Abstract
Previous studies have demonstrated abnormal H3K27 methylation status during clear cell renal cell carcinoma (ccRCC) carcinogenesis, and have suggested that the histone H3K27 demethylases, jumonji domain-containing protein 3 (JMJD3) and ubiquitously-transcribed TPR gene on the X chromosome, are important regulatory factors that alter H3K27 methylation status. The present study aimed to explore the prognostic value of JMJD3 in patients with ccRCC. A total of 331 ccRCC samples were stained for JMJD3 by immunohistochemistry. Stage, Size, Grade and Necrosis (SSIGN) and University of California Los Angeles Integrated Staging System (UISS) scores were applied to stratify risks. Survival analyses were performed through the Kaplan-Meier estimator method and Cox proportional hazard model. The results revealed that JMJD3 expression in ccRCC was significantly increased compared with that in the peritumoral tissue (P<0.001) and negatively associated with a number of other clinicopathological characteristics. Kaplan-Meier estimator and multivariate analyses revealed that decreased tumoral JMJD3 expression was associated with OS (hazard ratio, 2.141; P=0.003), and DFS prediction (hazard ratio, 1.737; P=0.033). In addition, following stratification of patients into three risk levels using the SSIGN and UISS scores, decreased tumoral JMJD3 expression was associated with shorter OS (P=0.003 for SSIGN and UISS scores) and DFS (P=0.007 for SSIGN and P=0.041 for UISS score) in the intermediate risk groups. The results from the present study suggest that JMJD3 is a novel prognostic marker for patients with ccRCC and is of particular significance in patients with intermediate-risk disease.
Keywords: clear cell renal cell carcinoma, jumonji domain-containing protein 3, overall survival, disease free survival, prognostic biomarker
Introduction
Renal cell carcinoma (RCC) is the eighth most common cancer in the USA (1). The numbers of new RCC cases and mortalities in the USA for 2015 were estimated to be 61,560 and 14,080, respectively (1). RCC is known for its multiresistance to conventional cancer therapies, and patients with localized diseases often experience recurrences following curative surgeries (2). Current clinical parameters, including Fuhrman grades (3) and tumor-necrosis-metastasis (TNM) stages (4) may be used to evaluate the prognosis of patients with RCC to a certain extent; however, these parameters are not reliable (5). Therefore, it is necessary to search for improved biomarkers for more accurate RCC prognosis prediction.
Epigenetic modifications that alter classical tumor suppressor genes are typical in inherited and sporadic RCC (6). Histone modifications are epigenetic modifications that are incorrectly added, interpreted or erased in a number of human cancers (7). Histone H3 lysine 27 tri-methylation (H3K27me3), a modification that inhibits the expression of certain target genes by altering the physical status of chromatin, is mediated by polycomb repressive complex 2 (PRC2), a multi-component enzymatic complex with a catalytic subunit known as histone-lysine N-methyltransferase EZH2 (EZH2) (8). H3K27 tri- and di-methylation is reversed by a subfamily of two related JmjC domain-containing histone demethylases, ubiquitously-transcribed TPR gene on the X chromosome (UTX) and jumonji domain-containing protein 3 (JMJD3), which have been identified as H3K27-specific demethylases that contribute to gene activation (9).
H3K27 methylation has been demonstrated to serve oncogenic functions through the inactivation of tumor suppressor genes in a number of types of human carcinoma, including renal cell carcinoma (8), and overexpression of EZH2 has been reported to contribute to tumor cell migration and invasion through epigenetic repression of E-cadherin in RCC (10). JMJD3 has been demonstrated to be involved in the development of certain types of cancer, including breast, prostate, colon and kidney cancer, and glioblastoma (11–14). Estrogen receptor-α-dependent recruitment of JMJD3 in hormone-dependent breast cancer cells leads to the demethylation of H3K27 and activation of B-cell lymphoma 2 transcription (11). In addition, deregulation of JMJD3 may contribute to gliomagenesis via inhibition of the tumor protein p53 (p53) signaling pathway, which results in the inhibition of terminal differentiation (13). Upregulated expression levels of JMJD3 are typical in RCC tissues in patients with early stage RCC with a good prognosis (14).
The results from the aforementioned studies suggest that there is an association between JMJD3 and RCC prognosis. Therefore, the aim of the present study was to evaluate the association between JMJD3 expression and the survival of patients with ccRCC following surgery. Immunohistochemical staining was performed to analyze the expression of JMJD3 in tissues from patients. In addition, the prognostic value of JMJD3 expression for overall survival (OS) and disease free survival (DFS) was evaluated in patients with clear cell (cc)RCC divided into different risk groups by Stage, Size, Grade and Necrosis (SSIGN) score and University of California Los Angeles Integrated Staging System (UISS) score (15–17).
Patients and methods
Patients
Between February 2005 and June 2007, 331 patients with ccRCC (validated pathologically by Dr Li Liu from Zhongshan Hospital of Fudan University) were enrolled from the Department of Urology, Zhongshan Hospital, Fudan University (Shanghai, China). Ethical approval was obtained from the Clinical Research Ethics Committee of Zhongshan Hospital, Fudan University and written consent was obtained from all patients. Patients' clinical and outcome data were updated every 3 months. Each patient's Fuhrman grade and TNM stage were verified according to the 2004 WHO criteria and the American Joint Committee on Cancer 2010 TNM classification, respectively (3,4). Patients' risk stratifications were performed using the SSIGN, SSIGN (localized) and UISS score, respectively (15–17). None of the patients in this cohort had received targeted therapies prior to or following surgery. Based on imaging tests (computed tomography and magnetic resonance imaging) or histopathology information, metastasis or recurrences were defined. A total of 86/331 enrolled patients succumbed to the disease during follow-up. Within the localized ccRCC patient group, 80 patients experienced relapse (recurrence or distant metastasis) during follow-up.
