3 indicate salivary samples in healthy controls, while EC-1, 2, 3, 4, 5, 6 and 7 indicate salivary samples in patients with esophageal cancer. miRNA, microRNA.
Screening of validated target mRNAs
According to the miRecords database, 43 validated target mRNAs corresponding to 7 upregulated miRNAs (including miR-144, miR-451 and miR-98) were acquired (Table II). No validated target gene for downregulated miRNAs was obtained.
Table II.
Target genes of differentially expressed miRNAs.
| miRNA | Target | Test methods |
|---|---|---|
| hsa-miR-144 | NOTCH1 | Luciferase reporter assay |
| hsa-miR-144 | FGA | Luciferase reporter assay |
| hsa-miR-144 | FGB | Luciferase reporter assay |
| hsa-miR-144 | FGG | ELISA; flow; luciferase reporter assay |
| hsa-miR-144 | PLAG1 | Luciferase reporter assay |
| hsa-miR-451 | AKT1 | RT-qPCR; western blotting |
| hsa-miR-451 | MMP9 | RT-qPCR; western blotting |
| hsa-miR-451 | MMP2 | RT-qPCR; western blotting |
| hsa-miR-451 | ABCB1 | RT-qPCR; luciferase reporter assay; western blotting |
| hsa-miR-451 | BCL2 | RT-qPCR; western blotting |
| hsa-miR-451 | CAB39 | RT-qPCR; luciferase reporter assay; western blotting; microarray |
| hsa-miR-451 | MIF | ELISA; luciferase reporter assay; microarray; RT-qPCR; western blotting |
| hsa-miR-451 | SSSCA1 | RT-qPCR; western blotting |
| hsa-miR-509-3p | NTRK3 | Luciferase reporter assay |
| hsa-miR-98 | E2F1 | Luciferase reporter assay |
| hsa-miR-98 | E2F2 | Northern blot |
| hsa-miR-98 | MYC | Luciferase reporter assay |
| hsa-miR-98 | ACADM | RT-qPCR |
| hsa-miR-98 | AMMECR1 | RT-qPCR |
| hsa-miR-98 | CBX5 | RT-qPCR |
| hsa-miR-98 | CCL5 | ELISA |
| hsa-miR-98 | CHRNB1 | RT-qPCR |
| hsa-miR-98 | CNOT4 | RT-qPCR |
| hsa-miR-98 | EZH2 | Northern blot |
| hsa-miR-98 | HMGA2 | Northern blot; RT-qPCR |
| hsa-miR-98 | HNRPDL | RT-qPCR |
| hsa-miR-98 | KLF13 | RT-qPCR |
| hsa-miR-98 | MEIS1 | RT-qPCR |
| hsa-miR-98 | NCOA3 | Luciferase reporter assay |
| hsa-miR-98 | SERP1 | RT-qPCR |
| hsa-miR-98 | SLC20A1 | RT-qPCR |
| hsa-miR-98 | SOCS4 | Luciferase reporter assay; northern blot; RT-qPCR; western blotting |
| hsa-miR-98 | THBS1 | RT-qPCR |
| hsa-miR-98 | TUSC2 | Luciferase reporter assay; western blotting |
| hsa-miR-98 | ZFP36L1 | RT-qPCR |
| hsa-miR-98 | ZMPSTE24 | RT-qPCR |
| hsa-miR-10b | PPARA | Luciferase reporter assay; RT-qPCR; western blotting |
| hsa-miR-10b | KLF4 | Luciferase reporter assay; western blotting |
| hsa-miR-10b | NCOR2 | RT-qPCR; western blotting |
| hsa-miR-10b | HOXD10 | Luciferase reporter assay |
| hsa-miR-10b | NF1 | GFP reporter assay; luciferase reporter assay; microarray; RT-qPCR |
| hsa-miR-486-5p | CD40 | Microarray; RT-qPCR |
| hsa-miR-363 | CDKN1A | RT-qPCR; luciferase reporter assay; western blotting |
miRNA/miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GFP, green fluorescent protein.
