Abstract
Cholangiocarcinoma (CCA) is a malignant transformation of biliary epithelial cells. It is a slow growing tumor, but is also highly metastatic with a poor prognosis. Bile acids are known to transactivate the epidermal growth factor receptor (EGFR) in cholangiocytes and induce cyclooxygenase-2 expression. The protein expression profiles of bile acid-treated CCA cells were studied using a proteomic approach. To elucidate the possible mechanisms involved in the bile acid-mediated enhancement of CCA cell migration, the effects of six bile acids, including cholic, deoxycholic, taurocholic, taurodeoxycholic, glycocholic and glycodeoxycholic acid, on the migration of CCA cells were examined in vitro using wound healing assays. Subsequently, the possible proteins involved in enhanced CCA cell migration were investigated using a proteomic approach. Changes to the protein expression profiles of CCA cells following bile acid treatment was examined using two-dimensional electrophoresis and mass spectrometry. The results demonstrated that cholic and deoxycholic acid significantly enhanced the migration of CCA cells, compared with the treated MMNK-1 control cells. CCA cells had 77 overexpressed protein spots following cholic acid treatment, and 50 protein spots following deoxycholic acid treatment, compared with the treated MMNK-1 control cells. Liquid chromatography tandem-mass spectrometry analysis revealed that coiled-coil domain containing 25 (CCDC25) was significantly overexpressed in cholic acid-treated CCA cells compared with in cholic acid-treated control cells. When the expression levels of CCDC25 were investigated using western blot analysis, CCDC25 was demonstrated to be highly expressed in CCA tissues, but not in the adjacent non-cancerous tissue samples. The identified proteins were further analyzed for protein-chemical interactions using STITCH version 3.1 software. CCDC25 protein was identified to be associated with Son of sevenless homolog 1 and growth factor receptor-bound protein 2, which are involved in EGFR signaling. The results of the present study demonstrated that following cholic acid treatment, CCDC25 is overexpressed in CCA cells, which is associated with significantly enhanced cell migration. This suggests that CCDC25 is a potential therapeutic target for the treatment of patients with CCA.
Keywords: cholangiocarcinoma, bile acids, migration, CCDC25
Introduction
Cholangiocarcinoma (CCA) is a malignant transformation of biliary epithelial cells. It is a slow growing tumor, but it is also highly metastatic with a poor prognosis (1). The highest incidence of this cancer has been identified in Asia, particularly in Northeast Thailand (2). In the majority of cases of CCA, the etiology is unknown. Chronic inflammation and cellular injury within bile ducts, together with the partial obstruction of bile flow result in high-risk conditions for CCA development (3). The tumor not only may develop mechanisms to survive in the toxic environment of bile, but may also utilize bile components to promote cell growth, survival and invasion (4–6). Bile acids are known to transactivate the epidermal growth factor receptor (EGFR) in cholangiocytes and induce cyclooxygenase-2 (COX-2) expression (4,5). These consequences contribute to promote angiogenesis and facilitate cellular growth of various cancer types (5). Furthermore, bile acids enhance the levels of a potent antiapoptotic protein, myeloid cell leukemia protein 1 (Mcl-1), in CCA cells (4).
Alteration of bile composition during chronic inflammation may be important for the carcinogenesis and progression of tumors of the biliary tract (7). Deoxycholic acid (DCA) exerts a pro-carcinogenic effect through the induction of EGFR activation, which in turn leads to stimulation of the mitogen activated protein kinase (MAPK) signaling cascade with associated activation of extracellular signal-regulated kinase 1/2, p38 and c-Jun N-terminal kinase proteins (4,8). Taurocholic acid (TCA) is known to stimulate the secretion of certain cytokines, including monocyte chemoattractant protein-1 (MCP-1) and interleukin-6 (IL-6), from cholangiocytes, which are involved in the inflammatory and fibrotic processes that occur during cholestatic diseases (9). Additionally, chenodeoxycholic acid (CDCA) and lithocholic acid (LCA) could promote CCA invasiveness via stimulating the Snail signaling pathway, leading to the downregulation of E-cadherin (10).
The present study aimed to investigate the effects of bile acids on the migration of human CCA cells. To elucidate the potential underlying mechanisms of bile acid-induced cell migration, the protein expression profiles of bile acid-treated CCA cells were studied using a proteomic approach.