Immunohistochemistry and evaluation
To confirm that each sample was representative of the whole ccRCC specimen, hematoxylin and eosin-stained slides were screened for optimal tumor content prior to construction of the tissue microarray (TMA) slides. Two cores from paired tumor and non-tumor renal tissue from the sample were extracted from the formalin-fixed, paraffin embedded surgical specimens using a 3.0 mm diameter punch. Immunohistochemical staining of JMJD3 was performed on the TMA slides as previously described (18) using a rabbit polyclonal anti-JMJD3 antibody (cat. no. ab38113; dilution; 1:100; Abcam, Cambridge, MA, USA). An Olympus CDD camera (Olympus Corporation, Tokyo, Japan) and a Nikon Eclipse Ti-S microscope (Nikon Corporation, Tokyo, Japan) were used to record the staining results. Samples were scanned at high-power magnification (×200) and images were captured using NIS-Elements software version 3.7 (Nikon Corporation). For each specimen, three images with the strongest staining were captured for analysis.
The aforementioned pathologist, who was unaware of the clinical features and outcomes of the patients represented by the slides, semi-quantitatively scored the staining intensity of JMJD3 in all specimens. Counting was performed using the H-Score algorithm (19). For H-score assessment, fields were at ×200 magnification and the staining intensity in the malignant cells was scored as 0, 1, 2, or 3 corresponding to the presence of negative, weak, intermediate, and strong brown staining, respectively. The total number of cells in each field and the number of cells stained at each intensity were counted. The average percentage of positive cells was calculated and the following formula was applied: H-score = (% of cells stained at intensity category 1×1) + (% of cells stained at intensity category 2×2) + (% of cells stained at intensity category 3×3). H-Scores between 0 and 300 were obtained, where 300 was equal to 100% of tumor cells stained strongly (3+). For interpretation, samples were divided into ‘low-’ and ‘high-’ expression level categories. The cutoff values for the high/low expression of JMJD3 were determined as 117.5 in tumor tissues and 89.5 in non-tumor tissues with X-tile software version 3.6.1 (Yale University, New Haven, CT, USA) using the minimum P-value method based on patients' overall survival (OS) information (20).
Survival assessment
The definition of disease free survival (DFS) is the time between curative surgery and identification of recurrence or metastasis. OS is defined as the interval from the day of surgery to mortality from any cause. The final follow-up time was Jan 30, 2015 for all available patients.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (version 6.0; GraphPad Software Inc., La Jolla, CA, USA) and SPSS (version 19.0; IBM Corp., Armonk, NY, USA) software. P<0.05 was considered to indicate a statistically significant difference. A paired Student's t-test was used to evaluate differences in JMJD3 expression between tumor and non-tumor tissue. Associations between JMJD3 expression and patients' clinicopathological features was evaluated using a χ2 test, Fisher's exact method, or Cochran-Mantel-Haenszel χ2 test. Cox univariate analyses were performed and those parameters with statistical significance were evaluated using a multivariate Cox proportional hazards model. A Kaplan-Meier estimator analysis and log-rank test were applied for DFS and OS evaluation.
Results
Patient characteristics
Patient clinicopathological characteristics are presented in Table I. The median follow-up time was 98.97 months (range, 2.63–120.47 months) and the median age was 55 years old (range, 23–86 years). The majority of patients were male (70.7%). In addition, the majority of patients suffered from Eastern Cooperative Oncology Group Performance Status (ECOG PS) 0 (67.4%) or 1 (20.2%), Fuhrman grade II (74.6%), or TNM stage I or III (63.1 and 25.1%, respectively) diseases. Regional lymph node metastasis was identified in only 2 individuals in this cohort and 16 patients developed distant metastatic disease prior to surgery.
Table I.
Clinicopathological characteristics of patients with clear cell renal cell carcinoma.
| Clinicopathological characteristic | n | % |
|---|---|---|
| All patients | 331 | 100 |
| Follow-up, months | ||
| Median (range) | 98.97 | |
| (2.63–120.47) | ||
| Age, years | ||
| Median (range) | 55 (23–86) | |
| Sex | ||
| Female | 97 | 29.3 |
| Male | 234 | 70.7 |
| ECOG PS | ||
| 0 | 223 | 67.4 |
| 1 | 67 | 20.2 |
| 2 | 32 | 9.7 |
| 3 | 9 | 2.7 |
| Fuhrman nuclear grade | ||
| 1 | 36 | 10.9 |
| 2 | 247 | 74.6 |
| 3 | 45 | 13.6 |
| 4 | 3 | 0.9 |
| Necrosis | ||
| Absent | 287 | 86.6 |
| Present | 44 | 13.3 |
| T classification | ||
| T1 | 215 | 65.0 |
| T2 | 25 | 7.6 |
| T3 | 87 | 26.3 |
| T4 | 4 | 1.2 |
| N classification | ||
| Nx | 283 | 85.5 |
| N0 | 46 | 13.9 |
| N1 | 2 | 0.6 |
| Distant metastasis | ||
| No | 315 | 95.2 |
| Yes | 16 | 4.8 |
| TNM stage | ||
| I | 209 | 63.1 |
| II | 20 | 6.0 |
| III | 83 | 25.1 |
| IV | 19 | 5.7 |
| Surgery | ||
| Nephron-sparing (open) | 37 | 11.2 |
| Nephron-sparing (laparoscopic) | 1 | 0.3 |
| Radical nephrectomy (open) | 268 | 81.0 |
| Radical nephrectomy (laparoscopic) | 25 | 7.5 |
ECOG PS, Eastern Cooperative Oncology Group performance status.
