Abstract
A number of recent studies have focused on the association between long non-coding RNAs (lncRNAs) and cancer. HOX transcript antisense RNA (HOTAIR), an lncRNA that functions as a transcriptional modulator, has been implicated in various fundamental biological activities. HOTAIR mediates the trimethylation of histone H3 at lysine 27 and the demethylation of histone H3 dimethyl Lys4 by recruiting the polycomb repressive complex 2 and the lysine-specific demethylase 1/co-repressor of RE1-silencing transcription factor (coREST)/REST complex to the target gene promoters, which leads to gene silencing. Overexpression of HOTAIR in hepatocellular carcinoma (HCC) is strongly associated with an unfavorable prognosis for patients with HCC. HOTAIR promotes the carcinogenic activity of HCC cells through the suppression of RNA binding motif protein 38, triggering the epithelial-mesenchymal transition, and by interacting with microRNAs that act as tumor suppressors. In the present review, the role of the lncRNA HOTAIR in HCC is examined. The potential use of HOTAIR as a biomarker to achieve more accurate prognostic predictions and as an effective therapeutic target for HCC is then discussed.
Keywords: long non-coding RNA, HOX transcript antisense RNA, hepatocellular carcinoma, PRC2, metastasis, recurrence, therapeutic target
1. Introduction
Hepatocellular carcinoma (HCC) is the fifth most common type of malignancy worldwide and is associated with the third highest number of cancer-related mortalities (1). Approximately 626,000 new cases are diagnosed each year, a large proportion of which are fatal (2). Although the diagnosis and treatment of HCC have improved over the years, the mortality rate of patients with HCC continues to be high (3). Tumor metastasis and recurrence remain the major obstacles to achieving higher long-term survival rates (4). It is therefore clinically important to elucidate the molecular pathogenesis of HCC, as this could assist in revealing effective therapeutic strategies and, ultimately, aid patients with HCC to achieve a favorable prognosis.
Conventional wisdom holds that protein-coding genes play dominant roles in gene regulation on the basis of the genetic central dogma: DNA encodes mRNA, which encodes protein. However, technological advances have revealed that only 1.5% of the human genome is protein coding, while the remaining ~98% is transcribed into RNAs that do not produce any proteins, termed non-coding RNAs (ncRNAs) (5). The ncRNAs are divided into long ncRNAs (lncRNAs; >200 nucleotides) and small ncRNAs (sncRNAs; <200 nucleotides) (6). sncRNAs such as microRNAs (miRNAs) have important roles in the initiation and development of various malignancies (such as breast cancer, lung cancer and gastric cancer) (7–10) and other diseases, including systemic lupus erythematosus, rheumatoid arthritis and multiple sclerosis (11–13). Recently, the association between human disease, including cancer, and lncRNAs has attracted increasing attention. Numerous investigations center around the functions of lncRNAs in the initiation and progression of HCC (2,14–19). These studies suggested that dysregulation of several lncRNAs was tightly associated with metastasis, disease recurrence and poorer clinical outcomes in patients with HCC (Table I) (2,14,19,20–34). HOX transcript antisense RNA (HOTAIR), a 2,158-nucleotide lncRNA derived from the antisense strand of homeobox gene C cluster (HOXC) (35), has been identified as an HCC-related lncRNA. In the present review, the expression and function of HOTAIR in HCC is investigated. Additionally, the clinical potential of HOTAIR as a biomarker for predicting HCC prognosis, and whether it could serve as a novel target for HCC therapy, is discussed.
Table I.
Summary of lncRNAs and their functions in HCC.
| lncRNA | Expression | Biological functions and clinical relevance | (Refs.) |
|---|---|---|---|
| HOTAIR | Upregulated | Lymph node metastasis; increased tumor size; tumor recurrence after LT; poorer DFS following surgical resection or LT; potential biomarker for prognosis and important target for HCC therapy | (2,14,19–21) |
| H19 | Upregulated | Suppresses HCC progression and metastasis | (22,23) |
| HULC | Upregulated | Potential biomarker for HCC diagnosis and prognostic prediction | (24,25) |
| MALAT1 | Upregulated | Promotes tumor progression; potential biomarker for predicting HCC recurrence | (26,27) |
| MEG3 | Downregulated | Suppresses cell growth; induces apoptosis | (28,29) |
| lncRNA-LET | Downregulated | Inhibits HCC metastasis; a tumor suppressor that could be a therapeutic target of HCC | (30) |
| MVIH | Upregulated | Promotes tumor growth and intrahepatic metastasis; predicts poor recurrence-free survival | (31) |
| HEIH | Upregulated | Prognostic factor for predicting tumor recurrence of HBV-related HCC | (32,33) |
| Dreh | Downregulated | Inhibits HCC growth and metastasis | (32) |
| MDIG | Upregulated | Indicates unfavorable prognosis | (34) |
DFS, disease-free survival; lncRNA, lon non-coding RNA; HCC, hepatocellular carcinoma; LT, liver transplantation; HBV, hepatitis B virus; H19, imprinted maternally expressed transcript; HULC, highly upregulated in liver cancer; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MEG3, maternally expressed gene 3; lncRNA-LET, lncRNA low expression in tumor; MVIH, lncRNA associated with microvascular invasion in HCC; HEIH, hepatocellular-carcinoma-upregulated EZH2-associated lncRNA; Dreh, downregulated expression by HBx; MDIG, mineral-dust-induced gene; HOTAIR, HOX transcript antisense RNA.