Materials and methods
Cell lines and cell culture
The KKU-M213 cell line, which was established from a primary tumor mass of intrahepatic CCA (11), was provided by the Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University (Khon Kaen, Thailand). An immortalized cholangiocyte cell line, MMNK-1, which was established by Maruyama et al (12), was used as the control. The two cell lines were cultured in Ham's F-12 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) culture medium supplemented with 10% fetal bovine serum (Gibco; Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin. The cells were incubated at 37°C in a 5% CO2 atmosphere. The two cell lines were confirmed to be Mycoplasma-free via using specific polymerase chain reaction.
Bile acid treatment
KKU-M213 and MMNK-1 cells were treated with 100 µM of each bile acid (10) as follows: Cholic acid (CA), DCA, TCA, taurodeoxycholic acid (TDCA), glycocholic acid (GCA) and glycodeoxycholic acid (GDCA). The bile acids were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). All of the bile acids used in the present study were kept as 10 mM stock solutions in dimethyl sulfoxide (DMSO) and stored at −20°C.
In vitro wound healing assay
The two cell lines were cultured at 37°C except otherwise stated, and were seeded into a 24-well culture plate at a density of 2×105 cells/well. The cells were serum-starved for 24 h to avoid possible serum effects (13). Then, they were pretreated with serum-free medium containing 100 µM of each bile acid for 24 h prior to wound formation. The ‘scratch’ wounds were created using a sterile 10 µl pipette tip (10). The old medium was discarded and the wells were washed twice with sterile PBS. A total of 200 µl/well of freshly prepared serum-free medium containing each bile acid was added. The cells were incubated with bile acids for 12 and 24 h at 37°C. The wound edges were imaged using a digital camera fitted to a light microscope with ×10 objective lens. The images were captured at 0, 12 and 24 h following wounding (10). The degree of wound closure was measured at three independent wound sites per each treatment group at 0, 12 and 24 h. The percentage of cell migration was calculated according to the following formula: [Distance of edge at 0–12 h (or 24 h)/distance of edge at 0 h] ×100. Values from three independent experiments were pooled and expressed as the mean ± standard deviation.
Two-dimensional gel electrophoresis (2-DE)
KKU-M213 and MMNK-1 cells treated with or without 100 µM CA or DCA were harvested by centrifugation at 500 × g for 5 min at 4°C and mixed with the equal concentration of lysis buffer consisting of 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 2% IPG buffer, 40 mM dithiothreitol (DTT), and inhibitors of protease and phosphatase, for 10 min on ice. Following centrifugation at 20,000 × g for 30 min at 4°C, the supernatant was collected and desalted using a 2-D Clean Up kit (GE Healthcare Life Sciences, Chalfont, UK). Protein concentrations of all samples were determined using Bradford reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of 10 µg protein sample in 125 µl 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 1% IPG buffer, 0.002% (w/v) bromophenol blue and 40 mM DTT was loaded onto a 7-cm-long, pH 4.0–7.0 IPG strip (GE Healthcare Life Sciences). Electrofocusing was performed according to the manufacturer's protocol. Following focusing, the strips were reduced with 130 mM DTT for 15 min at room temperature and then alkylated with 135 mM iodoacetamide for 15 min at room temperature in equilibration buffer [6 M urea, 75 mM Tris-HCl (pH 8.8), 29.3% glycerol, 2% SDS, 0.002% (w/v) bromophenol blue]. The second dimensional separation was performed in a SE260 Mini-Vertical Electrophoresis unit (GE Healthcare Life Sciences) with a constant current at 20 mA/gel for 90 min. Separated protein spots were visualized using silver staining. Images were collected and analyzed using ImageMaster™ 2D Platinum software (version 7.0; GE Healthcare Life Sciences). To determine the increase in intensity of the protein spots of CA- and DCA-treated CCA cells and MMNK-1 cells, the values of the untreated counterparts were subtracted. Subsequently, the statistical differences of the expression levels between CCA and MMNK-1 cells were assessed using a Student's t-test, and P<0.01 was considered to indicate a statistically significant difference.