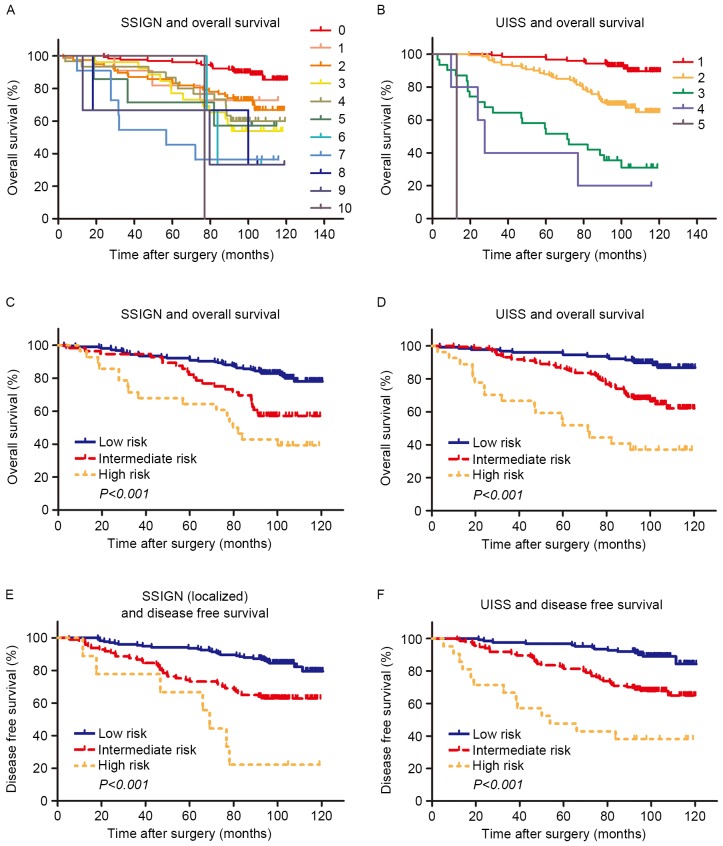
A number of models were used to stratify the survival risks of patients with ccRCC beyond TNM stage classification. The UISS score was used for OS and DFS analyses. For SSIGN stratifications, the classical SSIGN score was applied for OS evaluation and the SSIGN (localized) score was applied for DFS investigation. Owing to the limited number of samples and the complexity of the classical SSIGN score, the system was simplified into 3 risk groups (low, 0–2; intermediate, 3–4; high, 5–10). For similar reasons, when evaluating OS using the UISS risk group stratification, scores of 3–6 were combined into a single high-risk group. Kaplan-Meier estimator survival analyses are presented in Fig. 1. The SSIGN and the UISS score systems performed well (P<0.001 for both OS and PFS, Fig. 1C-F) in risk stratification (Fig. 1).
Figure 1.
Kaplan-Meier estimator analysis of overall survival and disease free survival in clear cell renal cell carcinoma according to the SSIGN/SSIGN (localized) and UISS score. Overall survival according to (A) SSIGN (B) UISS (C) SSIGN low/intermediate/low and (D) UISS low/intermediate/high score. Disease free survival according to (E) SSIGN (localized) and (F) UISS low/intermediate/high risk. SSIGN, Stage, Size, Grade and Necrosis; UISS, University of California Los Angeles Integrated Staging System.
JMJD3 expression in ccRCC tissue
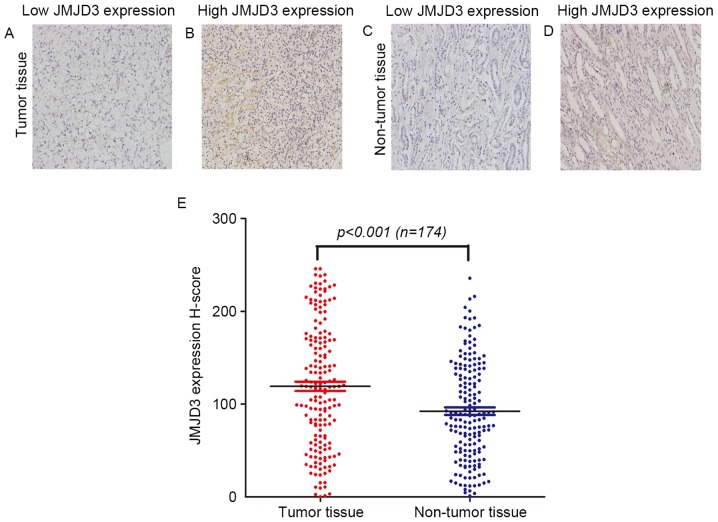
The expression of JMJD3 in tumor tissue and paired non-tumor tissue was investigated using immunohistochemical staining and categorized into low or high expression groups (Fig. 2A-D). In tumor tissue, positive JMJD3 staining was detected in the nucleus of ccRCC cells (Fig. 2B). In non-tumor renal tissue, JMJD3 was selectively expressed in the nucleus of nephric tubule cells (Fig. 2D). The H-score of JMJD3 expression was 119.0±66.7 (median, 118.42; range, 0.00–245.90) in tumor tissue and 92.3±53.5 (median, 86.31; range, 0.90–235.64) in non-tumor tissue, respectively (P<0.001; Fig. 2E).
Figure 2.
Immunohistochemical analysis of JMJD3 expression in human samples. Representative JMJD3 immunohistochemical staining in clear cell renal cell carcinoma with (A) low and (B) high JMJD3 expression, and non-tumor tissue with (C) low and (D) high JMJD3 expression (magnification, ×200). (E) H-score of JMJD3 expression in paired tumor and non-tumor tissue, compared using a t-test. JMJD3, jumonji domain-containing protein 3.
Associations between clinicopathological features and JMJD3 expression
The associations between patient clinicopathological characteristics and JMJD3 expression in tumor and non-tumor tissue are described in Table II. Analysis of 302 tumor tissue specimens revealed that JMJD3 expression was associated with ECOG PS (P=0.030) and tumor size (P=0.004) but not with with age (P=0.084). In addition, JMJD3 expression was associated with ECOG PS (P=0.017) and age (P=0.026) in non-tumor tissue samples (n=174). No associations between JMJD3 expression and other patient clinicopathological characteristics were identified.
