Abstract
CKLF-like MARVEL transmembrane domain-containing 5 (CMTM5) has been reported to function as a potential tumor suppressor in several human cancers. However, the involvement of CMTM5 in human renal cell carcinoma (RCC) remains unclear. The current study aimed to detect its expression pattern in RCC tissues and cells, and to determine its anti-proliferative functions in this malignancy. The mRNA and protein expression levels of CMTM5 in RCC tissues and cells were detected by reverse transcription-quantitative polymerase chain reaction, immunohistochemistry and western blotting. Following the transfection with CMTM5 lentivirus or control lenti-EGFP lentivirus into the RCC cell line ACHN, the viability, migration, apoptosis and cell cycle of these cells were detected by Cell Counting kit-8 assay, Transwell assay and flow cytometry, respectively. Compared with the adjacent non-malignant kidney tissue samples, CMTM5 expression was significantly downregulated in RCC tissues (P<0.05). In addition, enforced expression of CMTM5 could efficiently inhibit the cell growth of ACHN cells, which were arrested in G0/G1 phase. Furthermore, the migration and invasion of ACHN cells were also inhibited by restoration of CMTM5 expression. The present data suggest that CMTM5 may function as a tumor suppressor in human RCC by suppressing the viability of RCC cells, implying its potential as a therapeutic target for this malignancy.
Keywords: renal cell carcinoma, CMTM5, tumor suppressor
Introduction
Renal cell carcinoma (RCC) is a common malignant neoplasm in the urinary system and the sixth leading cause of cancer mortality in Western countries (1). It is estimated that 63,920 new cases of kidney cancer will occur in 2014 in the USA, with 13,860 mortalities due to this disease (1). Clear cell renal cell carcinoma (CCRCC), accounting for 70% of all RCC cases, and papillary (pRCC; 10–15% of all RCC cases) carcinomas, constitute the two most frequent subtypes of RCC (2). Although clinical management of RCC has changed in previous years, with increased incidental diagnosis and initiation of therapy in localized stages, 20–40% of patients with RCC succumb to cancer progression (3). Due to the high resistance of the tumors to chemotherapy and radiotherapy (4), it is important to identify novel approaches that can effectively inhibit RCC cell growth and metastasis. Molecularly-targeted therapies have shown potential in achieving these goals. CKLF-like MARVEL transmembrane domain-containing 5 (CMTM5) is a member of the CMTM protein family and was originally reported by the Human Genomics Center of Peking University (Beijing, China) in 2003 (5). CMTM5 is located at 14q11.2, and has been reported to act as a tumor-suppressor gene (TSG), since it was specifically downregulated in numerous human cancers, including cervical carcinoma, ovarian cancer, pancreatic cancer, myeloid leukemia and prostate cancer (6–11). However, the role of CMTM5 in human RCC remains unclear. The present study aimed to detect the expression pattern of CMTM5 and its association with clinicopathological characteristics in patients with RCC. The effect of CMTM5 on cell viability and apoptosis was also explored using an RCC cell line in vitro.
Materials and methods
Tissue samples and cancer cells
The present study was approved by the Human Study Ethics Committees of the Ministry of Public Health (the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China). All specimens were handled and made anonymous according to the ethical and legal standards. A tissue microarray (TMA) of 75 patients with RCC and adjacent normal kidney tissues was purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). The mean age of the patients with RCC was 58.7 years. Tissue samples were collected from 75 patients with RCC who underwent radical nephrectomy at the Department of Urology, the First Affiliated Hospital of Wenzhou Medical University between September 2006 and December 2008. None of the patients had received chemotherapy or radiotherapy prior to surgery. The RCC tissues consisted of 34 stage I, 23 stage II, 14 stage III and 4 stage IV, according to the World Health Organization criteria from 2004 (12). The clinicopathological features of this patient cohort are summarized in Table I. The metastatic clear renal cell adenocarcinoma CAKII, papillary renal cell carcinoma ACHN, the metastatic renal cell adenocarcinoma OS-RC-2 and the human kidney HK2 cell lines were purchased from the cell bank of the Chinese Academy of Sciences (Beijing, China), and were maintained in RPMI 1640 medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA USA) at 37°C with a 5% CO2 humidified atmosphere.
Table I.