In-gel tryptic digestion
The protein spots in 2-DE were manually punched with a pipette tip and kept in 20 µl sterile distilled water at 4°C. The gel pieces were transferred to a 96-well plate and each gel was processed for in-gel digestion. Briefly, the gel pieces were dehydrated twice in 100% acetronitrile (ACN) for 5 min each with agitation and dried at room temperature for 10 min. Subsequently, proteins were reduced with 10 mM DTT/10 mM ammonium bicarbonate (NH4HCO3) for 1 h at 56°C, and then alkylated with 100 mM iodoacetamide (IAA)/10 mM NH4HCO3 for 1 h at room temperature in the dark. The gel pieces were dehydrated twice in 100% ACN for 5 min each with agitation. Following reduction and alkylation, 20 µl of 10 ng/µl trypsin in 10 mM NH4HCO3 was added, and incubated for 20 min prior to adding 20 µl of 30% ACN, and then incubated overnight at room temperature. The tryptic peptides were extracted from the gel three times with 30 µl of 50% ACN/0.1% formic acid for 10 min at room temperature. Finally, the tryptic peptide mixtures were dried and kept at 40°C overnight. Samples were resuspended in 0.1% formic acid/water and centrifuged at 10,000 × g for 10 min. Samples were applied to liquid chromatography tandem-mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS analysis
LC-MS/MS analyses of the digested peptide mixtures were performed using a Waters® SYNAPT™ HDMS™ system (Waters Corporation, Milford, MA, USA). The 1D-nanoLC was performed with a Waters® nanoACQUITY UPLC® system (Waters Corporation) at 0°C. A total of 4 µl tryptic digests were injected onto the reversed phase analytical chromatography column (20 cm × 75 µm) packed with a 1.7 µm Bridged Ethyl Hybrid C18 material (Waters Corporation). Peptides were eluted with a linear gradient from 2–40% acetonitrile developed over 60 min at a flow rate of 350 nl/min. This was followed by a 15 min period of 80% acetonitrile to clean the column prior to returning to 2% acetonitrile for the next sample.
The MS/MS data were used to identify proteins from the primary protein sequence database using Mascot software version 2.3 (Matrix Science, Ltd., London, UK) (14). The data were searched against the National Center for Biotechnology Information database for protein identification (http://www.ncbi.nlm.nih.gov). Database interrogations included taxonomy (Homo sapiens), enzyme (trypsin), variable modifications (carbamidomethyl, oxidation of methionine residues), mass values (monoisotopic), protein mass (unrestricted), peptide mass tolerance (1.2 Da), fragment mass tolerance (±0.6 Da), peptide charge state (1+, 2+ and 3+) and maximum number of missed cleavages (1). Protein hits with the highest Molecular Weight Search score with accurate molecular mass and isoelectric point values according to the 2D-gel appearance were reported.
Tumor samples
CCA tissue samples, including cancerous and adjacent non-cancerous tissue samples, were obtained from 3 patients (a 44-year-old female, 47- and 49-year-old males) immediately following surgical operation on September 2010, January and December 2011, respectively, which were provided by the Liver Flukes and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University. The inclusion criteria of the 3 patients were CCA without lymph node metastasis and long (53–64 weeks) survival. Tissue samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until use. A part of each frozen tissue section was fixed in 4% buffered formalin, and processed for histological examination using hematoxylin and eosin staining. In this study, CCA specimen from only 3 cases were used, because this is our first trial to examine the expression level of CCDC25 in CCA; therefore, the present study decided to use a minimum number of freshly prepared samples. The present study was approved by the Khon Kaen University Ethics Committee for Human Research (no. HE571416). Written informed consent was obtained from all participating patients.