Table II.
Clinicopathological characteristics of patients according to JMJD3 expression in tumor and non-tumor tissue.
| JMJD3 expression in tumor tissue (n=302) | JMJD3 expression in non-tumor tissue (n=174) | |||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristic | High | Low | P-value | High | Low | P-value |
| All patients | 160 | 142 | 96 | 78 | ||
| Age, yearsa | 0.084b | 0.026b | ||||
| ≤55 | 88 | 63 | 64 | 39 | ||
| >55 | 72 | 79 | 32 | 39 | ||
| Sex | 0.667b | 1.000b | ||||
| Female | 46 | 45 | 29 | 24 | ||
| Male | 114 | 97 | 67 | 54 | ||
| ECOG PS | 0.030c | 0.017c | ||||
| 0 | 121 | 90 | 75 | 46 | ||
| 1 | 28 | 29 | 17 | 19 | ||
| 2 | 10 | 17 | 3 | 11 | ||
| 3 | 1 | 6 | 1 | 2 | ||
| Fuhrman nuclear grade | 0.118c | 0.161c | ||||
| 1 | 23 | 11 | 15 | 5 | ||
| 2 | 119 | 106 | 73 | 61 | ||
| 3 | 16 | 24 | 7 | 11 | ||
| 4 | 2 | 1 | 1 | 1 | ||
| Necrosis | 0.465b | 1.000b | ||||
| Absent | 149 | 128 | 92 | 75 | ||
| Present | 11 | 14 | 4 | 3 | ||
| T classification | 0.108c | 0.479c | ||||
| T1 | 107 | 85 | 73 | 51 | ||
| T2 | 7 | 17 | 2 | 3 | ||
| T3 | 44 | 38 | 20 | 23 | ||
| T4 | 2 | 2 | 1 | 1 | ||
| N classification | 1.000b | 0.917b | ||||
| N0 | 159 | 141 | 96 | 77 | ||
| N1 | 1 | 1 | 0 | 1 | ||
| Distant metastasis | 0.273b | 1.000b | ||||
| No | 156 | 134 | 93 | 76 | ||
| Yes | 4 | 8 | 3 | 2 | ||
| TNM stage | 0.132c | 0.587c | ||||
| I | 105 | 83 | 71 | 51 | ||
| II | 7 | 14 | 2 | 3 | ||
| III | 43 | 36 | 20 | 22 | ||
| IV | 5 | 9 | 3 | 2 | ||
| Tumor size, cma | 0.004b | 0.856b | ||||
| ≤4 | 106 | 70 | 70 | 55 | ||
| >4 | 54 | 72 | 26 | 23 | ||
Split at median
χ2 test or Fisher's exact test
Cochran-Mantel-Haenszel χ2 test. ECOG PS, Eastern Cooperative Oncology Group performance status; TNM, tumor-necrosis-metastasis.
Low expression of JMJD3 is associated with poor OS and DFS
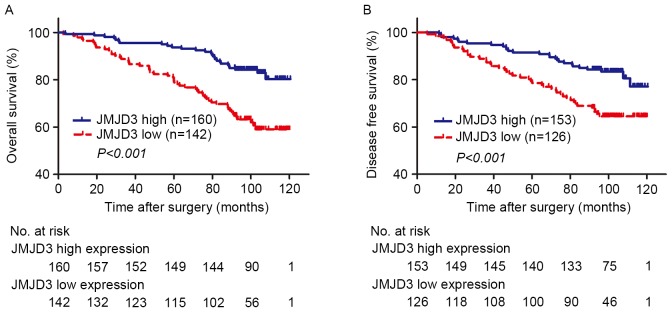
Kaplan-Meier estimator analyses were performed to assess the association between OS or DFS and JMJD3 expression in tumor tissue. X-tile software was used to determine the H-score cutoff points for low and high expression using the minimum P-value method. A total of 23 patients were excluded due to preoperative distant metastasis, regional lymph node metastasis or mortality from other causes. Low expression of JMJD3 in tumor tissue was significantly associated with poor OS (Fig. 3A; P<0.001) and DFS (Fig. 3B, P<0.001).
Figure 3.
Kaplan-Meier estimator analysis of overall survival and disease free survival in clear cell renal cell carcinoma according to tumoral JMJD3 expression. (A) Overall survival according to tumoral JMJD3 expression. (B) Disease free survival according to tumoral JMJD3 expression. JMJD3, jumonji domain-containing protein 3; no. at risk, number of patients at risk. No. at risk indicates the number of patients at risk.