Associations between CMTM5 expression and various clinicopathological parameters of human renal cell carcinoma.
| Clinicopathological parameters | Cases, n | CMTM5-low, n (%) | CMTM5-high, n (%) | P-value |
|---|---|---|---|---|
| Sex | 0.80 | |||
| Male | 50 | 32 (42.7) | 18 (24.0) | |
| Female | 25 | 17 (22.7) | 8 (10.7) | |
| Age (years) | 0.32 | |||
| <60 | 45 | 27 (36.0) | 18 (24.0) | |
| ≥60 | 30 | 22 (29.3) | 8 (10.7) | |
| Clinical stage | 0.58 | |||
| I–II | 57 | 36 (48.0) | 21 (28) | |
| III–IV | 18 | 13 (17.3) | 5 (6.7) | |
| Histological grade | 0.63 | |||
| I–II | 40 | 25 (33.3) | 15 (20.0) | |
| III–IV | 35 | 24 (32.0) | 11 (14.7) |
CMTM5, CKLF-like MARVEL transmembrane domain-containing 5.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR analysis was performed to detect the expression of CMTM5 mRNA in RCC and adjacent normal kidney tissues, as well as in RCC and human kidney cells. Briefly, the 20-µl RT reaction system (K1622, Fermentas; Thermo Fisher Scientific, Inc.) containing 1 µg RNA and diethyl pyrocarbonate-treated water was placed in an RNase-free PCR tube. The components were mixed and heated to 65°C for 5 min before being transferred to an ice bath. Then, 4.1 µl 5X reaction buffer, 0.6 µl RiboLock RNase inhibitor (20 U/µl), 2 µl deoxynucleotide triphosphate mix (10 mM of each deoxynucleotide triphosphate) and 1.1 µl ReverTraAce were added (K1622; Fermentas; Thermo Fisher Scientific, Inc.). The mixture was heated to 72°C for 60 min, followed by 72°C for 10 min. The complementary DNAs were either stored at −20°C or immediately used for qPCR, which was performed using the AceQ qPCR SYBR-Green Master mix, Vazyme Biotech Co., Ltd., China) on the ABI 2720 Thermal Cycler; Thermo Fisher Scientific, Inc.). The CMTM5 primers used for qPCR were forward, 5′-TGATGCCTTCAAGATCTACCG-3′ and reverse, 5′-TCCACAGATAAGTCCCCAGTG-3′. As an internal control, β-actin was used, and its primers were forward, 5′-GGCACTCTTCCAGCCTTCC-3′ and reverse, 5′-GAGCCGCCGATCCACAC-3′. The thermocycler conditions were as follows: Step 1: 95°C for 2 min; then Step 2: 60–95°C for 2 min for 40 cycles; and then Step 3: 72°C for 2 min for 40 cycles. The 2−ΔΔCq method was used to quantify the results (13).
Western blot analysis
Total ACHN cell protein was extracted using radioimmunoprecipitation protein lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) with added 1% protease inhibitor cocktail and 1 mM phenylmethylsulfonyl fluoride (PMSF). The protein concentration was measured by BCA Protein Assay kit (Beyotime Institute of Biotechnology). The 50 µg protein samples were separated by electrophoresis in 10% SDS-PAGE and then transferred onto a nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). The membranes were probed with ananti-CMTM5 antibody (dilution 1:500; kindly provided by Professor Wenling Han, Peking University) at 4°C overnight, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; sc2030; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at room temperature for 1 h. Then, the protein bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.). An anti-a-tubulin antibody (dilution 1:3,000; sc-73242, Santa Cruz Biotechnology, Inc.) was used as a loading control.
Cell transfection
Recombinant pLenti6.3-CMTM5-IRES-EGFP lentivirus and negative control lentivirus Lenti-EGFP were purchased from Shanghai R&S Biotechnology Co., Ltd. (Shanghai, China). ACHN cells (1×105 cells/dish) were seeded in a 24-well plate. The Lenti-EGFP and CMTM5 lentiviruses were infected into ACHN cells using Polo3000 (Shanghai R&S Biotechnology Co., Ltd.), according to the manufacturer's protocol. ACHN cells were infected with lentiviruses (1×109 TU/ml; Shanghai R&S Biotechnology Co., Ltd.) in the presence of 8 µg/ml Polybrene (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The efficiency of transfection was confirmed as >70% of cells were expressing EGFP.