Western blot analysis
To validate CCDC25 protein expression, three paired cancerous and adjacent non-cancerous tissue samples were examined using western blot analysis with an anti-human CCDC25 antibody at 1:500 dilution (cat no. orb2517; Biorbyt, Ltd., Cambridge, UK). Briefly, ~100–200 mg frozen CCA and adjacent noncancerous tissue samples were washed in ice-cold PBS (0.1 M sodium phosphate, 0.15 M sodium chloride, pH 7.2) to remove any blood clots and connective tissue. Samples were incubated with lysis buffer consisting of 7 M urea, 2 M thiourea, 4% CHAPS, 2% IPG buffer, 40 mM DTT, and inhibitors of protease and phosphatase for 10 min on ice. Following centrifugation at 20,000 × g for 30 min at 4°C, the supernatant was collected. A total of 50 µg protein was separated using a 12.5% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (GE Healthcare Life Sciences) for 1 h at room temperature. Subsequently, the membrane was blocked with 5% skim milk powder in Tris-buffered saline with 0.1% Tween-20 (TBS-T, pH 7.4) for 1 h at room temperature. The membrane was then incubated overnight at 4°C with a rabbit polyclonal antibody directed against human CCDC25. Following washing with 0.1% TBS-T, the membrane was incubated for 1 h at room temperature with a horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat no. NA934-1ML; GE Healthcare Life Sciences) diluted (1:5,000) in 5% skim milk/TBS-T. The chemiluminescence was detected using an ECL plus system (GE Healthcare Life Sciences) and then visualized using the ImageQuant™ LAS mini 400. A primary antibody directed against β-actin at 1:500 dilution (cat no. orb10033; Biorbyt, Ltd.) was used as the control.
Bioinformatics using STITCH
The potential interactions of the identified proteins were analyzed using STITCH (version 3.1; http://stitch.embl.de) (14). The input page icon was selected followed by ‘multiple names’. The list of proteins names, including CCDC25, was entered and ‘Homo sapiens’ was selected as the organism. The page depicted ‘list names’ followed by the confidence view. Stronger associations were presented as thicker lines. Protein-protein interactions were indicated as solid lines, chemical-protein interactions as dashed lines and interactions between chemicals as dotted lines. High confidence interactions were identified if they possessed a score >0.7 (15).
Statistical analysis
Data are presented as the mean ± standard error of the mean following three independent experiments using Microsoft Excel 2010 software (Microsoft Corporation, Redmond, WA, USA). Comparison of data between the groups was analyzed using Student's t-tests for parametric data, and Mann-Whitney U tests for nonparametric data. P<0.05 was considered to indicate a statistically significant difference. The GraphPad Prism v.5 software (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses.
Results
CA and DCA treatment enhances CCA cell migration
Using the wound-healing assay, the migration of KKU-M213 cells was compared with that of MMNK-1 cells. Without bile acid stimulation, the degree of migration of KKU-M213 and MMNK-1 cells at 12 h and also 24 h were not significantly different from each other (Table I). Following treatment of KU-M213 and MMNK-1 cells with various bile acids, cell motility was assessed using a wound-healing assay. Both CA and DCA were demonstrated to enhance cell migration, with a significantly higher efficacy on KKU-M213 cells compared with MMNK-1 cells at 12 (P=0.021 and 0.002) and 24 h (P=0.001 and 0.001), respectively (Table I). The other four types of bile acids did not significantly induce cell migration following 24 h of incubation.
Table I.
Effect of bile acids on migration of KKU-M213 and MMNK-1 cells in wound healing assays.
| % Migrationa (mean ± standard error of the mean) | P-valueb | ||||
|---|---|---|---|---|---|
| Bile acid treatment | 12 h | 24 h | Migration comparison | 12 h | 24 h |
| None | KKU-M213<MMNK-1 | ||||
| KKU-M213 | 15.5±2.0 | 167±1.9 | 0.466 | 0.147 | |
| MMNK-1 | 19.3±5.3 | 24.4±3.3 | |||
| CA | KKU-M213>MMNK-1 | ||||
| KKU-M213 | 27.3±5.6 | 36.7±5.6 | 0.021 | 0.010 | |
| MMNK-1 | 10.0±3.3 | 22.8±2.0 | |||
| DCA | KKU-M213>MMNK-1 | ||||
| KKU-M213 | 23.6±4.3 | 32.0±4.3 | 0.002 | 0.001 | |
| MMNK-1 | 3.1±1.7 | 9.1±2.7 | |||
| TCA | KKU-M213<MMNK-1 | ||||
| KKU-M213 | 20.0±4.9 | 34.1±5.0 | 0.003 | 0.101 | |
| MMNK-1 | 38.1±2.3 | 44.9±3.6 | |||
| TDCA | KKU-M213<MMNK-1 | ||||
| KKU-M213 | 21.2±2.3 | 35.8±3.2 | 0.022 | 0.953 | |
| MMNK-1 | 32.3±3.2 | 36.5±2.3 | |||
| GCA | KKU-M213<MMNK-1 | ||||
| KKU-M213 | 17.9±3.2 | 22.4±4.3 | <0.001 | <0.001 | |
| MMNK-1 | 36.8±2.9 | 52.6±3.4 | |||
| GDCA | KKU-M213<MMNK-1 | ||||
| KKU-M213 | 21.0±3.8 | 33.5±4.5 | 0.072 | 0.543 | |
| MMNK-1 | 28.2±3.3 | 36.5±4.3 | |||
Cells were treated with 100 µM of each bile acid type.