2. Identification and characterization of lncRNA HOTAIR
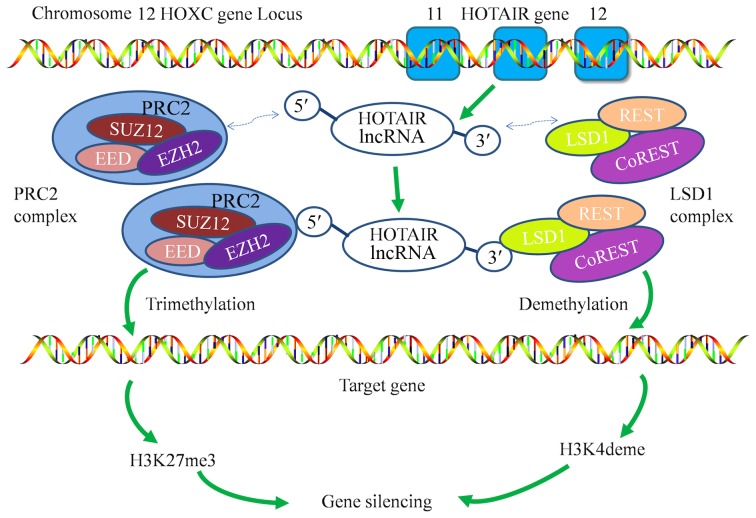
HOTAIR was originally discovered by Rinn et al through tiling microarray analysis in 2007 (36). A later study reported that HOTAIR is found only in mammals and can possess divergent sequences, although the structure is conserved (37). HOTAIR is transcribed from the HOXC locus, situated between HOXC12 and HOXC11 on chromosome 12q13.13 (Fig. 1), a region that evolved more quickly than nearby HOXC genes (37). HOTAIR is the first identified trans-acting lncRNA that functions as a modulator of gene expression. For instance, when co-expressed with the HOXC gene on chromosome 12, HOTAIR represses the expression of HOXD on chromosome 2 through recruitment of and interaction with the polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1)/co-repressor of RE1-silencing transcription factor (coREST)/REST complexes (36,38,39).
Figure 1.
Mechanism of the gene-silencing action of HOTAIR. HOTAIR lncRNA is transcribed from the HOXC locus located at chromosome 12q13.13, in a position flanked by HOXC12 and HOXC11. HOTAIR functions as a molecular scaffold, interacting with the PRC2 and LSD1 complexes via its 5′ and 3′ ends and recruiting them to its target gene promoters. This induces the trimethylation of H3K27 (H3K27me3) and the demethylation of H3K4 (H3K4deme), ultimately resulting in gene silencing. HOTAIR, HOX transcript antisense RNA; lncRNA, long non-coding RNA; HOXC, homeobox C cluster; PRC2, polycomb repressive complex 2; SUZ12, suppressor of zeste 12 protein homolog; EED, embryonic ectoderm development; EZH2, enhancer of zeste homolog 2; REST, RE1-silencing transcription factor; LSD1, lysine-specific histone demethylase 1; coREST, REST corepressor 1; H3K27me3, trimtethylated lysine 27 of histone H3; H3K4deme, demethylated lysine 4 of histone H3.