Immunohistochemistry (IHC)
The subcellular localization and expression level of CMTM5 protein in TMA (tissue thickness, 4 µm) was analyzed by the streptavidin-biotin complex method. Upon deparaffinization in xylene and rehydration in a 100-95-75–0% graded ethanol series, antigens were retrieved in citrate (0.1 M, pH 6.0) for 15 min at 95°C. The TMA samples were incubated with 3% hydrogen peroxide for 20 min at room temperature, washed with PBS, and then incubated with 3% goat serum (Shanghai Mengry Bio-Technology Co., Ltd., Shanghai, China) for 30 min at room temperature to reduce non-specific binding. Subsequently, the TMA were incubated with a primary polyclonal rabbit anti-human CMTM5 antibody (dilution 1:1,000; kindly provided by Professor Wenling Han), or with goat serum instead of anti-CMTM5 antibody as a negative control, at 4°C overnight, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; SC2030; Santa Cruz Biotechnology, Inc.) for 30 min at room temperature. The immunostaining of CMTM5 protein was visualized with 3,3′-diaminobenzidine (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) subsequent to washing with PBS. The IHC results were evaluated by two independent investigators who were blinded to the clinical background of the specimens. The immunoreactive score of CMTM5 protein expression was semi-quantified by staining intensity and percentage of tumor cell positivity. The percentage of positive cells was scored as follows: 0 (0%), 1 (<10%), 2 (11–50%), 3 (51–80%) and 4 (>80%). The staining intensity was scored as follows: Below the level of detection =0; weak expression =1; moderate expression =2; and intense expression =3. The percentage of positive cells was multiplied with the staining intensity to obtain the immunoreactive score (scale, 0–12). Scores >6 were considered to indicate high expression, while scores =0–6 were considered to indicate low expression.
Cell viability analysis
The anti-proliferative effects of CMTM5 were determined by Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer's protocol. Following transfection with CMTM5-overexpressing lentivirus (CMTM5 OE) and negative control lentivirus (NC) ACHN cells were seeded in 96-well microplates at a density of ~3,000 cells per well for 24, 48 and 72 h, and then incubated at 37°C for another 2 h with CCK-8 (10 µl per well). The absorption of each well at 450 nm was measured under a microplate reader.
Cell apoptosis and cell cycle analyses
Cell apoptosis was measured by flow cytometry using the Annexin V-FITC/PI Apoptosis Detection kit (BD556547; BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer's protocol. At 72 h post-transfection, ACHN cells were harvested and incubated with fluorescein isothiocyanate (FITC)-conjugated Annexin V for 30 min at room temperature in the dark. Then samples were analyzed on a FACScan flow cytometer (BD Biosciences). The cell cycle distribution was analyzed 48 h after transfection. ACHN cells were washed in PBS, trypsinized, washed again in PBS and fixed in 70% ethanol at −20°C. Subsequently, 10 µg/ml propidium iodide (PI) was added to each sample, and the cell cycle was analyzed with a FACScan flow cytometer. The obtained data were analyzed using CellQuest software (version 7.5.3, BD Biosciences).
In vitro migration and invasion assays
Migration and invasion assays were performed using Transwell chamber inserts (Corning Incorporated, Corning, NY, USA) without or with Matrigel (for invasion) according to the manufacturer's protocol. An equal number of ACHN cells infected with CMTM5 OE and NC (4.0×105) and cells were seeded onto the inserts. The lower chamber was incubated in RPMI 1640 medium containing 20% FBS. The cells were incubated for 24 h before the inserts on the upper chamber were removed. Cells that had migrated to or invaded the lower membrane were stained with 0.1% crystal violet and counted under a light microscope in five random fields.
Statistical analysis
A χ2 test was used to analyze the association between CMTM5 expression and different clinicopathological characteristics. For the in vitro studies, the statistical significance of the differences between groups was determined using one-way analysis of variance. Data were presented as the mean ± standard error. All tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference. SPSS 19.0 package (IBM Corp., Armonk, NY, USA) was used for all statistical evaluations. All experiments were performed in triplicate.