Interaction network analysis
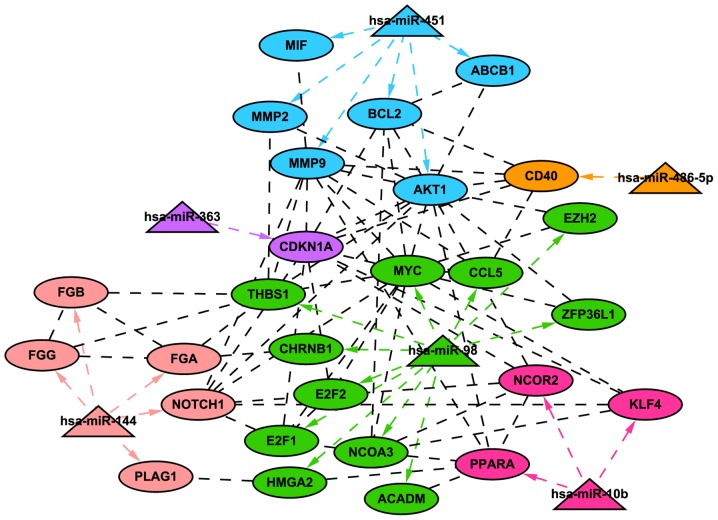
A total of 6 miRNAs and 27 validated target genes were mapped into the network using STRING and Cytoscape. In the network, 5 genes including Notch homolog 1 (NOTCH1), fibrinogen γ chain (FGG), fibrinogen α chain (FGA), and fibrinogen β chain (FGB) were modulated by miR-144; while 6 genes including AKT serine/threonine kinase 1 (AKT1), matrix metalloproteinase (MMP)9, and MMP2 were modulated by miR-451. A total of 11 genes, including v-myc avian myelocytomatosis viral oncogene homolog (c-MYC; MYC), E2F transcription factor (E2F)1, and E2F2 were modulated by miR-98. A total of 3 genes, including peroxisome proliferator-activated receptor α (PPARA) and Kruppel-like factor 4 (KLF4) were modulated by miR-10b. TNF receptor superfamily member 5 (CD40) and cyclin-dependent kinase inhibitor 1A (CDKN1A) were modulated by miR-485-5p and miR-363, respectively (Fig. 2).
Figure 2.
miRNA-target regulatory network. The triangle nodes represent miRNAs, while oval nodes represent target genes. The different miRNAs and their target genes are marked with different colors. miRNA/miR, microRNA.
Function and pathway enrichment analysis
To gain additional insight into the underlying functions of all target genes and the target genes in the network, GO and KEGG enrichment analyses were performed. The results demonstrated that the top 3 enriched GO terms were cell cycle (E2F1, E2F2 and MYC), blood vessel development (AKT1, NOTCH1 and MMP2) and vasculature (AKT1, NOTCH1 and MMP2; Table III); and the top 3 enriched KEGG pathways were cell cycle (E2F1, E2F2 and MYC), complement and coagulation cascades (FGG, FGA and FGB) and epidermal growth factor receptor signaling pathway (AKT1, CDKN1A and MYC; Table IV).
Table III.
Enriched GO terms of target genes in the microRNA-target regulatory network.
| Term | Count | P-value | Genes |
|---|---|---|---|
| GO: 0007049~cell cycle | 10 | 4.27×10−4 | E2F1, E2F2, MYC, AKT1, SSSCA1, TUSC2, CDKN1A, BCL2, HMGA2, THBS1 |
| GO: 0001568~blood vessel development | 6 | 8.23×10−4 | AKT1, NOTCH1, MMP2, ZFP36L1, NF1, THBS1 |
| GO: 0001944~vasculature development | 6 | 9.18×10−4 | AKT1, NOTCH1, MMP2, ZFP36L1, NF1, THBS1 |
| GO: 0022402~cell cycle process | 8 | 1.39×10−3 | E2F1, MYC, AKT1, SSSCA1, CDKN1A, BCL2, HMGA2, THBS1, |
| GO: 0043067~regulation of programmed cell death | 9 | 2.65×10−3 | MYC, AKT1, NOTCH1, MMP9, CDKN1A, BCL2, NF1, THBS1, MIF |
| GO: 0010941~regulation of cell death | 9 | 2.71×10−3 | MYC, AKT1, NOTCH1, MMP9, CDKN1A, BCL2, NF1, THBS1, MIF |
| GO: 0051726~regulation of cell cycle | 6 | 3.09×10−3 | E2F1, E2F2, MYC, AKT1, CDKN1A, BCL2 |
| GO: 0022403~cell cycle phase | 6 | 7.93×10−3 | E2F1, AKT1, SSSCA1, CDKN1A, BCL2, HMGA2 |
| GO: 0008283~cell proliferation | 6 | 9.80×10−3 | E2F1, MYC, TUSC2, BCL2, CD40, MIF |
GO, gene ontology.
Table IV.