Percentage migration = [distance of edge at 0–12 h (or 24 h)/distance of edge at 0 h] ×100.
The difference in percentage migration between KKU-M213 and MMNK-1 cells. The Mann-Whitney U test was used to determine significant differences between the groups. <, Slower migration rate; >, faster migration rate; CA, cholic acid; DCA, deoxycholic acid; TCA, taurocholic acid; TDCA, taurodeoxycholic acid; GCA, glycocholic acid; GDCA, glycodeoxycholic acid.
Effect of bile acids on protein expression in CCA cells
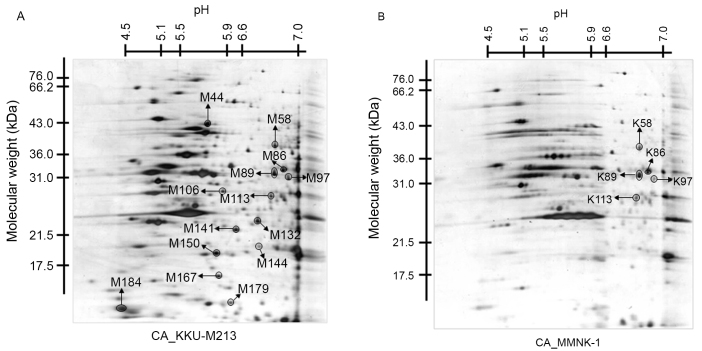
To assess whether the CA- or DCA-induced enhancement of cell migration is associated with changes in protein expression, protein expression profiles were assessed using 2-DE. The protein profile of KKU-M213 cells in the 2D images was compared with that of MMNK-1 cells under CA- (Fig. 1) and DCA-treated conditions (data not presented). Prior to comparison, all spots were subtracted by each untreated condition to select candidate spots for matched spots analysis.
Figure 1.
Two-dimensional electrophoresis patterns of protein spots of (A) CA-treated KKU-M213 cells, compared with (B) CA-treated MMNK-1 cells. The x-axis represents the isoelectric point values and the y-axis represents the molecular weight (kDa). CA, cholic acid.
Following spot matching analysis of the CA-treated group, 19 protein spots were identified to be overexpressed and 19 protein spots were underexpressed in KKU-M213 cells, compared with in the MMNK-1 cells. Furthermore, 77 protein spots were observed only in KKU-M213 cells. In the DCA-treated group, 31 protein spots were identified to be overexpressed and 37 protein spots were underexpressed in KKU-M213 cells, compared with those in the MMNK-1 cells. Additionally, 50 protein spots were detected only in KKU-M213 cells. The protein spots that were expressed exclusively in KKU-M213 cells, or those that demonstrated ≥2-fold higher differences in density in the same spot in MMNK-1 cells in triplicate gels (14 spots for CA- and 13 spots in the DCA-treated condition), were excised and subjected to in-gel digestion for further MS analysis.
Following MS analysis, only 1/27 spots exhibited a protein score >56 (significantly different hit, P<0.05), which was CCDC25 in the spot M97. As illustrated in Fig. 2, this spot was significantly expressed in KKU-M213 cells treated with CA (Fig. 1). The protein scores of all other spots were <56. The expression levels of CCDC25 in the tissue lysates of CCA and adjacent non-cancerous tissue samples obtained from three patients were examined using western blot analysis. High expression of the CCDC25 protein was observed in the CCA tissue, but not in the adjacent non-cancerous tissues (Fig. 2).
Figure 2.