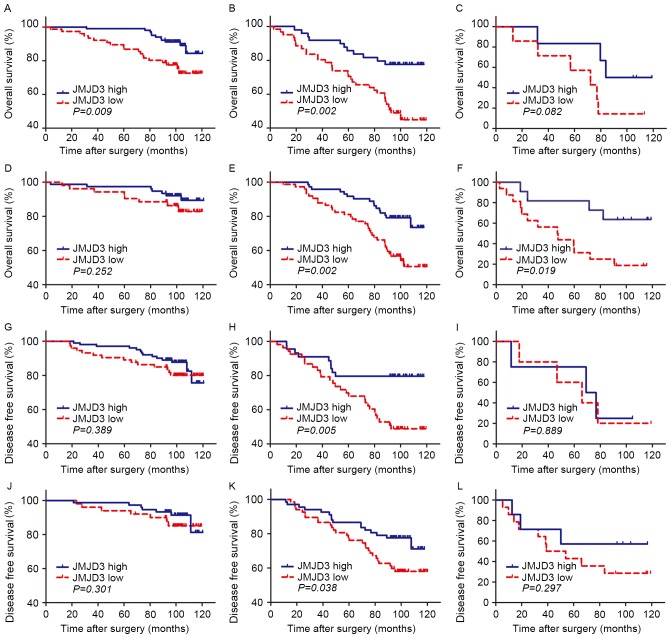
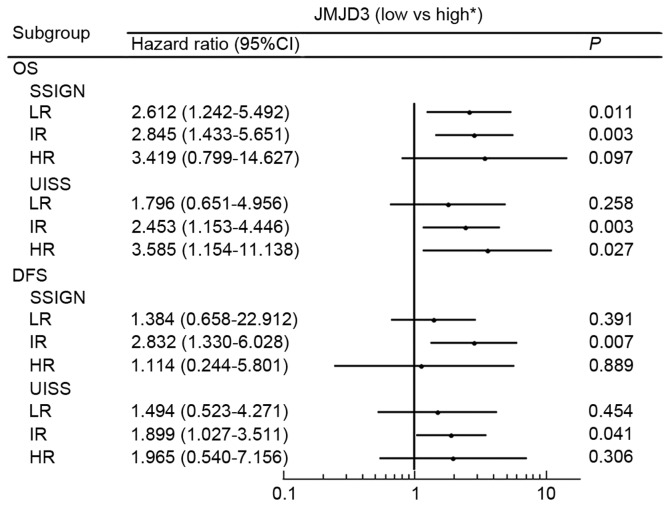
To further explore the prognostic value of tumor JMJD3 expression in different risk groups, the SSIGN and UISS score systems were used to group patients into low, intermediate and high risk groups for OS and DFS (Fig. 4). For the SSIGN low risk group, an association was observed between low JMJD3 expression and poor prognosis for OS (P=0.009; Fig. 4A) but not for DFS (P=0.389; Fig. 4G). An association was observed between low JMJD3 expression and poor prognosis in the SSIGN intermediate risk groups for OS and DFS (OS, P=0.002; DFS, P=0.005; Fig. 4B and H, respectively). In the SSIGN high risk group, no association was observed between low JMJD3 expression and poor prognosis for OS or DFS (P=0.082 and P=0.889, respectively; Fig. 4C and I). No association was observed between low JMJD3 expression and poor prognosis in the UISS low risk groups (OS, P=0.0252; DFS, P=0.301; Fig. 4D and J, respectively). Low JMJD3 expression was associated with a shorter survival time in the UISS intermediate risk groups for OS and DFS (OS, P=0.002; DFS, P=0.038; Fig. 4E and K, respectively). In the UISS high risk group, an association was observed between low JMJD3 expression and poor prognosis for OS (P=0.019; Fig. 4F) but not for DFS (P=0.889; Fig. 4I).
Figure 4.
Kaplan-Meier estimator analyses of overall survival and disease free survival in clear cell renal cell carcinoma according to tumoral JMJD3 expression stratified by the SSIGN/SSIGN (localized) and UISS score. Overall survival of patients in the SSIGN (A) low, (B) intermediate and (C) high risk groups. Overall survival of patients in the UISS (D) low, (E) intermediate and (F) high risk groups. Disease free survival of patients in the SSIGN (localized) (G) low, (H) intermediate and (I) high risk groups. Disease free survival of patients in the UISS (J) low, (K) intermediate and (L) high risk groups. JMJD3, jumonji domain-containing protein 3; SSIGN, Stage, Size, Grade and Necrosis; UISS, University of California Los Angeles Integrated Staging System.
Univariate and multivariate analyses
Univariate survival analyses of JMJD3 expression in different risk groups revealed similar results to the Kaplan-Meier estimator analyses (Tables III and IV; Fig. 5). In addition, JMJD3 expression was assessed in peritumoral specimens for prognostic value in different SSIGN or UISS risk groups, but no association was observed (Table III). The univariate analyses highlighted the prognostic value of intratumoral JMJD3 expression for OS and DFS (P<0.001 and P=0.001, respectively; n=302; Table V). Univariate analysis of peritumoral JMJD3 expression revealed a similar result (OS, P=0.006; DFS, P=0.012; n=174; Table V). In the multivariate analysis, low expression of JMJD3 in tumor tissue increased the risk of mortality by >2 (hazard ratio (HR) 2.141, 95% CI 1.304–3.517, P=0.003; Table VI), and was correlated with a shorter DFS (HR 1.737, 95% CI 1.047–2.882, P=0.033) (Table VI). These results suggested that tumoral JMJD3 expression is an independent prognostic marker for ccRCC when adjusted to age, ECOG PS, Fuhrman grade, necrosis, T stage and distant metastasis.
Table III.
Univariate analyses of JMJD3 expression associated with survival in different SSIGN & SSIGN (local) risk groups.
| Overall survival | Disease free survival | ||||||
|---|---|---|---|---|---|---|---|
| Risk group | Clinicopathological characteristics | Hazard ratio | 95% CI | P-valuea | Hazard ratio | 95% CI | P-valuea |
| Low risk | Intratumoral JMJD3 (low vs. high) | 2.612 | 1.242–5.492 | 0.011 | 1.384 | 0.658–2.912 | 0.391 |
| Peritumoral JMJD3 (low vs. high) | 1.920 | 0.828–4.449 | 0.128 | 2.247 | 0.917–5.504 | 0.077 | |
| Intermediate risk | Intratumoral JMJD3 (low vs. high) | 2.845 | 1.433–5.651 | 0.003 | 2.832 | 1.330–6.028 | 0.007 |
| Peritumoral JMJD3 (low vs. high) | 2.059 | 0.790–5.363 | 0.139 | 1.624 | 0.648–4.072 | 0.301 | |
| High risk | Intratumoral JMJD3 (low vs. high) | 3.419 | 0.799–14.627 | 0.097 | 1.114 | 0.244–5.081 | 0.889 |
| Peritumoral JMJD3 (low vs. high) | 55.578 | 0.001–5456325.509 | 0.493 | – | – | – | |
Data obtained from the Cox proportional hazards model. SSIGN, stage, size, grade and necrosis; CI, confidence interval; JMJD3, jumonji domain-containing protein 3.