Enriched Kyoto Encyclopedia of Genes and Genomes pathways of target genes in the microRNA-target regulatory network.
| Term | Pathway | Count | P-value | Genes |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 3 | 5.49×10−9 | E2F1, E2F2, MYC |
| hsa04610 | Complement and coagulation cascades | 3 | 1.43×10−4 | FGG, FGA, FGB |
| hsa04012 | ErbB signaling pathway | 3 | 1.52×10−3 | AKT1, CDKN1A, MYC |
| hsa04620 | Toll-like receptor signaling pathway | 3 | 2.15×10−3 | AKT1, CD40, CCL5 |
ErbB, epidermal growth factor receptor.
Discussion
miRNA expression is known to be associated with various types of human cancer (29). In the present study, the salivary miRNA expression profile of EC deposited by Xie et al (18) in GEO database was analyzed, to screen for biomarkers for therapeutic intervention. In concordance with the study of Xie et al (18), the present study also demonstrated that miR-144, miR-451 and miR-10b were significantly upregulated in saliva samples from patients with EC and healthy controls. In addition, miR-144 was revealed to potentially be involved in EC progression by directly targeting 5 mRNAs, including NOTCH1, FGG, FGA and FGB. NOTCH 1 is a member of the NOTCH family. NOTCH signaling serves an important function in a variety of developmental processes by regulating cell differentiation, proliferation and apoptosis (30). NOTCH signaling has been demonstrated to be upregulated in EC, and may be used as a therapeutic target for EC (31,32). FGG, FGA and FGB belong to the fibrinogen family. Fibrinogen is a glycoprotein in vertebrates that assists with the formation of blood clots and functions as a common component of hemostasis (33). Takeuchi et al (34) identified that plasma fibrinogen concentration was significantly associated with chemoradiotherapy responsiveness in patients with ESCC. Matsuda et al (35) suggested that the plasma fibrinogen level may be used as a biomarker for postoperative recurrence in advanced EC. The fibrinogen level is correlated with tumor metastasis and recurrence in patients with ESCC (34,35). Thus, by targeting NOTCH1, FGG, FGA and FGB, miR-144 may be involved in cell invasion and metastasis of EC.
miR-451 was revealed to target 6 mRNAs in the present study, including AKT1, MMP9 and MMP2. The protein kinase B (AKT) family has been demonstrated to integrate extracellular signals with cell processes including proliferation, differentiation, migration and survival (36). Previous data have demonstrated that the phosphoinositide 3-kinase/protein kinase B/mechanistic target of rapamycin pathway is frequently activated in numerous types of human cancer, including colorectal cancer (37). Cancer cells are well known to exhibit a higher proliferation rate compared with normal cells, and cancer cells frequently fail to undergo apoptosis (38). Activated AKT may regulate its downstream targets to increase cell proliferation and decrease apoptosis (39). AKT activation was described as an early cellular response to carcinogen exposure, and may be a key step in environmental carcinogenesis (40). In addition, MMP2 and MMP9 were demonstrated to be overexpressed in ESCC and to be associated with esophageal tumorigenesis (41). Therefore, the increased expression of MMPs may promote tumor proliferation and inhibit apoptosis in the development of EC (42).
The present study revealed that miR-10b may regulate 3 mRNAs (including PPARA and KLF4). PPARA, also termed NR1C1, is a transcription factor that is a member of the peroxisome proliferator-activated receptor (PPAR) group. PPARs have been demonstrated to stimulate tumor cell proliferation and induce neoangiogenesis in different types of gastrointestinal cancer, including EC, liver and pancreatic cancer (43). KLF4, as a known tumor suppressor, may regulate cell proliferation, differentiation, development and apoptosis (44). KLF4 has been suggested to suppress EC cell migration and invasion, as a direct target of miR-10b (45). Therefore, the associated miRNAs, including miR-144 and miR-451 and miR-10b, may serve important functions in cellular processes including cell differentiation, proliferation, metastasis and apoptosis in EC by regulating NOTCH, FGG, FGA, FGB, AKT1, MMP9, MMP2, PPARA and KLF4.