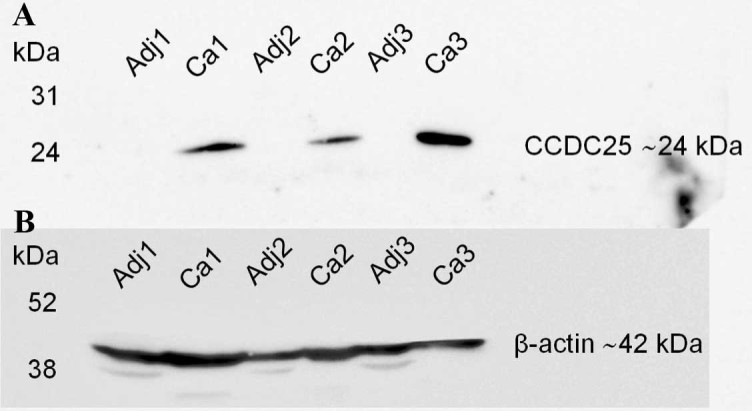
Expression levels of CCDC25 protein in CCA and adjacent non-cancerous tissue samples. (A) The CCDC25 protein in tissue lysates was detected using western blot analysis. High expression levels of CCDC25 were observed in CCA (Ca1-3) tissue but not in the adjacent (Adj1-3) non-cancerous tissue samples. (B) β-actin was used as the control. The y-axis represents the molecular weight (kDa). CCA, cholangiocarcinoma; CCDC25, coiled-coil domain containing 25.
Protein profiles identify two putative migration-associated EGFRs
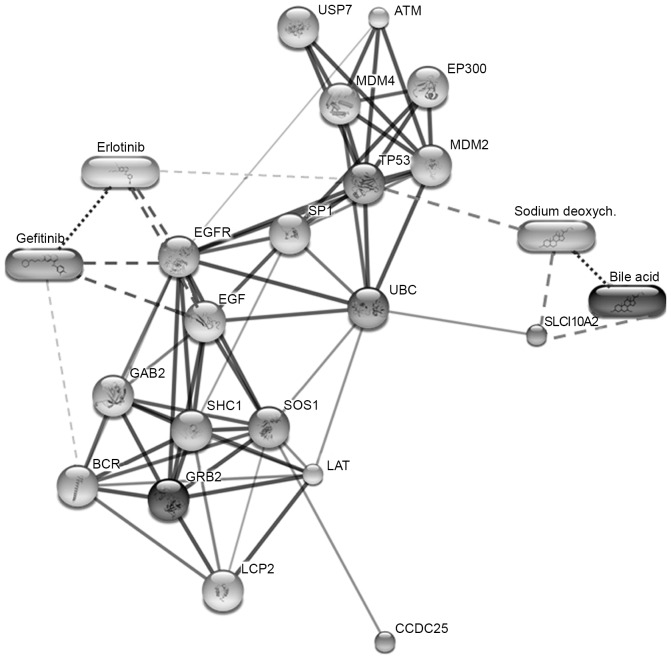
Association or interaction of the putative migration-associated proteins, including EGF, EGFR, growth factor receptor-bound protein 2 (GRB2), CCDC25, Son of sevenless homolog 1 (SOS1) and Src homology 2 domain containing transforming protein 1, were predicted using STITCH software (version 3.1). CCDC25 was identified to be associated with SOS1 and GRB2. These two proteins are predicted to associate with EGFR signaling (15). Therefore, CCDC25 may be a novel initiator for EGF signaling pathways involved in cancer cell migration (Fig. 3).
Figure 3.
Protein-ligand interaction map of the six identified proteins predicted using STITCH v.3.1. Stronger associations are presented as thicker lines. Protein-protein interactions are indicated by solid lines, chemical-protein interactions by dashed lines and interactions between chemicals by dotted lines.
Discussion
The present results demonstrated that CA and DCA enhance the migration of CCA cells (KKU-M213) but not of control (MMNK-1) cells. Stimulation of cancer progression by bile acids requires numerous signaling pathways, including phosphoinositide-3-kinase, protein kinase C, MAPK and COX-2 (3). Pai et al (16) reported that DCA activated the β-catenin signaling pathway, which increased the growth and invasion of colon cancer cells. Debruyne et al (17) demonstrated that bile acids are able to stimulate the invasion and haptotaxis of human colorectal cancer cells through the activation of multiple oncogenic signaling pathways. Unconjugated CDCA and LCA at concentrations of 100 µM may activate the transcription of the Snail gene, which downregulates the expression of E-cadherin and, as a consequence, promote the invasion of human hepatocellular carcinoma cells (9). These results imply that the stimulatory effects of bile acids on cancer cells are variable depending on the combination of various forms of bile acids and cancer cells.