Results
Downregulation of CMTM5 mRNA and protein expression in human RCC tissues
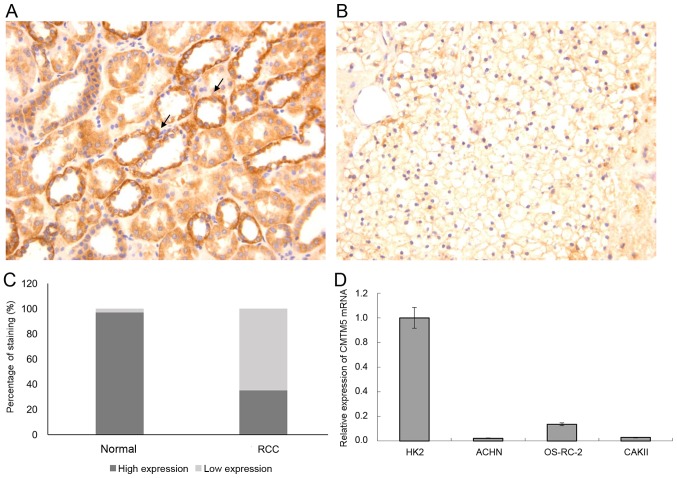
The expression patterns of CMTM5 protein in RCC and adjacent non-cancerous kidney tissues were examined using a TMA by IHC. As shown in Fig. 1A, the positive immunostaining of CMTM5 protein was mainly localized in the membrane and cytoplasm of renal epithelial cells, and was detected in 97% (73/75) of normal kidney tissues. However, CMTM5 protein was either absent or weakly expressed in 65% (48/75) of RCC tissues (Fig. 1B). There was a significant difference in the percentage of positive immunostaining expression of CMTM5 protein between RCC (26/75, 35%) and adjacent normal kidney tissues (73/75, 97%) (P<0.01; Fig. 1C).
Figure 1.
Representative immunohistochemical staining of CMTM5 protein in tissue microarray. (A) Positive immunostaining of CMTM5 protein was mainly localized in the membrane and cytoplasm of renal epithelial cells in adjacent non-cancerous kidney tissues (magnification, ×200). The representative areas with positive immunostaining are indicated by arrows. (B) CMTM5 protein was either absent or weakly expressed in RCC tissues (magnification, ×200). (C) Percentage of CMTM5 immunostaining in RCC and normal adjacent renal tissues. (D) Relative expression of CMTM5 mRNA in metastatic renal cell adenocarcinoma cell lines ACHN and CAKII and metastatic clear renal cell adenocarcinoma cell line OS-RC-2, as well as the human kidney HK2 cell line. CMTM5, CKLF-like MARVEL transmembrane domain-containing 5; RCC, renal cell carcinoma.
Next, the expression levels of CMTM5 mRNA were detected in metastatic renal cell adenocarcinoma cell lines ACHN and CAKII and metastatic clear renal cell adenocarcinoma cell line OS-RC-2. Compared with that in the normal renal tubular epithelial cell line HK2, the expression level of CMTM5 mRNA was significantly downregulated in metastatic renal cell adenocarcinoma cell lines ACHN and CAKII and metastatic clear renal cell adenocarcinoma cell line OS-RC-2 (Fig. 1D). The ACHN cell line was selected for further functional experiments as it exhibited the lowest expression of CMTM5 mRNA.
Furthermore, no statistically significant associations were identified between various clinicopathological features, including sex, age, clinical staging or histological grade, and the expression of CMTM5 protein (Table I).