The human HOTAIR gene contains six exons; exons 1 to 5 (range, 33–140 bp) are short, whereas exon 6 (1,817 bp) is long and is classified into domains A and B (37). Although the overall sequences of HOTAIR are poorly conserved, the 5′ and 3′ binding domains of HOTAIR, which bind to PRC2 and LSD1/CoREST/REST complexes, respectively, may possess relatively constant sequences and structures (40). The PRC2 complex is made up of suppressor of zeste 12 homolog (SUZ12), enhancer of zeste homolog 2 (EZH2) and embryonic ectoderm development (14). EZH2 is a methyltransferase that requires the presence of the other subunits to be functional (41). EZH2 mediates the transcriptional silencing via the trimethylation of histone H3Lys27 (giving H3K27me3) (42). Within the LSD1/CoREST/REST complex, LSD1 also acts as a transcriptional repressor. However, LSD1 also functions as a histone demethylase that mediates transcriptional repression via the demethylation of dimethyl histone H3 Lys4 (H3K4me2) (Fig. 1) (43). In brief, HOTAIR coordinates the functions of the two chromatin-modification complexes (44) and alters the expression of multiple genes that are associated with diverse biological functions (35).
3. Expression of HOTAIR in HCC
The expression of HOTAIR in tumor tissue is markedly higher than in the adjacent healthy tissue in HCC patients (2,19,35,45). The abnormal expression of HOTAIR in HCC is associated with lymph node metastasis, larger tumor size, tumor recurrence after liver transplantation (LT) and shorter disease-free survival (DFS) after surgical resection or LT (2,14,19,21).
It is well known that the expression of the v-myc avian myelocytomatosis viral oncogene homolog (MYC) oncogene, a powerful cancer-promoting gene, affects the expression of HOTAIR. MYC is implicated in tumorigenesis in a diverse range of tissues, regulating gene expression and cell proliferation (46). For instance, a previous experimental study demonstrated that expression of HOTAIR is stimulated by c-Myc through interaction with a putative E-box element located upstream of the HOTAIR gene in gallbladder cancer cells (47). Since c-Myc has a pivotal role in hepatocarcinogenesis (47,48), the part it plays in HCC necessitates investigation.
A recent study reported that HOTAIR is regulated by IκB kinase (IKK), made up of IKKα, IKKβ and IKKγ subunits, in a H3k27me3-dependent manner (49). The three subunits of IKK affected the growth of liver cancer stem cells (LCSCs) through regulation of HOTAIR expression. Specifically, IKKα and IKKβ upregulated the expression of HOTAIR, whereas IKKγ downregulated it. A reverse interaction was also observed, with the carcinogenic effects of three core IKK subunits being activated by HOTAIR (49).
More recent studies have demonstrated that miRNAs serve notable roles in HOTAIR regulation (50,51). Unlike HOTAIR, miR-141 exerts a tumor suppressive function that inhibits the malignancy of cancer cells in several types of cancer (35). HOTAIR expression levels were found to be inversely associated with those of miR-141 in renal carcinoma cells (51). miR-141 suppressed the expression of HOTAIR in an argonaute-2-dependent manner, which ultimately resulted in reduced cell proliferation and tumor invasion (51). Although these findings were observed in other cancer types, the miRNA-141-mediated degradation of HOTAIR in HCC necessitates further investigation (52–55).
Osteopontin (OPN) is a phosphoglycoprotein that has been reported to be able to upregulate HOTAIR in cancer cells, with the knockdown of OPN markedly diminishing the level of HOTAIR expression (56). The CD44 receptor is a positive regulator of OPN that is also implicated in the regulation of OPN-mediated HOTAIR expression. Previously considered to be a tumor suppressor, expression of interferon regulatory factor-1 (IRF1) was verified to be elevated in cancer cells, with its overexpression decreasing the levels of HOTAIR expression (56). In fact, OPN promoted HOTAIR expression via the inhibition of IRF1 and its signaling pathway (56). Since OPN has been identified as a promoter of HCC progression and metastasis (57,58), the mechanism of OPN-induced HOTAIR expression in HCC requires further investigation.
Transforming growth factor-β (TGF-β) stimulates expression of HOTAIR and triggers EMT (59). Similarly, type I collagen (Col-1), which is often overexpressed in lung cancer, could also promote HOTAIR expression (60). Since TGF-β and Col-1 are powerful triggers of EMT in HCC (61,62), their further investigation may lead to novel insights into the regulatory role of HOTAIR in HCC.
4. Functions of HOTAIR in HCC
HOTAIR enhancement is associated with invasion, progression, metastasis and poorer clinical outcomes in HCC patients (Table I) (2,14,19–21). In HCC cells, HOTAIR modulates various genes and molecules that play critical roles in cancer migration and invasion. A recent experimental study found that knockdown of HOTAIR by short interfering RNAs upregulated RNA binding motif protein 38 (RBM38) expression in HCC cells (16). Moreover, the study also reported that the level of RBM38 expression in HCC tissue was markedly lower than that in healthy tissue in the same patients. RBM38-knockdown could restore the inhibition of malignancy that was mediated by the downregulation of HOTAIR (16). Therefore, it could be concluded that HOTAIR mediates the migratory and invasive phenotypes of HCC cells through the suppression of RBM38, indicating that HOTAIR and RBM38 serve notable roles in HCC development (16).
