 Multivalent carbohydrate photoaffinity probes were developed based on gold nanoparticles (AuNPs) to provide a streamlined approach toward identification of carbohydrate-binding proteins.
Multivalent carbohydrate photoaffinity probes were developed based on gold nanoparticles (AuNPs) to provide a streamlined approach toward identification of carbohydrate-binding proteins.
Abstract
Multivalent carbohydrate photoaffinity probes were developed based on gold nanoparticles (AuNPs) to provide a streamlined approach toward identification of carbohydrate-binding proteins. By using AuNPs as scaffolds, a carbohydrate ligand and a photoreactive group could be readily assembled on a probe in a modular fashion, which greatly accelerated the process of optimizing the probe design. The novel AuNP-based probes serve dual functions by facilitating photoaffinity labeling and by directly enriching the crosslinked proteins by centrifugation. We demonstrated that their ability to enhance the affinity and to stringently remove nonspecific proteins allowed selective photoaffinity labeling and isolation of a low affinity carbohydrate-binding protein in cell lysate.
Introduction
Carbohydrate–protein interactions play important roles in various biological processes such as endocytosis, cell recognition, cell adhesion and differentiation.1 Identification of carbohydrate-binding proteins therefore is a critical first step for elucidating carbohydrate functions at the molecular level as well as for developing new therapeutics. However, many of them remain uncharacterized due to lack of appropriate tools. Carbohydrate-binding proteins often display low affinity and selectivity,2 which makes them difficult to isolate by conventional methods such as affinity chromatography.3 In particular, direct capture of carbohydrate-binding proteins out of the complex cellular proteome is inefficient, and co-purification of nonspecific proteins compromises their effective separation. To overcome these problems, multivalent photoaffinity probes have been developed, in which carbohydrate ligands and photoreactive groups were multivalently displayed on a judicious scaffold.4a This approach combines the multivalent effect for promoting the typically weak carbohydrate–protein interaction,5 and the utility of photoaffinity labelling (PAL)6 in facilitating the purification of the labile carbohydrate–protein complexes by covalent crosslinking. Various scaffold designs for multivalent photoaffinity probes have been shown to improve the capturing efficiency of carbohydrate-binding proteins.4 However, the issue of co-purification of nonspecific proteins has not been addressed. For a more efficient exploration of carbohydrate-binding proteins, a new design of multivalent photoaffinity probes is desired, which implements both affinity enhancement and highly efficient purification of specific binding proteins.
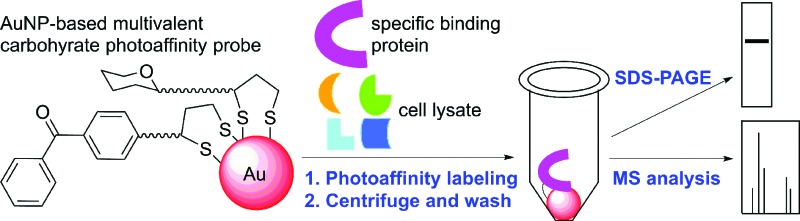
In this study, we developed multivalent carbohydrate photoaffinity probes based on gold nanoparticles (AuNPs), which are capable of stringently removing nonspecific proteins as well as promoting affinity enhancement in order to achieve the efficient and selective capture of specific carbohydrate-binding proteins. They represent novel bifunctional chemical tools, which can perform both photoaffinity labeling and affinity purification in one-pot. AuNPs have been widely employed for mimicking the multivalent presentation of cell surface carbohydrates in applications such as drug delivery systems, sensors, bioimaging reagents and affinity probes.7–9 AuNPs offer attractive scaffolds for photoaffinity probes for several reasons: their surface functionalization is facile, modular and covalent, which allows multiple functionalities to be presented in a well-defined geometry.10 The ease of multifunctionalization on AuNPs is a particular advantage in the straightforward optimization of the probe design, which is often a laborious and time-consuming step in the PAL studies.
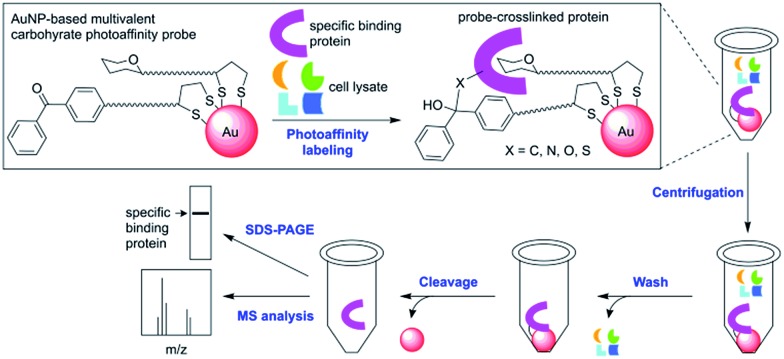
As outlined in Scheme 1, our new PAL approach using AuNP probes proceeds in two stages: first, the AuNP probes serve to photocrosslink specific carbohydrate-binding proteins in solution similar to the conventional molecular photoaffinity probes. In the second stage, they function as solid affinity reagents for separating the probe-crosslinked proteins from the unreacted proteins in solution and the noncovalently bound proteins on the AuNP surfaces through repeated cycles of centrifugation, removal of supernatant solution and washing with a harsh protein denaturing buffer. While AuNPs are dispersive in water, their property as metallic scaffolds enables their separation from a solution simply by centrifugation. The probe-crosslinked proteins are cleaved from the AuNPs, then detected either by MS analysis or by gel-based fluorescent imaging analysis. A common problem with separating the photoaffinity-labeled proteins using the conventional affinity resins is co-purification of the nonspecifically bound proteins, which complicates the subsequent identification of true binding proteins.3 In the cases with low-affinity carbohydrate-binding proteins, there is an additional difficulty that the reaction conditions with high concentration of photoaffinity probes is required to ensure maximum binding and this often leads to increased nonselective crosslinking of abundant proteins.11 The AuNP-based photoaffinity probes therefore, should help address these issues by increasing the selectivity of PAL through affinity enhancement and by efficiently removing the nonspecifically bound proteins using a stringent washing procedure.