CMTM5 suppresses RCC cell proliferation
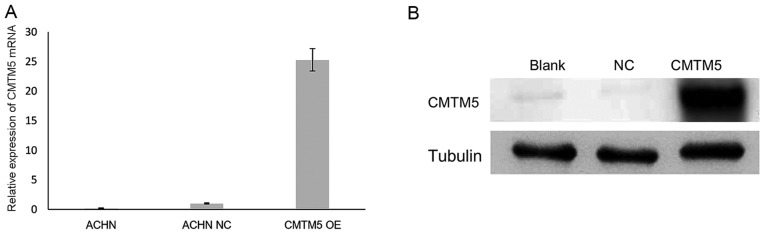
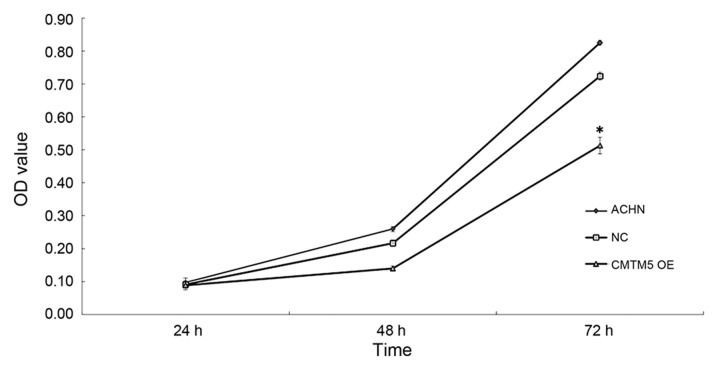
The downregulation of CMTM5 in RCC suggested that CMTM5 may act as a TSG in this cancer type. To investigate its anti-proliferative effect, the restoration of CMTM5 mRNA and protein expression in ACHN cells was confirmed by RT-qPCR and western blotting, respectively (Fig. 2). In the CCK-8 assay, the viability of ACHN-CMTM5 cells was decreased significantly compared with that of the untreated and null-infected controls at 72 h (all P<0.05; Fig. 3).
Figure 2.
Restoration of CMTM5 expression in ACHN cells. (A) Relative expression of CMTM5 mRNA in ACHN cells with different treatment. ACHN NC refers to ACHN cells transfected with the lenti-EGFP, while CMTM5 OE refers to ACHN cells infected with CMTM5 overexpressing lentivirus. (B) Expression of CMTM5 proteinin ACHN cells with different treatment; α-tubulin was used as an internal control. CMTM5, CKLF-like MARVEL transmembrane domain-containing 5; NC, negative control; OE, overexpression.
Figure 3.
Restoration of CMTM5 expression suppresses the viability of renal cell carcinoma cells, as determined by Cell Counting kit-8 assay. ACHN NC refers to ACHN cells infected with the lenti-EGFP while CMTM5 OE refers to ACHN cells infected with the CMTM5 overexpressing lentivirus. Cell growth curves were determined by absorbance at 450 nm. The results were expressed as the mean ± standard error of ≥3 independent experiments. *P<0.05. CMTM5, CKLF-like MARVEL transmembrane domain-containing 5; NC, negative control; OE, overexpression; OD, optical density.
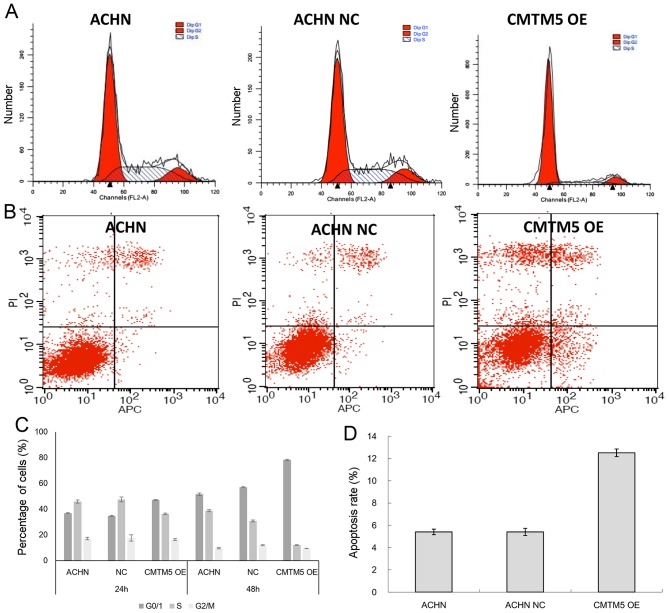
Restoration of CMTM5 expression induces cell cycle arrest in G0/G1 phase and apoptosis
The cell cycle analysis results generated by flow cytometry revealed that the CMTM5-infected group contained more cells in G0/G1 phase than the control Lenti-EGFP infected cells (cell viability index, 42.88 vs. 21.68%, respectively) (P<0.05; Fig. 4A and C). The apoptotic effect of CMTM5 in RCC cells was evaluated by Annexin V-FITC/PI staining assay. Compared with that of control Lenti-EGFP infected cells, the average apoptosis rate of CMTM5-ACHN cells was significantly higher at 72 h post-transfection (5.41 vs. 12.53%, respectively; Fig. 4B and D). These results suggest that the growth inhibition caused by CMTM5 may be mediated by G0/G1 arrest.