A recent study observed that HOTAIR exerted oncogenic effects in LCSCs. A negative correlation between the expression of HOTAIR and SET domain-containing 2 (SETD2) was also found in this study (63). Furthermore, the study demonstrated that SETD2 abrogates the oncogenic activity of HOTAIR, thus exerting a tumor suppressive function. Taking these observations together, HOTAIR serves an oncogenic role through decreasing the expression of SETD2 in LCSCs (63).
It is well accepted that autophagy exerts important effects on the initiation and development of human cancer (64). A recent study suggested that HOTAIR was overexpressed in HCC and that its dysregulation could promote autophagy (65). Moreover, it was also observed that increases in the expression of autophagy-related gene 3 (ATG3) and ATG7 were accompanied by HOTAIR overexpression. In summary, overexpression of HOTAIR triggers autophagy through the upregulation of ATG3 and ATG7, thus eventually facilitating HCC progression (65).
HOTAIR exerts biological functions through recruiting the PRC2 and LSD1 complexes to the target genes and repressing their expression (35). The PRC2 complex has been observed to be essential for silencing certain tumor suppressive genes (45). Several studies concerning HCC demonstrated that the subunits of the PRC2 complex are hyperexpressed in HCC and could mediate hepatocarcinogenesis (66–68). EZH2 was found to be upregulated in HCC cells; its overexpression could inactivate several tumor suppressive miRNAs. Moreover, high expression of EZH2 was closely associated with a negative clinical outcome in patients with HCC (66,67). SUZ12, another component of the PRC2 complex, serves an important role in H3-K27 methylation and gene silencing (68). SUZ12 was also found to be upregulated in liver tumors and has been considered as a novel target for HCC therapy (68). Besides the PRC2 complex, HOTAIR also facilitates the progression of HCC through LSD1. LSD1 is an important regulator of hepatic carcinogenesis whose upregulation is closely associated with advanced stages of HCC (69). B-cell lymphoma 2 (Bcl-2) and c-Myc are oncogenes that can stimulate the initiation and development of cancer. Data suggest that the downregulation of Bcl-2 and c-Myc is followed by LSD1 silencing, indicating that LSD1 may function as a tumor promoter through the stimulation of Bcl-2 and c-Myc expression (69). Further evidence has suggested that LSD1 overexpression is a poor prognostic predictor for patients with primary HCC (70).
The invasive and metastatic phenotypes of HCC are attributed to the progression of EMT (62). A recent study observed that HOTAIR overexpression in normal liver stem cells (NLSCs) could result in their malignant transformation (71). Ye et al (71) suggested that HOTAIR could promote the initiation of HCC by inducing EMT. During EMT, HOTAIR promoted tumor progression by regulating EMT-related genes (72,73). For example, HOTAIR stimulated EMT by suppressing miR-331-3p, which is an inhibitor of the human epidermal growth factor receptor 2 (HER2)/AKT/heat-shock factor 1/Slug pathway (74). HOTAIR has also been reported to stimulate EMT through suppression of the EMT inhibitor Wnt inhibitory factor 1 in esophageal squamous cell carcinoma cells (75,76). HOTAIR has been shown to downregulate miR-7, a tumor suppressor that has been reported to reverse EMT in breast cancer cells (77). In addition to suppressing EMT inhibitors, HOTAIR triggers EMT by facilitating the expression of EMT effector molecules. For instance, HOTAIR may be involved in the expression of matrix metalloproteinases (MMPs), which are critical regulators of the invasive and metastatic phenotypes of HCC cells (19,78–80). However, future investigation is required to support this viewpoint.
HOTAIR serves a considerable role in hepatitis B virus (HBV)-mediated hepatic tumorigenesis by enhancing the ubiquitination of SUZ12 and zinc finger protein 198 (ZNF198) in the presence of polo-like kinase 1 (Plk1) (81). SUZ12 is an important component of the PRC2 complex that can repress transcription. Another complex that represses transcription, LSD1/Co-REST/histone deacetylase 1, is stabilized by ZNF198. In cells infected with HBV, negative regulation of ZNF198 and SUZ12 results in the epigenetic reprogramming of the cell. In brief, HOTAIR facilitates the ubiquitination of Plk1-phosphorylated SUZ12 and ZNF198 and accelerates their proteasomal degradation during HBV-induced liver carcinogenesis (81).