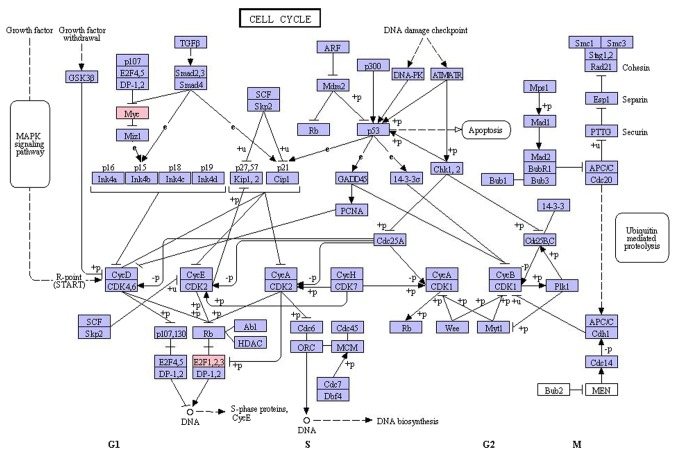
Compared with the aforementioned previous study, among the upregulated miRNAs, miR-98 and miR-363 were novel (18). miR-98 belongs to the lethal-7 (let-7) family of miRNAs (let-7f, let-7d, miR-98 and let-7g). The miRNA let-7 may regulate key differentiation processes during development (46). The expression of let-7 has been suggested to predict the response to chemotherapy in EC (47). miR-98 may target almost half of the validated target genes in the network, including MYC, E2F1, and E2F2. MYC, a well-known transcription factor, exhibited a high degree of connectivity in the regulatory network of the present study. E2F1 and E2F2 are transcription factors and members of the E2F family. MYC and E2Fs have been identified as associated with the cell cycle (48). MYC overexpression enhances and MYC downregulation inhibits cell cycle progression (48). E2F1 expression is positively associated with cell proliferation in EC (49). E2F2 has been demonstrated to negatively regulate a subset of genes involved in the processes of DNA metabolism and cell cycle control (50). In addition, E2F1 and E2F2 are expressed in a cell cycle-regulated manner, exhibiting their highest levels in the late G1 and S phase (51). Cell cycle deregulation is a common feature of human tumors (52). The results of the present study also demonstrated that MYC, E2F1 and E2F2 were enriched in the cell cycle, which was the most significantly enriched GO function and KEGG pathway, suggesting that MYC, E2F1 and E2F2 serve critical functions in the cell cycle (Fig. 3). Therefore, by targeting MYC, E2F1 and E2F2, miR-98 may be involved in the cell cycle of EC cells, and act as novel biomarker for EC.
Figure 3.
Detailed molecular network of pathways of the cell cycle (Kyoto Encyclopedia of Genes and Genomes ID, has04110). Red genes are the significant genes involved in the pathway. G1, G1 phase; S, S phase; G2, G2 phase; M, mitotic phase.
miR-363 was identified to be downregulated in head and neck squamous cell carcinoma (HNSCC) tissues, and to affect HNSCC cell invasion and metastasis (53). Hsu et al (54) indicated that miR-363 served a key function in the increment of gastric carcinogenesis via targeting c-Myc promotor binding protein 1, a negative regulator of MYC. In the present study, miR-363 was suggested to directly target CDKN1A, also termed p21, WAF1 or CIP1. The cyclin-dependent kinase inhibitor CDKN1A is a regulator of cell cycle progression at the G1 and S phases (55). Therefore, miR-363 may be involved in EC by regulating its target gene, and may be used as a novel biomarker for EC.
In addition, in contrast to the study of Xie et al (18), the present study identified that miR-486-5p was significantly upregulated in EC samples (P=0.01052). This may be caused by the different preprocessing methods used in these studies. The present study also identified that miR-486-5p may modulate EC by targeting CD40. CD40, a member of the TNF-receptor superfamily, was originally identified on the surface of B cells and was suggested to regulate B cell growth and differentiation (56). The interaction of CD40 with the CD40 ligand was revealed to modulate immune cells in the lymph nodes and to promote growth of local squamous cell cancer of the head and neck by protecting tumor cells (57,58). miR-486-5p was dysregulated in EC saliva samples and was demonstrated to regulate CD40 in the present study. However, additional studies are needed to verify the involvement of miR-486-5p in EC.
In conclusion, the present study demonstrated that the biomarkers (miR-144, and miR-451 and miR-10b) screened by Xie et al (18) may serve an important function in the regulation of EC by targeting NOTCH, fibrinogen, AKT1, MMPs, PPARA and KLF4. In addition, 2 novel biomarkers, miR-98 and miR-363, were identified. These are associated with the EC and modulate a serial of cell cycle-associated genes, including MYC, E2F1, E2F2 and CDKN1A. Additional experiments are needed to confirm the expression of miR-486-5p and its involvement in EC, due to the discordant conclusions between the present study and a previous study (18).