Figure 4.
CMTM5 induces cell cycle arrest and apoptosis. (A) The histograms show the percentages of cells in each cell cycle phase, as determined by the gating of the flow cytometer. (B) ACHN cells infected with NC and CMTM5-OE were labeled with fluoresce in isothiocyanate-Annexin V/PI, and apoptosis was assessed by flow cytometry. (C) Percentages of cells in each cell cycle phase. (D) Apoptosis rate of cells infected with NC and CMTM5-OE after 72 h. ACHN NC refers to ACHN cells infected with the lenti-EGFP, while CMTM5 OE refers to ACHN cells infected with CMTM5 overexpressing lentivirus. CMTM5, CKLF-like MARVEL transmembrane domain-containing 5; CMTM5-OE, CMTM5 over-expression lentivirus; NC, negative control; OE, overexpression; PI, propidium iodide.
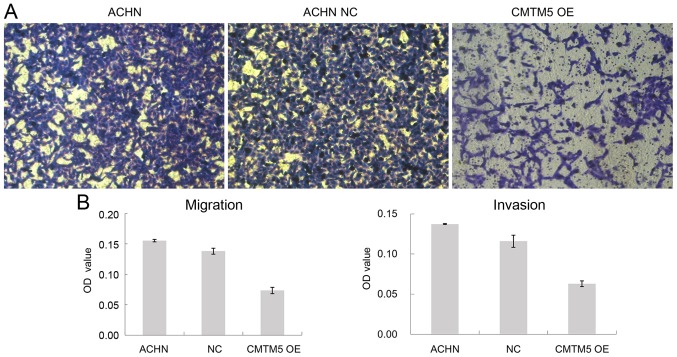
Restoration of CMTM5 expression reduces migration and invasion of RCC
Cell migration and invasion are both key steps for tumor development and metastasis. Using a Transwell assay, it was observed that the restoration of CMTM5 expression reduced the penetration rate of ACHN cells through the polycarbonate membrane compared with that of the Lenti-EGFP-infected ACHN cells in both migration and invasion assays (both P<0.05; Fig. 5).
Figure 5.
Effects of CMTM5 on cell migration and invasion, as detected by Transwell assay. (A) Crystal violet staining of the CMTM5-ACHN cells that passed through the polycarbonate membranes (magnification, ×100). (B) The number of invasded and migrated cells was determined by absorbance at 450 nm. ACHN NC refers to ACHN cells infected with lenti-EGFP, while CMTM5 OE refers to ACHN cells infected with CMTM5 over-expression lentivirus. CMTM5, CKLF-like MARVEL transmembrane domain-containing 5; NC, negative control; OE, overexpression; OD, optical density.
Discussion
In the present study, downregulation of CMTM5 at the mRNA and protein level was observed in RCC tissues and cell lines. Functional analyses demonstrated the tumor-suppressive roles of CMTM5 in RCC cells in vitro, including inhibition of cell proliferation, promotion of cell apoptosis, and suppression of cell migration and invasion.
DNA methylation is one of the most common epigenetic modifications in the regulation of gene expression (14). CMTM5 inactivated by promoter CpG methylation has been detected in numerous carcinomas (6,7,15). Our group also investigated whether CMTM5 was silenced by promoter methylation in RCC by bisulfite genomic sequencing (data not shown). However, similarly to the case of CMTM3, no promoter methylation of CMTM5 was observed in RCC cell lines (16). It has been previously indicated that the expression of CMTM5 in multiple other types of tumor was downregulated in RCC mainly by different epigenetic or genetic mechanisms (6–11).