HOTAIR promotes tumorigenesis via interaction with miRNAs. As aforementioned, the level of HOTAIR expression is modulated by several miRNAs with tumor suppressor effects, including miR-141 (51). However, HOTAIR also modulates miRNA levels, recognizing and then degrading target miRNAs (82). For instance, a miR-130a binding site has been found in the HOTAIR lncRNA and was proven to be important for the HOTAIR-mediated regulation of miR-130a (47). In gallbladder cancer tissue, miR-130a expression was found to be negatively associated with HOTAIR expression. This suggests that HOTAIR partly exerts its carcinogenic effect by downregulating miR-130a (47). A previous study demonstrated that HOTAIR functions as a competitive endogenous RNA (ceRNA), competing with HER2 for miR-331-3p binding sites and thus ultimately alleviating the miR-331-3p-mediated repression of oncogenic HER2 expression in gastric cancer cells (83). Another, more recent, study suggested that the dysregulated expression of HOTAIR is also involved in hematological malignancies (84). HOTAIR was again found to act as a ceRNA, binding miR-193a and thereby regulating the expression of proto-oncogene c-KIT in acute myeloid leukemia cells (84). Although these findings have not all been reported in HCC, the interaction between HOTAIR and miRNAs should be investigated in HCC, as miR-130a and miR-193a-3p serve as tumor suppressors in the disease (85,86).
5. Clinical values of HOTAIR in HCC
HOTAIR is considered to be an effective molecular marker in HCC: its reinforced expression in HCC tissues is closely associated with lymph node metastasis, larger tumor size, tumor recurrence after LT and inferior DFS after surgical resection or LT (Table I). In a cohort of 50 patients with HCC that underwent surgical resection, 3-year recurrence-free survival rates in patients with low HOTAIR expression were noticeably higher than those in patients with high HOTAIR expression. The study also suggested that HOTAIR could be employed as a marker for predicting lymph node metastasis in HCC (19). In another study of 64 patients with HCC, the overall survival rate of the 13 patients with high HOTAIR expression was markedly lower than that of the 51 patients with low HOTAIR expression. A positive association between hyperexpression of HOTAIR and larger tumor size was also observed in this study (14). In addition, a recent study of 60 patients with HCC who underwent LT demonstrated that patients who overexpressed HOTAIR were more susceptible to tumor recurrence after LT. Yang et al (2) proposed that HOTAIR can be employed as a predictive biomarker for predicting tumor relapse after LT.
As with cancer-specific miRNAs, HOTAIR, as an lncRNA, is stable and detectable in various sorts of biological specimens, including serum and urine (3,87). lncRNAs can be determined at low cost and levels are easily assessed by simple methodologies such as quantitative PCR, which are already in routine clinical practice (87). Therefore, lncRNAs, including HOTAIR, could be potentially employed as fluid-based non-invasive markers for clinical use (3).
An increasing number of studies have reported that HOTAIR is a promising therapeutic target in HCC. For example, under the positive regulation of HOTAIR, vascular endothelial growth factor (VEGF) and MMP-9 could promote the progression of HCC. Therefore, treatments aimed at HOTAIR could inhibit HCC growth (19,45). Drugs directly targeting VEGF or MMP-9 may also be effective for HCC therapy (45). As aforementioned, HOTAIR mediates migratory and invasive phenotypes of HCC cells via the inhibition of RBM38, with downregulation of HOTAIR leading to a marked reduction in cell motility. These observations indicate that HOTAIR is a potential target for HCC therapy and that RMB38 serves as a suppressed target of HOTAIR (16). Moreover, in a previous study, inhibiting HOTAIR function decreased the viability and invasiveness of HCC cells and increased their sensitivity to TNF-α-mediated apoptosis and chemotherapeutic drugs (2). An independent study has shown that HOTAIR plays a considerable role in hepatic tumorigenesis by downregulating miR-218 expression and inactivating the P14ARF and P16Ink4a signal pathways (88). HOTAIR silenced the expression of miR-218 by directly recruiting EZH2 to its promoter. Antitumor drugs for HCC could therefore be designed to directly disrupt the HOTAIR-EZH2-miR-218 negative regulatory axis (88). As has been discussed, PRC2 and LSD1 complexes were overexpressed in HCC and acted as partners of HOTAIR in the development of HCC. They could therefore be considered to be latent targets of HCC treatment (66,69).