Acknowledgements
The present study was supported by Natural Science Foundation of Liaoning Province (grant no. 2015 020561).
References
- 1.Whiteman DC. Esophageal cancer: Priorities for prevention. Curr Epidemiol Rep. 2014;1:138–148. doi: 10.1007/s40471-014-0015-3. [DOI] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zeng H, Fan Y, Qiao Y, Zhou Q. Esophageal cancer incidence and mortality in China, 2010. Thorac Cancer. 2014;5:343–348. doi: 10.1111/1759-7714.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei WQ, Yang J, Zhang SW, Chen WQ, Qiao YL. Esophageal cancer mortality trends during the last 30 years in high risk areas in China: Comparison of results from national death surveys conducted in the 1970's, 1990's and 2004–2005. Asian Pac J Cancer Prev. 2011;12:1821–1826. [PubMed] [Google Scholar]
- 4.Yang M, Liu R, Sheng J, Liao J, Wang Y, Pan E, Guo W, Pu Y, Yin L. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol Rep. 2013;29:169–176. doi: 10.3892/or.2012.2105. [DOI] [PubMed] [Google Scholar]
- 5.Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19:1292–1300. doi: 10.2174/138161213804805775. [DOI] [PubMed] [Google Scholar]
- 6.Yao WJ, Wang YL, Lu JG, Guo L, Qi B, Chen ZJ. MicroRNA-506 inhibits esophageal cancer cell proliferation via targeting CREB1. Int J Clin Exp Pathol. 2015;8:10868–10874. [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Lucas K, Raikhel AS. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol. 2013;43:24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 11.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725–10734. doi: 10.1074/jbc.M110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YH, Wong DT. Saliva: An emerging biofluid for early detection of diseases. Am J Dent. 2009;22:241–248. [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: Current state and future applications. Clin Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Elashoff D, Oh M, Sinha U, St John MA, Zhou X, Abemayor E, Wong DT. Serum circulating human mRNA profiling and its utility for oral cancer detection. J Clin Oncol. 2006;24:1754–1760. doi: 10.1200/JCO.2005.03.7598. [DOI] [PubMed] [Google Scholar]
- 17.Mbulaiteye SM, Walters M, Engels EA, Bakaki PM, Ndugwa CM, Owor AM, Goedert JJ, Whitby D, Biggar RJ. High levels of Epstein-Barr virus DNA in saliva and peripheral blood from Ugandan mother-child pairs. J Infect Dis. 2006;193:422–426. doi: 10.1086/499277. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z, Chen G, Zhang X, Li D, Huang J, Yang C, Zhang P, Qin Y, Duan Y, Gong B, Li Z. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One. 2013;8:e57502. doi: 10.1371/journal.pone.0057502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita A, Sato JR, Lde O Rodrigues, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [DOI] [Google Scholar]
- 22.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohl M, Wiese S, Warscheid B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(Web Server issue):W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramaniam D, Ponnurangam S, Ramamoorthy P, Standing D, Battafarano RJ, Anant S, Sharma P. Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS One. 2012;7:e30590. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters JH, Avisar N. The molecular pathogenesis of Barrett's esophagus: Common signaling pathways in embryogenesis metaplasia and neoplasia. J Gastrointest Surg. 2010;14(Suppl 1):81–87. doi: 10.1007/s11605-009-1011-7. [DOI] [PubMed] [Google Scholar]
- 32.Mendelson J, Song S, Li Y, Maru DM, Mishra B, Davila M, Hofstetter WL, Mishra L. Dysfunctional transforming growth factor-β signaling with constitutively active Notch signaling in Barrett's esophageal adenocarcinoma. Cancer. 2011;117:3691–3702. doi: 10.1002/cncr.25861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi H, Ikeuchi S, Kitagawa Y, Shimada A, Oishi T, Isobe Y, Kubochi K, Kitajima M, Matsumoto S. Pretreatment plasma fibrinogen level correlates with tumor progression and metastasis in patients with squamous cell carcinoma of the esophagus. J Gastroenterol Hepatol. 2007;22:2222–2227. doi: 10.1111/j.1440-1746.2006.04736.x. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda S, Takeuchi H, Fukuda K, Nakamura R, Takahashi T, Wada N, Kawakubo H, Saikawa Y, Omori T, Kitagawa Y. Clinical significance of plasma fibrinogen level as a predictive marker for postoperative recurrence of esophageal squamous cell carcinoma in patients receiving neoadjuvant treatment. Dis Esophagus. 2014;27:654–661. doi: 10.1111/dote.12115. [DOI] [PubMed] [Google Scholar]