The evidence of CMTM5 downregulation in RCC cells prompted us to investigate the biological function of CMTM5 on RCC cell lines in vitro. Previous studies revealed that CMTM family proteins, including CMTM3, CMTM4 andCMTM5, act as TSGs that can inhibit cell growth and induce apoptosis (5,15–18). The present study investigated the role of CMTM5 on RCC cell growth and apoptosis by restoration of CMTM5 expression in the RCC cell line ACHN. The growth of ACHN-CMTM5 cells was significantly inhibited, and the cell apoptotic rate was markedly increased, following the transfection of the CMTM5 vector. In addition, it was observed that the restoration of CMTM5 expression suppressed both the motility and invasiveness of ACHN cells, which is in line with previous findings in other tumors (6,7,10,11). In the present study, the ACHN line was selected for functional studies as the CMTM5 mRNA expression level was the lowest of the cell lines tested. However, a previous study by Brodaczewska et al (2) has indicated that the ACHN cell line is not suitable for studies of clear cell RCC, which accounts for 70–75% of all RCC cases. Therefore, further studies in clear cell RCC are required in the future to further validate the data of the present study.
The molecular mechanisms underlying the tumor-suppressive effects of CMTM5 on RCC cell growth and migration remain unclear. As a member of the CMTM family, the CMTM5 protein has a predicted MARVEL domain, which is a common feature of the occludin, physin, gyrin, MAL and CMTM families, and is a membrane-associating domain that may form part of the machinery of membrane apposition events (19). For instance, CMTM3, another member of the CMTM family, may inhibit tumor growth and invasion through promoting the transcription of p21, downregulating the expression of matrix metalloproteinase 2 and phosphorylating extracellular signal-regulated kinase 1/2 (Erk1/2) (16,20). CMTM5 induces the apoptosis of pancreatic cancer cells not only by activation of caspases 3, 8 and 9, but also by exerting synergistic effects with tumor necrosis factor-α (6). CMTM7 can repress oncogenic epidermal growth factor receptor (EGFR) signaling through promoting EGFR internalization (21). CMTM8 is also a regulator of the Erk1/2 signaling pathway (22). Thus, it can be proposed that the mechanism of action of CMTM5 in RCC cells may be associated with membrane protein sorting and trafficking, such as EGFR and vascular endothelial growth factor receptor signaling, which are both approved targets for the treatment of advanced RCC (23,24). Further evaluation of this hypothesis will provide additional information on molecular-targeted therapy for RCC.
In conclusion, the present data suggest that CMTM5 may function as a tumor suppressor in human RCC by suppressing the viability of RCC cells, implying its potential as a therapeutic target for this malignancy.
Acknowledgements
The present study was supported by the Wenzhou Natural Science Foundation (grant no. Y20140579). The authors thank Professor Wenling Han for generously providing the anti-CMTM5 antibody, as well as the Department of Pathology of The First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China), for their support in IHC and staining evaluation.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cell cancer research. Mol Cancer. 2016;15:83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leibovich BC, Lohse CM, Crispen PL, Boorjian SA, Thompson RH, Blute ML, Cheville JC. Histological subtype is an independent predictor of outcome for patients with renal cell carcinoma. J Urol. 2010;183:1309–1315. doi: 10.1016/j.juro.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Grimm MO, Wolff I, Zastrow S, Frohner M, Wirth M. Advances in renal cell carcinoma treatment. Ther Adv Urol. 2010;2:11–17. doi: 10.1177/1756287210364959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao W, Wang J, Li H, Guan W, Xia D, Yu G, Xiao H, Lang B, Ma X, Liu J, et al. Fibulin-1 is down-regulated through promoter hypermethylation and suppresses renal cell carcinoma progression. J Urol. 2013;190:291–301. doi: 10.1016/j.juro.2013.01.098. [DOI] [PubMed] [Google Scholar]