6. Conclusion and future perspectives
HCC is one of the most pressing health problems around the world; it is aggressively malignant and has a high risk of recurrence. Despite the use of diverse treatment methods, including chemotherapy, surgery and LT, the clinical prognosis remains poor. Improvements in the speed of diagnosis and the discovery of effective therapeutic targets for HCC are therefore essential if this prognosis is to be improved. As has been mentioned, HOTAIR is abnormally expressed in HCC and serves a key role in the progression of HCC. HOTAIR recruits the PRC2 and LSD1 complexes to their target gene promoters, inducing H3K27 trimethylation and H3K4me2 demethylation, which eventually leads to gene silencing (Fig. 1). HOTAIR expression is modulated by various molecules, including miR-141, c-Myc, IKK and OPN. HOTAIR promotes invasive and aggressive phenotypes of HCC cells by diverse mechanisms, which include suppression of RBM38, reduction of SETD2 expression, stimulation of autophagy, induction of EMT and interaction with different tumor suppressor miRNAs, including miR-130a.
Numerous studies have demonstrated that HOTAIR can be employed as a novel prognostic molecular marker in HCC, as its enforced expression is strongly associated with lymph node metastasis, larger tumor size, tumor recurrence following LT and inferior DFS after surgical resection or LT. In addition, HOTAIR expression levels can be easily determined non-invasively, making its clinical application feasible.
As a promising therapeutic target of HCC, HOTAIR could be blocked in diverse ways. Firstly, drugs that mask the certain binding sites could disrupt the interplay between HOTAIR and its molecule partners, which include the PCR2 and LSD1/CoREST/REST complexes, thus silencing HOTAIR. Moreover, HOTAIR could be targeted for degradation by specific miRNAs such as miR-141. Finally, antitumor agents could be designed that are targeted at molecules that participate in the HOTAIR pathway in HCC, such as VEGF and MMP-9. This could be an indication of the future directions of novel drug development in HCC therapy.
In summary, HOTAIR is emerging as a novel prognostic molecular marker and as an efficient therapeutic target for HCC. However, further investigation of the underlying molecular mechanism behind dysregulated HOTAIR expression, and how it drives HCC progression, is required to maximize the clinical potential of HOTAIR.
Acknowledgements
This study was supported by the Natural Science Foundation of Zhejiang Province (grant no. LY15H160021) and the Traditional Chinese Medicine Scientific Research Fund Project of Zhejiang Province (grant no. 2017ZA079).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:423–434. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 4.Hao K, Luk JM, Lee NP, Mao M, Zhang C, Ferguson MD, Lamb J, Dai H, Ng IO, Sham PC, Poon RT. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Song YX, Ma B, Wang JJ, Sun JX, Chen XW, Zhao JH, Yang YC, Wang ZN. Regulatory roles of non-coding RNAS in colorectal cancer. Int J Mol Sci. 2015;16:19886–19919. doi: 10.3390/ijms160819886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13:7. doi: 10.1186/1475-2867-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Wu J, Zheng J, Li Y, Yang T, Hu G, Dai J, Yang Q, Dai L, Jiang Y. Altered miRNA expression profiles and miR-1a associated with urethane-induced pulmonary carcinogenesis. Toxicol Sci. 2013;135:63–71. doi: 10.1093/toxsci/kft131. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. Eur J Cell Biol. 2013;92:123–128. doi: 10.1016/j.ejcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X, Huang K, Tong Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol Cancer Res. 2013;11:182–193. doi: 10.1158/1541-7786.MCR-12-0534. [DOI] [PubMed] [Google Scholar]
- 11.Lin CP, Choi YJ, Hicks GG, He L. The emerging functions of the p53-miRNA network in stem cell biology. Cell cycle. 2012;11:2063–2072. doi: 10.4161/cc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roderburg C, Luedde M, Cardenas D Vargas, Vucur M, Mollnow T, Zimmermann HW, Koch A, Hellerbrand C, Weiskirchen R, Frey N, et al. miR-133a mediates TGF-β-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J Hepatol. 2013;58:736–742. doi: 10.1016/S0168-8278(13)60603-0. [DOI] [PubMed] [Google Scholar]
- 13.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Zhang P, Wang L, Piao HL, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin (Shanghai) 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, Zheng S. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toffanin S, Villanueva A, Llovet JM. miRNA delivery: Emerging therapy for hepatocellular carcinoma. Gastroenterology. 2010;138:1202–1204. doi: 10.1053/j.gastro.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371–1383. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 19.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 20.Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Greene CM, Gray SG, Lawless MW. Long noncoding RNAs in liver cancer: What we know in 2014. Expert Opin Ther Targets. 2014;18:1207–1218. doi: 10.1517/14728222.2014.941285. [DOI] [PubMed] [Google Scholar]