Scheme 1. A streamlined photoaffinity labeling approach toward identification of carbohydrate-binding proteins by using AuNP-based multivalent carbohydrate probes.
Results and discussion
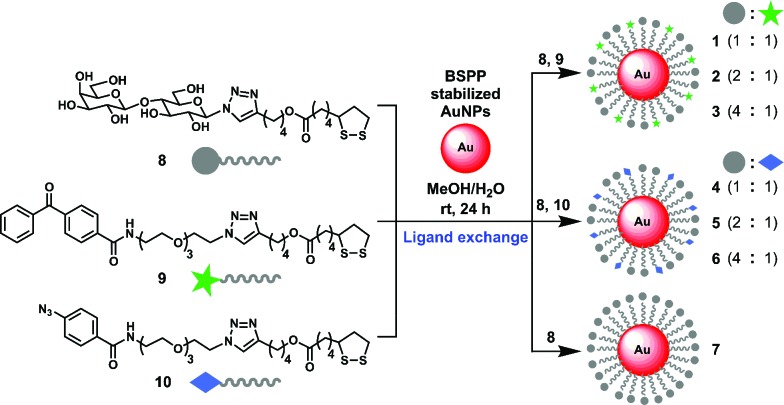
To explore the utility of the AuNP-based probes, we designed probes 1–6 (Scheme 2) displaying β-d-lactose derivative (Lac) as a model system and benzophenone or aryl azide as a photoreactive group. β-d-Lactoside is an important carbohydrate motif in glycoproteins and glycolipids. It can be specifically recognized by several well characterized carbohydrate binding proteins such as peanut agglutinin (PNA, K d = 400 μM),12 Erythrina cristagalli lectin (ECA, K d = 323 μM)13 and Ricinus communis agglutinin (RCA, K d = 37 μM).14 The Lac and photoreactive moieties were each conjugated to lipoic acid to stably functionalize AuNPs through bivalent S–Au bonds.15 AuNPs with a diameter of 5 nm were used so that they are small enough to provide good water-dispersion with minimum light scattering effects, and large enough for efficient centrifugal separation. To investigate the effect of different photoreactive group and the ligand density on the efficiency of PAL, AuNP-based probes were designed to present Lac ligand and a photoreactive group at three different molar ratios (1 : 1, 1, 4; 2 : 1, 2, 5; and 4 : 1, 3, 6; Scheme 2).
Scheme 2. Modular one-step assembly of multivalent carbohydrate photoaffinity probes on AuNPs.
Based on the ligand exchange method,16 lipoic acid conjugated Lac 8 and photoreactive groups (9, 10) mixed at a desired ratio were assembled on a AuNP scaffold by treating with AuNPs stabilized with bis(p-sulfonatophenyl)phenylphosphine dipotassium salt (BSPP) to give probe 1–6. As a reference, Lac functionalized probe 7, which only presents Lac ligand 8 was also prepared. The functionalization of Lac and photoreactive group on AuNPs was confirmed by UV-Vis, SDS-PAGE and MALDI-TOF MS analysis (Fig. S1 and S2†). The representative ligand density was determined for probe 5 by first cleaving the ligands from the AuNPs by thiolate exchange reaction,17 and then quantitated by LC-MS analysis, which gave 152 molecules of Lac and 70 molecules of aryl azide group per AuNP.