- 6.Guo X, Li T, Wang Y, Shao L, Zhang Y, Ma D, Han W. CMTM5 induces apoptosis of pancreatic cancer cells and has synergistic effects with TNF-alpha. Biochem Biophys Res Commun. 2009;387:139–142. doi: 10.1016/j.bbrc.2009.06.148. [DOI] [PubMed] [Google Scholar]
- 7.Li P, Liu K, Li L, Yang M, Gao W, Feng J, Lv Y, Qu X, Kong B. Reduced CMTM5 expression correlates with carcinogenesis in human epithelial ovarian cancer. Int J Gynecol Cancer. 2011;21:1248–1255. doi: 10.1097/IGC.0b013e3182259c31. [DOI] [PubMed] [Google Scholar]
- 8.Niu J, Li H, Zhang Y, Li J, Xie M, Li L, Qin X, Qin Y, Guo X, Jiang Q, et al. Aberrant expression of CKLF-like MARVEL transmembrane member 5 (CMTM5) by promoter methylation in myeloid leukemia. Leuk Res. 2011;35:771–776. doi: 10.1016/j.leukres.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang Y, Zhang Y, Ng KM, Han W, Ma D, Tao Q. Cmtm5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res. 2007;13:5756–5762. doi: 10.1158/1078-0432.CCR-06-3082. [DOI] [PubMed] [Google Scholar]
- 10.Shao L, Guo X, Plate M, Li T, Wang Y, Ma D, Han W. CMTM5-v1 induces apoptosis in cervical carcinoma cells. Biochem Biophys Res Commun. 2009;379:866–871. doi: 10.1016/j.bbrc.2008.12.126. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Yuan Y, Zhang Y, Li J, Liu Z, Zhang X, Sheng Z, Xu T, Wang X. CMTM5 is reduced in prostate cancer and inhibits cancer cell growth in vitro and in vivo. Clin Transl Oncol. 2015;17:431–437. doi: 10.1007/s12094-014-1253-z. [DOI] [PubMed] [Google Scholar]
- 12.Eble JN, Sauter G, Epstein JI, Sesterhenn IA, editors. IARC Press; Lyon: 2004. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs; p. 7. [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Nan X, Li X, Chen Y, Zhang J, Sun L. CMTM5 exhibits tumor suppressor activity through promoter methylation in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2014;447:304–310. doi: 10.1016/j.bbrc.2014.03.158. [DOI] [PubMed] [Google Scholar]
- 16.Xie J, Yuan Y, Liu Z, Xiao Y, Zhang X, Qin C, Sheng Z, Xu T, Wang X. CMTM3 is frequently reduced in clear cell renal cell carcinoma and exhibits tumor suppressor activities. Clin Transl Oncol. 2014;16:402–409. doi: 10.1007/s12094-013-1092-3. [DOI] [PubMed] [Google Scholar]
- 17.Su Y, Lin Y, Zhang L, Liu B, Yuan W, Mo X, Wang X, Li H, Xing X, Cheng X, et al. CMTM3 inhibits cell migration and invasion and correlates with favorable prognosis in gastric cancer. Cancer Sci. 2014;105:26–34. doi: 10.1111/cas.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plate M, Li T, Wang Y, Mo X, Zhang Y, Ma D, Han W. Identification and characterization of CMTM4, a novel gene with inhibitory effects on hela cell growth through inducing G2/M phase accumulation. Mol Cells. 2010;29:355–361. doi: 10.1007/s10059-010-0038-7. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. Marvel: A conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:599–601. doi: 10.1016/S0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Xie J, Wu J, Li W, Nie L, Sun X, Sun X, Tang A, Li X, Liu R, et al. CMTM3 inhibits human testicular cancer cell growth through inducing cell-cycle arrest and apoptosis. PloS One. 2014;9:e88965. doi: 10.1371/journal.pone.0088965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Li J, Su Y, Fan Y, Guo X, Li L, Su X, Rong R, Ying J, Mo X, et al. A novel 3p22.3 gene CMTM7 represses oncogenic egfr signaling and inhibits cancer cell growth. Oncogene. 2014;33:3109–3118. doi: 10.1038/onc.2013.282. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Mendoza MC, Pei X, Ilter D, Mahoney SJ, Zhang Y, Ma D, Blenis J, Wang Y. Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem. 2012;287:11850–11858. doi: 10.1074/jbc.M111.258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark PE. The role of VHL in clear-cell renal cell carcinoma and its relation to targeted therapy. Kidney Int. 2009;76:939–945. doi: 10.1038/ki.2009.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Liu B, Dai Z, Tao Y. Knockdown of VEGF receptor-1 (VEGFR-1) impairs macrophage infiltration, angiogenesis and growth of clear cell renal cell carcinoma (crcc) Cancer Biol Ther. 2011;12:872–380. doi: 10.4161/cbt.12.10.17672. [DOI] [PMC free article] [PubMed] [Google Scholar]