- 22.Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 24.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 27.Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Zhang X, Zhang M, Zhu JD, Zhang YL, Lin Y, Wang KS, Qi XF, Zhang Q, Liu GZ, et al. Up-regulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma. Carcinogenesis. 2007;28:1094–1103. doi: 10.1093/carcin/bgl215. [DOI] [PubMed] [Google Scholar]
- 29.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 32.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 34.Chen B, Yu M, Chang Q, Lu Y, Thakur C, Ma D, Yi Z, Chen F. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget. 2013;4:1427–1437. doi: 10.18632/oncotarget.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Gene Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J, Hao X. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 41.Wu H, Zeng H, Dong A, Li F, He H, Senisterra G, Seitova A, Duan S, Brown PJ, Vedadi M, et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One. 2013;8:e83737. doi: 10.1371/journal.pone.0083737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Gene Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao Y, Li J, Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15:18985–18999. doi: 10.3390/ijms151018985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: Potential prospects for combined targeted therapeutics (review) Int J Oncol. 2012;41:393–406. doi: 10.3892/ijo.2012.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD, Qin YY, Gong W, Quan ZW. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD, Wen WH, Wang Z, Wang T, Zhao J, et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology. 2014;59:1850–1863. doi: 10.1002/hep.26720. [DOI] [PubMed] [Google Scholar]
- 49.An J, Wu M, Xin X, Lin Z, Li X, Zheng Q, Gui X, Li T, Pu H, Li H, Lu D. Inflammatory related gene IKKα, IKKβ, IKKγ cooperates to determine liver cancer stem cells progression by altering telomere via heterochromatin protein 1-HOTAIR axis. Oncotarget. 2016;7:50131–50149. doi: 10.18632/oncotarget.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, Saini S, Chang I, Tanaka Y, Enokida H, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V, Chang I, Tanaka Y, Enokida H, Nakagawa M, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550–12565. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu SM, Ai HW, Zhang DY, Han XQ, Pan Q, Luo FL, Zhang XL. MiR-141 targets ZEB2 to suppress HCC progression. Tumour Biol. 2014;35:9993–9997. doi: 10.1007/s13277-014-2299-9. [DOI] [PubMed] [Google Scholar]
- 53.Xue J, Niu YF, Huang J, Peng G, Wang LX, Yang YH, Li YQ. miR-141 suppresses the growth and metastasis of HCC cells by targeting E2F3. Tumour Biol. 2014;35:12103–12107. doi: 10.1007/s13277-014-2513-9. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Ding Y, Huang J, Wang S, Ni W, Guan J, Li Q, Zhang Y, Ding Y, Chen B, Chen L. MiR-141 suppresses the migration and invasion of HCC cells by targeting Tiam1. PLoS One. 2014;9:e88393. doi: 10.1371/journal.pone.0088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin L, Liang H, Wang Y, Yin X, Hu Y, Huang J, Ren T, Xu H, Zheng L, Chen X. microRNA-141 inhibits cell proliferation and invasion and promotes apoptosis by targeting hepatocyte nuclear factor-3β in hepatocellular carcinoma cells. BMC cancer. 2014;14:879. doi: 10.1186/1471-2407-14-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang G, Zhang S, Gao F, Liu Z, Lu M, Peng S, Zhang T, Zhang F. Osteopontin enhances the expression of HOTAIR in cancer cells via IRF1. Biochim Biophys Acta. 2014;1839:837–848. doi: 10.1016/j.bbagrm.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 57.Qin L. Osteopontin is a promoter for hepatocellular carcinoma metastasis: A summary of 10 years of studies. Front Med. 2014;8:24–32. doi: 10.1007/s11684-014-0312-8. [DOI] [PubMed] [Google Scholar]
- 58.Dong Q, Zhu X, Dai C, Zhang X, Gao X, Wei J, Sheng Y, Zheng Y, Yu J, Xie L, et al. Osteopontin promotes epithelial-mesenchymal transition of hepatocellular carcinoma through regulating vimentin. Oncotarget. 2016;7:12997–13012. doi: 10.18632/oncotarget.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alves C Padua, Fonseca AS, Muys BR, de Barros E Lima Bueno R, Bürger MC, De Souza JE, Valente V, Zago MA, Silva WA., Jr Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem cells. 2013;31:2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- 60.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 61.Yang MC, Wang CJ, Liao PC, Yen CJ, Shan YS. Hepatic stellate cells secretes type I collagen to trigger epithelial mesenchymal transition of hepatoma cells. Am J Cancer Res. 2014;4:751–763. [PMC free article] [PubMed] [Google Scholar]