In the present study, CA and DCA were observed to alter the protein expression patterns of CCA cells and cholangiocytes. CA and DCA are unconjugated forms; unconjugated hydrophobic bile acids possess higher affinities for nuclear receptors, such as the farnesoid X receptor, compared with conjugated hydrophilic bile acids (10), thus may have an effect on cancer progression. It is hypothesized that unconjugated bile acids, which can enter the cells via passive diffusion through the plasma membrane, can directly activate nuclear receptors and modulate the transcriptional activity of stimulating protein1 (Sp1) on the Snail promotor (10). Certain reports have indicated that unconjugated bile acids may induce cell apoptosis, particularly at high concentrations (>100 µM) (18–20).
In the present study, a proteomic approach revealed that CA and DCA upregulate the protein expression of KKU-M213 cells more compared with that of MMNK-1 cells. Among the upregulated proteins in KKU-M213 cells stimulated by CA, only CCDC25 was expressed significantly higher in KKU-M213 cells, compared with in control MMNK-1 cells. When CCDC25 protein expression in the CCA tissue and adjacent non-cancerous tissue samples was examined using western blot analysis, CCDC25 was highly expressed in CCA but not in the adjacent non-cancerous tissue of all three samples. The function of CCDC25 remains poorly understood. Previously, it has been reported that the deletion of a cluster of six genes on chromosome 8p, including CCDC25, was found to be associated with poor prognosis of hepatocellular carcinoma patients (21).
In the present study, the identified proteins and biomolecules of interest were submitted to the STITCH database to investigate potential cellular functions, and identify the associated functional interactions among proteins in the cell (15). The results revealed strong interactions between CCDC25 and SOS1 or GRB2, which are also highly associated with EGFR signaling. Previous studies revealed that DCA at 200 µM induced COX-2 protein expression via the activation of EGFR (3) and could also increase cellular Mcl-1 protein expression by inhibiting its degradation via activating the EGFR/RAF proto-oncogene serine/threonine-protein kinase signaling cascade in human CCA cell lines (4). Further studies to investigate the molecular mechanisms underlying the upregulation of CCD25 expression by unconjugated bile acids and the interactions between CCDC25, SOS1 and GRB2 are warranted. In conclusion, the results of the present study demonstrated that CA can act as a stimulator of cell migration and can increase the expression levels of CCD25 in a human CCA cell line. This molecule is hypothesized to be involved in cell migration via the EGFR signaling pathway. The inhibition of this protein may be an alternative therapeutic option for the treatment of patients with CCA in the future.
Acknowledgements
The authors would like to thank the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences for providing supporting instruments, and the Liver Flukes and Cholangiocarcinoma Research Center, Faculty of Medicine (both Khon Kaen University) for providing cell lines and tissue samples from patients with CCA. In addition, the authors would like to thank Professor Yukifumi Nawa for manuscript editing, and Ms Janthima Jaresitthikunchai, Ms Naumon Phaonakrop, Ms Daraporn Chua-on and Ms Chatthawee Charadchaiyanun for technical support throughout the proteome and western blotting analyses. The present study was supported by Khon Kaen University (grant no. KKU580602).