Table IV.
Univariate analyses of JMJD3 expression associated with survival in different UISS & UISS (local) risk groups.
| Overall survival | Disease free survival | ||||||
|---|---|---|---|---|---|---|---|
| Risk group | Clinicopathological characteristics | Hazard ratio | 95% CI | P-valuea | Hazard ratio | 95% CI | P-valuea |
| Low risk | Intratumoral JMJD3 (low vs. high) | 1.796 | 0.651–4.956 | 0.258 | 1.494 | 0.523–4.271 | 0.454 |
| Peritumoral JMJD3 (low vs. high) | 1.409 | 0.395–5.021 | 0.597 | 1.844 | 0.532–6.399 | 0.335 | |
| Intermediate risk | Intratumoral JMJD3 (low vs. high) | 2.453 | 1.153–4.446 | 0.003 | 1.899 | 1.027–3.511 | 0.041 |
| Peritumoral JMJD3 (low vs. high) | 2.122 | 0.915–4.921 | 0.079 | 1.650 | 0.722–3.773 | 0.235 | |
| High risk | Intratumoral JMJD3 (low vs. high) | 3.585 | 1.154–11.138 | 0.027 | 1.965 | 0.540–7.156 | 0.306 |
| Peritumoral JMJD3 (low vs. high) | 1.682 | 0.349–8.119 | 0.517 | 1.129 | 0.224–5.680 | 0.883 | |
Data obtained from the Cox proportional hazards model. UISS, University of California Los Angeles Integrated Staging System; CI, confidence interval; JMJD3, jumonji domain-containing protein 3.
Figure 5.
Hazard ratios for overall survival probabilities and disease free survival probabilities based on JMJD3 expression levels in patient subgroups stratified by the SSIGN/SSIGN (localized) and UISS score. *Reference group. OS, overall survival; DFS, disease free survival; LR, low risk; IR, intermediate risk; HR, high risk; CI, confidence interval; JMJD3, jumonji domain-containing protein 3.
Table V.
Univariate analyses of characteristics associated with overall survival and disease free survival.
| Overall survival | Disease free survival | |||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristics | Hazard ratio | 95% CI | P-valuea | Hazard ratio | 95% CI | P-valuea |
| Age, yearsb | ||||||
| >55 vs. ≤55 | 1.196 | 1.068–1.340 | 0.002 | 1.743 | 1.085–2.798 | 0.021 |
| Sex | ||||||
| Male vs. female | 1.069 | 0.664–1.720 | 0.785 | 0.925 | 0.563–1.521 | 0.759 |
| ECOG PS | <0.001 | 0.001 | ||||
| 0 | 1.000 | Reference | – | 1.000 | Reference | – |
| 1 | 1.864 | 1.096–3.170 | 0.022 | 1.871 | 1.069–3.275 | 0.028 |
| 2 | 2.805 | 1.507–5.219 | <0.001 | 3.011 | 1.576–5.753 | 0.001 |
| 3 | 13.134 | 5.529–31.202 | <0.001 | 5.039 | 1.212–20.957 | 0.026 |
| Fuhrman grade | 0.003 | 0.002 | ||||
| 1 | 1.000 | Reference | – | 1.000 | Reference | – |
| 2 | 3.345 | 1.048–10.671 | 0.041 | 3.036 | 0.947–3.036 | 0.062 |
| 3 | 7.126 | 2.107–24.104 | 0.002 | 7.425 | 2.174–25.361 | 0.001 |
| 4 | 4.971 | 0.517–47.829 | 0.165 | 0.000 | – | 0.977 |
| Necrosis | ||||||
| Present vs. absent | 2.440 | 1.347–4.421 | 0.003 | 2.132 | 1.059–4.293 | 0.034 |
| T classification | <0.001 | <0.001 | ||||
| T1 | 1.000 | Reference | – | 1.000 | Reference | – |
| T2 | 4.383 | 2.353–8.163 | <0.001 | 4.252 | 2.143–8.438 | <0.001 |
| T3 | 2.393 | 1.468–3.901 | <0.001 | 2.421 | 1.447–4.049 | 0.001 |
| T4 | 11.618 | 3.555–37.975 | <0.001 | 43.220 | 5.437–343.597 | <0.001 |
| N classification (n=48) | ||||||
| N1 vs. N0 | 1.800 | 0.250–12.946 | 0.559 | – | – | – |
| Distant metastasis | ||||||
| Yes vs. no | 6.916 | 3.560–13.436 | <0.001 | – | – | – |
| Tumor size | ||||||
| >4 cm vs. ≤4 cm | 2.079 | 1.342–3.221 | <0.001 | 2.391 | 1.492–3.830 | <0.001 |
| TNM stage | <0.001 | <0.001 | ||||
| I | 1.000 | Reference | – | 1.000 | Reference | – |
| II | 3.964 | 1.989–7.898 | <0.001 | 4.252 | 2.143–8.438 | <0.001 |
| III | 2.622 | 1.571–4.374 | <0.001 | 2.421 | 1.447–4.049 | 0.001 |
| IV | 13.180 | 6.742–25.768 | <0.001 | 43.220 | 5.437–343.597 | <0.001 |
| Intratumoral JMJD3 (n=302) | ||||||
| Low vs. high | 2.327 | 1.474–3.674 | <0.001 | 2.212 | 1.369–3.572 | 0.001 |
| Peritumoral JMJD3 (n=174) | ||||||
| Low vs. high | 2.381 | 1.287–4.403 | 0.006 | 2.204 | 1.187–4.093 | 0.012 |
Data obtained from the Cox proportional hazards model.