- 62.van Zijl F, Zulehner G, Petz M, Schneller D, Kornauth C, Hau M, Machat G, Grubinger M, Huber H, Mikulits W. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol. 2009;5:1169–1179. doi: 10.2217/fon.09.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu H, Lu D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6:27847–27864. doi: 10.18632/oncotarget.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/C6MB00114A. [DOI] [PubMed] [Google Scholar]
- 66.Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, Wong CM. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–631. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- 67.Gao SB, Xu B, Ding LH, Zheng QL, Zhang L, Zheng QF, Li SH, Feng ZJ, Wei J, Yin ZY, et al. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol. 2014;61:832–839. doi: 10.1016/j.jhep.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 68.Kirmizis A, Bartley SM, Farnham PJ. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol Cancer Ther. 2003;2:113–121. [PubMed] [Google Scholar]
- 69.Zhao ZK, Dong P, Gu J, Chen L, Zhuang M, Lu WJ, Wang DR, Liu YB. Overexpression of LSD1 in hepatocellular carcinoma: A latent target for the diagnosis and therapy of hepatoma. Tumour Biol. 2013;34:173–180. doi: 10.1007/s13277-012-0525-x. [DOI] [PubMed] [Google Scholar]
- 70.Zhao ZK, Yu HF, Wang DR, Dong P, Chen L, Wu WG, Ding WJ, Liu YB. Overexpression of lysine specific demethylase 1 predicts worse prognosis in primary hepatocellular carcinoma patients. World J Gastroenterol. 2012;18:6651–6656. doi: 10.3748/wjg.v18.i45.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye P, Wang T, Liu WH, Li XC, Tang LJ, Tian FZ. Enhancing HOTAIR/MiR-10b drives normal liver stem cells toward a tendency to malignant transformation through inducing epithelial- to-mesenchymal transition. Rejuvenation Res. 2015;18:332–340. doi: 10.1089/rej.2014.1642. [DOI] [PubMed] [Google Scholar]
- 72.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 73.Ono H, Motoi N, Nagano H, Miyauchi E, Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N, Ishikawa Y. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med. 2014;3:632–642. doi: 10.1002/cam4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou X, Chen J, Tang W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim Biophys Sin (Shanghai) 2014;46:1011–1015. doi: 10.1093/abbs/gmu104. [DOI] [PubMed] [Google Scholar]
- 75.Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Irigoyen O, Latasa MU, Carotti S, Uriarte I, Elizalde M, Urtasun R, Vespasiani-Gentilucci U, Morini S, Benito P, Ladero JM, et al. Matrix metalloproteinase 10 contributes to hepatocarcinogenesis in a novel crosstalk with the stromal derived factor 1/C-X-C chemokine receptor 4 axis. Hepatology. 2015;62:166–178. doi: 10.1002/hep.27798. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Shen Y, Cao B, Yan A, Ji H. Elevated expression levels of androgen receptors and matrix metalloproteinase-2 and −9 in 30 cases of hepatocellular carcinoma compared with adjacent tissues as predictors of cancer invasion and staging. Exp Ther Med. 2015;9:905–908. doi: 10.3892/etm.2014.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang H, Diab A, Fan H, Mani SK, Hullinger R, Merle P, Andrisani O. PLK1 and HOTAIR accelerate proteasomal degradation of SUZ12 and ZNF198 during Hepatitis B virus-induced liver carcinogenesis. Cancer Res. 2015;75:2363–2374. doi: 10.1158/0008-5472.CAN-14-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin Zhou, Wu JB, Tang LY, Gao SM. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 85.Li B, Huang P, Qiu J, Liao Y, Hong J, Yuan Y. MicroRNA-130a is down-regulated in hepatocellular carcinoma and associates with poor prognosis. Med Oncol. 2014;31:230. doi: 10.1007/s12032-014-0230-2. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y, Ren F, Luo Y, Rong M, Chen G, Dang Y. Down-regulation of MiR-193a-3p dictates deterioration of HCC: A Clinical real-time qRT-PCR study. Med Sci Monit. 2015;21:2352–2360. doi: 10.12659/MSM.894077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kladi-Skandali A, Michaelidou K, Scorilas A, Mavridis K. Long noncoding RNAs in digestive system malignancies: A novel class of cancer biomarkers and therapeutic targets? Gastroenterol Res Pract. 20152015:319861. doi: 10.1155/2015/319861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, Liang WC, Wang SS, Ko CH, Waye MM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]