References
- 1.Malaguarnera G, Paladina I, Giordano M, Malaguarnera M, Bertino G, Berretta M. Serum markers of intrahepatic cholangiocarcinoma. Dis Markers. 2013;34:219–228. doi: 10.1155/2013/196412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazaridis KN, Gores GJ. Cholangiocarcinoma. Gastroenterology. 2005;128:1655–1667. doi: 10.1053/j.gastro.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 3.Sirica AE. Cholangiocarcinoma: Molecular targeting strategies for chemoprevention and therapy. Hepatology. 2005;41:5–15. doi: 10.1002/hep.20537. [DOI] [PubMed] [Google Scholar]
- 4.Yoon JH, Higuchi H, Werneburg NW, Kaufmann SH, Gores GJ. Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology. 2002;122:985–993. doi: 10.1053/gast.2002.32410. [DOI] [PubMed] [Google Scholar]
- 5.Yoon JH, Werneburg NW, Higuchi H, Canbay AE, Kaufmann SH, Akgul C, Edwards SW, Gores GJ. Bile acids inhibit Mcl-1 protein turnover via an epidermal growth factor receptor/Raf-1-dependent mechanism. Cancer Res. 2002;62:6500–6505. [PubMed] [Google Scholar]
- 6.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1:11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 7.Gores GJ. Cholangiocarcinoma: Current concepts and insights. Hepatology. 2003;37:961–969. doi: 10.1053/jhep.2003.50200. [DOI] [PubMed] [Google Scholar]
- 8.Yoon JH, Canbay AE, Werneburg NW, Lee SP, Gores GJ. Oxysterols induce cyclooxygenase-2 expression in cholangiocytes: Implications for biliary tract carcinogenesis. Hepatology. 2004;39:732–738. doi: 10.1002/hep.20125. [DOI] [PubMed] [Google Scholar]
- 9.Lamireau T, Zoltowska M, Levy E, Yousef I, Rosenbaum J, Tuchweber B, Desmoulière A. Effects of bile acids on biliary epithelial cells: Proliferation, cytotoxicity, and cytokine secretion. Life Sci. 2003;72:1401–1411. doi: 10.1016/S0024-3205(02)02408-6. [DOI] [PubMed] [Google Scholar]
- 10.Fukase K, Ohtsuka H, Onogawa T, Oshio H, Ii T, Mutoh M, Katayose Y, Rikiyama T, Oikawa M, Motoi F, et al. Bile acids repress E-cadherin through the induction of Snail and increase cancer invasiveness in human hepatobiliary carcinoma. Cancer Sci. 2008;99:1785–1792. doi: 10.1111/j.1349-7006.2008.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sripa B, Leungwattanawanit S, Nitta T, Wongkham C, Bhudhisawasdi V, Puapairoj A, Sripa C, Miwa M. Establishment and characterization of an opisthorchiasis-associated cholangiocarcinoma cell line (KKU-100) World J Gastroenterol. 2005;11:3392–3397. doi: 10.3748/wjg.v11.i22.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruyama M, Kobayashi N, Westerman KA, Sakaguchi M, Allain JE, Totsugawa T, Okitsu T, Fukazawa T, Weber A, Stolz DB, et al. Establishment of a highly differentiated immortalized human cholangiocyte cell line with SV40T and hTERT. Transplantation. 2004;77:446–451. doi: 10.1097/01.TP.0000110292.73873.25. [DOI] [PubMed] [Google Scholar]
- 13.Werneburg NW, Yoon JH, Higuchi H, Gores GJ. Bile acids activate EGF receptor via a TGF-alpha-dependent mechanism in human cholangiocyte cell lines. Am J Physiol Gastrointest Liver Physiol. 2003;285:G31–G36. doi: 10.1152/ajpgi.00536.2002. [DOI] [PubMed] [Google Scholar]
- 14.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn M, Szklarczyk D, Pletscher-Frankild S, Blicher TH, von Mering C, Jensen LJ, Bork P. STITCH 4: Integration of protein-chemical interactions with user data. Nucleic Acids Res. 2014;42(Database Issue):D401–D407. doi: 10.1093/nar/gkt1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15:2156–2163. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debruyne PR, Bruyneel EA, Karaguni IM, Li X, Flatau G, Müller O, Zimber A, Gespach C, Mareel MM. Bile acids stimulate invasion and haptotaxis in human colorectal cancer cells through activation of multiple oncogenic signaling pathways. Oncogene. 2002;21:6740–6750. doi: 10.1038/sj.onc.1205729. [DOI] [PubMed] [Google Scholar]
- 18.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15:1677–1689. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell AA, LaRue JM, Batta AK, Martinez JD. Bile acid hydrophobicity is correlated with induction of apoptosis and/or growth arrest in HCT116 cells. Biochem J. 2001;356:481–486. doi: 10.1042/bj3560481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milovic V, Teller IC, Murphy GM, Caspary WF, Stein J. Deoxycholic acid stimulates migration in colon cancer cells. Eur J Gastroenterol Hepatol. 2001;13:945–949. doi: 10.1097/00042737-200108000-00012. [DOI] [PubMed] [Google Scholar]
- 21.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(e12):957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]