Split at median. ECOG PS, Eastern Cooperative Oncology Group performance status; CI, confidence interval; TNM, tumor-node-metastasis; JMJD3, jumonji domain-containing protein 3.
Table VI.
Multivariate analyses of characteristics associated with overall survival and disease free survival.
| Overall survival | Disease free survival | |||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristics | Hazard ratio | 95% CI | P-valuea | Hazard ratio | 95% CI | P-valuea |
| Age, yearsb | ||||||
| >55 vs. ≤55 | 1.518 | 0.928–2.485 | 0.097 | 1.292 | 0.783–2.132 | 0.315 |
| ECOG PS | 0.001 | 0.032 | ||||
| 0 | 1.000 | Reference | – | 1.000 | Reference | – |
| 1 | 1.708 | 0.966–3.019 | 0.066 | 1.659 | 0.923–2.983 | 0.091 |
| 2 | 2.313 | 1.207–4.434 | 0.012 | 2.543 | 1.311–4.932 | 0.006 |
| 3 | 5.046 | 1.962–12.977 | 0.001 | 1.921 | 0.422–8.737 | 0.398 |
| Fuhrman grade | 0.414 | 0.049 | ||||
| 1 | 1.000 | Reference | – | 1.000 | Reference | – |
| 2 | 2.382 | 0.738–7.691 | 0.147 | 2.365 | 0.729–7.668 | 0.151 |
| 3 | 2.906 | 0.789–10.701 | 0.109 | 4.834 | 1.342–17.416 | 0.016 |
| 4 | 1.601 | 0.134–19.165 | 0.710 | – | – | – |
| Necrosis | ||||||
| Present vs. absent | 1.487 | 0.731–3.024 | 0.273 | 1.136 | 0.508–2.544 | 0.756 |
| T stage | 0.032 | <0.001 | ||||
| T1 | 1.000 | Reference | – | 1.000 | Reference | – |
| T2 | 2.411 | 1.243–4.677 | 0.009 | 2.861 | 1.401–5.842 | 0.004 |
| T3 | 1.747 | 1.035–2.947 | 0.037 | 2.394 | 1.397–4.104 | 0.001 |
| T4 | 3.308 | 0.623–17.563 | 0.160 | 42.448 | 4.850–371.518 | 0.001 |
| Distant metastasis | ||||||
| Yes vs. no | 3.524 | 1.519–8.175 | 0.003 | – | – | – |
| Intratumoral JMJD3 (n=302) | ||||||
| Low vs. high | 2.141 | 1.304–3.517 | 0.003 | 1.737 | 1.047–2.882 | 0.033 |
Data obtained from the Cox proportional hazards model.
Split at median. ECOG PS, Eastern Cooperative Oncology Group performance status; CI, confidence interval.
Discussion
JMJD3 is encoded by the KDM6B gene, which is located on chromosome 17p13.1 (21). Together with UTX, it forms the KDM6 subfamily, whose primary function is to reverse gene silencing induced by H3K27 methylation through the demethylation of H3K27me3 and H3K27me2 (22). H3K27 methylation, primarily catalyzed by EZH2, has been demonstrated to serve oncogenic functions in a number of human carcinomas, including renal cell carcinoma (8). Reduced expression of JMJD3 has been demonstrated in certain types of cancer, which implies that JMJD3 is involved in cancer generation, progression and suppression (23).
Results from the present study demonstrated that low expression of JMJD3 is an independent poor prognostic factor for patients with ccRCC. The incorporation of JMJD3 expression as a factor into the SSIGN/UISS score system may improve its predictive accuracy. Notably, improvement in prediction was primarily observed in patients classified into SSIGN/UISS intermediate risk groups. Therefore, taking into account JMJD3 staining in tissues from patients in the SSIGN/UISS intermediate risk groups may better inform postoperative management.
Data from the present study indicated that JMJD3 serves a function in ccRCC progression. Although low JMJD3 expression was demonstrated to correlate with poor prognosis, JMJD3 expression was revealed to be elevated in tumor tissues compared with non-tumor tissues from the same patient. In addition, a previous study demonstrated that EZH2 is upregulated in ccRCC (10). One potential reason for the concurrent upregulation of a histone methyltransferase and histone demethylase in ccRCC tissue is that JMJD3 and EZH2 are upregulated in different cells types, which is consistent with the substantial intratumoral heterogeneity of ccRCC (24). Alternatively, a compensation mechanism may be adopted if JMJD3 and EZH2 are upregulated in the same ccRCC cells. When EZH2 is upregulated during tumor progression, the H3K27 methylation level would be elevated. As a result, a series of tumor suppressor genes would be downregulated and oncogenes upregulated. JMJD3 may be upregulated to compensate for these changes in gene expression.
The molecular mechanisms underlying the involvement of JMJD3 in RCC suppression remain unclear. One potential explanation is oncogene-induced senescence (OIS). OIS acts as a tumor suppressor mechanism by driving stable growth arrest of cancer progenitor cells harboring the initial oncogenic mutation (25). P53/cyclin-dependent kinase inhibitor 1A and 2A (p16INK4a) signaling axes are effector pathways of replicative senescence, leading to the hypophosphorylated form of retinoblastoma protein which in turn represses the activity of cell cycle progression-associated transcription factors (26). JMJD3 has been reported to bind to the cyclin-dependent kinase inhibitor protein-tumor suppressor ARF-p16INK4a locus, inhibiting EZH2 (PRC subunit) occupancy and decreasing H3K27me3 content, resulting in the increased expression of the three proteins and the promotion of cell senescence (27). In RCC, JMJD3 has been reported to activate p16INK4a gene expression, resulting in inhibition of RCC development via induction of cell cycle arrest (14). Data from the present study revealed that downregulation of JMJD3 is an independent prognostic factor of RCC, indicating its effect against H3K27 methylation and tumor development.