- 36.Freeman-Cook KD, Autry C, Borzillo G, Gordon D, Barbacci-Tobin E, Bernardo V, Briere D, Clark T, Corbett M, Jakubczak J, et al. Design of selective, ATP-competitive inhibitors of Akt. J Med Chem. 2010;53:4615–4622. doi: 10.1021/jm100769x. [DOI] [PubMed] [Google Scholar]
- 37.Eide PW, Cekaite L, Danielsen SA, Eilertsen IA, Kjenseth A, Fykerud TA, Ågesen TH, Bruun J, Rivedal E, Lothe RA, Leithe E. NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cell Signal. 2013;25:12–18. doi: 10.1016/j.cellsig.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3:51–58. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber SM, Bornstein S, Li Y, Malkoski SP, Wang D, Rustgi AK, Kulesz-Martin MF, Wang XJ, Lu SL. Tobacco-specific carcinogen nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induces AKT activation in head and neck epithelia. Int J Oncol. 2011;39:1193–1198. doi: 10.3892/ijo.2011.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111:81–90. doi: 10.1172/JCI200316147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samantaray S, Sharma R, Chattopadhyaya TK, Gupta SD, Ralhan R. Increased expression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2004;130:37–44. doi: 10.1007/s00432-003-0500-4. [DOI] [PubMed] [Google Scholar]
- 42.Groblewska M, Siewko M, Mroczko B, Szmitkowski M. The role of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia Histochem Cytobiol. 2012;50:12–19. doi: 10.5603/FHC.2012.0002. [DOI] [PubMed] [Google Scholar]
- 43.Pazienza V, Vinciguerra M, Mazzoccoli G. PPARs signaling and cancer in the gastrointestinal system. PPAR Res. 2012;2012:560846. doi: 10.1155/2012/560846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/S1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peter ME. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugimura K, Miyata H, Tanaka K, Hamano R, Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori M, Doki Y. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res. 2012;18:5144–5153. doi: 10.1158/1078-0432.CCR-12-0701. [DOI] [PubMed] [Google Scholar]
- 48.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki K, Hasegawa M, Ohoka I, Hanami K, Asoh A, Nagao T, Sugano I, Ishida Y. Increased E2F-1 expression via tumour cell proliferation and decreased apoptosis are correlated with adverse prognosis in patients with squamous cell carcinoma of the oesophagus. J Clin Pathol. 2005;58:904–910. doi: 10.1136/jcp.2004.023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laresgoiti U, Apraiz A, Olea M, Mitxelena J, Osinalde N, Rodriguez JA, Fullaondo A, Zubiaga AM. E2F2 and CREB cooperatively regulate transcriptional activity of cell cycle genes. Nucleic Acids Res. 2013;41:10185–10198. doi: 10.1093/nar/gkt821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xanthoulis A, Tiniakos DG. E2F transcription factors and digestive system malignancies: How much do we know? World J Gastroenterol. 2013;19:3189–3198. doi: 10.3748/wjg.v19.i21.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 53.Sun Q, Zhang J, Cao W, Wang X, Xu Q, Yan M, Wu X, Chen W. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol. 2013;45:513–520. doi: 10.1016/j.biocel.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Hsu KW, Wang AM, Ping YH, Huang KH, Huang TT, Lee HC, Lo SS, Chi CW, Yeh TS. Downregulation of tumor suppressor MBP-1 by microRNA-363 in gastric carcinogenesis. Carcinogenesis. 2014;35:208–217. doi: 10.1093/carcin/bgt285. [DOI] [PubMed] [Google Scholar]
- 55.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 56.Kehry MR. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J Immunol. 1996;156:2345–2348. [PubMed] [Google Scholar]
- 57.Posner MR, Cavacini LA, Upton MP, Tillman KC, Gornstein ER, Norris CM., Jr Surface membrane-expressed CD40 is present on tumor cells from squamous cell cancer of the head and neck in vitro and in vivo and regulates cell growth in tumor cell lines. Clin Cancer Res. 1999;5:2261–2270. [PubMed] [Google Scholar]
- 58.Cao W, Cavacini LA, Tillman KC, Posner MR. CD40 function in squamous cell cancer of the head and neck. Oral Oncol. 2005;41:462–469. doi: 10.1016/j.oraloncology.2004.11.005. [DOI] [PubMed] [Google Scholar]