JMJD3 has been demonstrated to be involved in tumor generation, progression and suppression, making it a potentially attractive drug target. Kruidenier et al (28) identified compounds that inhibit UTX and JMJD3. The KDM6 inhibitor demonstrated high substrate selectivity and inhibited transcription of a number of pro-inflammatory genes in vitro by altering the H3K27me3 level of certain KDM6 target genes (29). Therefore, KDM6 inhibitors may be of use to treat patients.
There are several limitations of the present study that merit further discussion. First, the number of patients enrolled in this study was limited. A larger, multi-centered, prospective study is required for validation of these results. Second, non-ccRCC patients were excluded from the present study due to the limited number of non-ccRCC samples. Analysis of the prognostic effect of JMJD3 in papillary RCC and chromophobe RCC is required. Third, although two tissue cores from the same tumor allowed the immunohistochemical data to be verified, substantial intratumoral heterogeneity of RCC may influence the findings of the present study. Fourth, functional studies are required to elucidate the molecular mechanisms underlying the association between JMJD3 expression and RCC. Finally, the prognostic significance of other demethylases, including UTX, requires further investigation.
In conclusion, low JMJD3 expression in ccRCC tissue is an independent poor prognostic factor in patients with ccRCC, particularly for patients with SSIGN/UISS intermediate-risk disease. These findings provide a novel independent predictor of clinical outcome and may improve the postoperative management of patients with ccRCC, allowing the personalization of treatment.
Acknowledgements
The present study was supported by the National Basic Research Program of China (grant no. 2012CB822104), the National Natural Science Foundation of China (grant nos. 31100629, 31270863, 31300671, 81372755, 31470794, 81401988, 81402082, 81402085, 81471621, 81472227, 81472376, 31570803 and 81572352), the Program for New Century Excellent Talents in University (grant no. NCET-13-0146) and the Shanghai Rising-Star Program (grant no. 13QA1400300).
Glossary
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- RCC
renal cell carcinoma
- JMJD3
jumonji domain-containing protein 3
- UTX
ubiquitously-transcribed TPR gene on the X chromosome
- H3K27me3
Histone H3 lysine 27 tri-methylation
- EZH2
enhancer of zeste homolog 2
- SSIGN
stage, size, grade and necrosis
- UISS
University of California Los Angeles Integrated Staging System
- OS
overall survival
- DFS
disease free survival
- ECOG PS
Eastern Cooperative Oncology Group Performance Status
- CI
confidence interval
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 3.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 5.Volpe A, Patard JJ. Prognostic factors in renal cell carcinoma. World J Urol. 2010;28:319–327. doi: 10.1007/s00345-010-0540-8. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman AM, Cairns P. Epigenetics of kidney cancer and bladder cancer. Epigenomics. 2011;3:19–34. doi: 10.2217/epi.10.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi P, Allis CD, Wang GG. Covalent histone modifications-miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat. 2014;46:209–222. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Xu Z, Zhong L, Wang H, Jiang S, Long Q, Xu J, Guo J. Enhancer of zeste homolog 2 (EZH2) promotes tumour cell migration and invasion via epigenetic repression of E-cadherin in renal cell carcinoma. BJU Int. 2016;117:351–362. doi: 10.1111/bju.12702. [DOI] [PubMed] [Google Scholar]
- 11.Svotelis A, Bianco S, Madore J, Huppé G, Nordell-Markovits A, Mes-Masson AM, Gévry N. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ERα ligand dependency. EMBO J. 2011;30:3947–3961. doi: 10.1038/emboj.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 13.Ene CI, Edwards L, Riddick G, Baysan M, Woolard K, Kotliarova S, Lai C, Belova G, Cam M, Walling J, et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One. 2012;7:e51407. doi: 10.1371/journal.pone.0051407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Guo X, Wang Y, Qiu W, Chang Y, Zhang A, Duan X. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. BMC Cancer. 2012;12:470. doi: 10.1186/1471-2407-12-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED, Weaver AL, Parker AS, Zincke H. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials. Cancer. 2003;97:1663–1671. doi: 10.1002/cncr.11234. [DOI] [PubMed] [Google Scholar]
- 16.Zisman A, Pantuck AJ, Dorey F, Said JW, Shvarts O, Quintana D, Gitlitz BJ, deKernion JB, Figlin RA, Belldegrun AS. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–1657. doi: 10.1200/JCO.2001.19.6.1649. [DOI] [PubMed] [Google Scholar]
- 17.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: The SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Xu J, Zhou L, Yun X, Chen L, Wang S, Sun L, Wen Y, Gu J. Hepatitis B virus large surface antigen promotes liver carcinogenesis by activating the Src/PI3K/Akt pathway. Cancer Res. 2011;71:7547–7557. doi: 10.1158/0008-5472.CAN-11-2260. [DOI] [PubMed] [Google Scholar]
- 19.Budwit-Novotny DA, McCarty KS, Cox EB, Soper JT, Mutch DG, Creasman WT, Flowers JL, McCarty KS., Jr Immunohistochemical analyses of estrogen receptor in endometrial adenocarcinoma using a monoclonal antibody. Cancer Res. 1986;46:5419–5425. [PubMed] [Google Scholar]
- 20.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 21.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature. 2006;440:1045–1049. doi: 10.1038/nature04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases; Proc Natl Acad Sci USA; 2007; pp. 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aird KM, Zhang R. Nucleotide metabolism, oncogene-induced senescence and cancer. Cancer Lett. 2015;356:204–210. doi: 10.1016/j.canlet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 27.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Shi Y. A new horizon for epigenetic medicine? Cell Res. 2013;23:326–328. doi: 10.1038/cr.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]