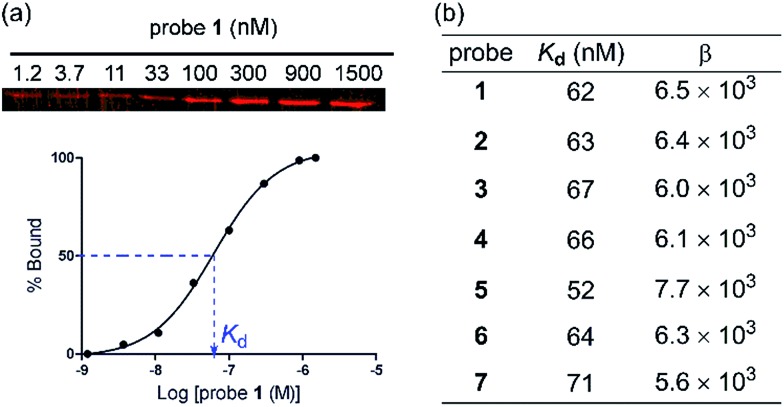
To evaluate the protein binding activity of probe 1–7, we first developed a novel binding assay involving an affinity pull-down assay and a gel-based fluorescent imaging analysis (Fig. 1a). Due to the photoreactive property of the probes, it was preferred that the binding assay does not involve spectrophotometric monitoring of the binding interaction. The assay also needed to be conducted at sufficiently low protein concentrations to avoid binding induced agglutination. Varied concentrations of probe 1–6 were each allowed to bind PNA at low concentration (91 nM) then were centrifuged to separate the probe–PNA complex from free PNA. The bound PNA was eluted from AuNPs by treatment with 2-mercaptoethanol to be resolved by SDS-PAGE and quantitated by fluorescence imaging after staining with a fluorescent protein dye. All the probes efficiently bound PNA in a dose dependent fashion with similar K d values (Fig. 1b) as that of the reference probe 7 bearing only Lac ligand 8 (K d = 71 nM). The differences in the Lac ligand density apparently did not affect the binding affinity of the multivalent probes 1–7. These results indicated that the effective local concentration of the Lac ligand in probe 1–7 were above that required to gain the multivalent effect.18 The multivalent effect of a probe can be described by an enhancement factor β, where β = (K d(monomeric ligand)/K d(multivalent ligand)).19 Based on the reported K d value of 400 μM for d-lactose,11 we obtained β in the rage of 5.6 × 103–7.7 × 103. Therefore, we demonstrated that the AuNP scaffold serves to increase the binding affinity of probe 1–6 regardless of different ligand densities.
Fig. 1. (a) A plot showing the binding of probe 1 to PNA (91 nM) in PBS evaluated by an affinity pull-down assay and gel-based fluorescence imaging. (b) Dissociation constants (K d) and multivalency enhancement factors (β) for probe 1–7.
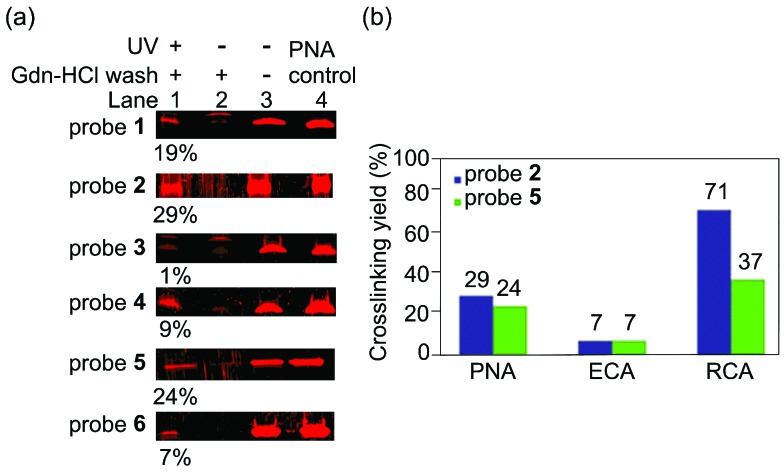
Next, we assessed the effect of the photoreactive group and the ligand density on the photocrosslinking efficiency toward PNA. In a typical PAL reaction, a photoaffinity probe was incubated with a protein to bind at 4 °C for 2 h then was irradiated at 365 nm at 0 °C for 1 h. After the PAL step, the unreacted protein was removed by centrifugation and repeated washing with 3 M guanidinium hydrochloride (Gdn-HCl) buffer, which is a strong protein denaturing agent. The resultant probe-crosslinked protein was cleaved from AuNPs by 2-mercaptoethanol then was analysed by SDS-PAGE. The probe-crosslinked protein was quantitated by fluorescence imaging analysis to provide the crosslinking yield.20 Among the six probes tested, probe 2 and 5 with a 2 : 1 ratio of Lac and a photoreactive group provided the optimal crosslinking efficiency (Fig. 2a, 2: 29%, 5: 24%). These data thus show that the different photoreactive groups (benzophenone and aryl azide) reacted with PNA with similar crosslinking efficiency. In contrast, the ligand density significantly affected the outcomes of PAL. Since the density of Lac ligand did not affect the apparent binding affinity of the probes (Fig. 1b), different outcomes of the PAL experiments suggested that the local concentration of the photoreactive group on AuNPs is important for the photocrosslinking efficiency. Two opposing factors can be considered to explain these data; the higher effective concentration of the photoreactive group is expected to increase the crosslinking yield, while too high local concentration may be unfavourable due to self-reaction or self-quenching of the photoreactive group.
Fig. 2. (a) Photoaffinity labeling of probe 1–6 (100 nM) and PNA (100 nM) in PBS. Lane 1: crosslinked-PNA enriched by Gdn-HCl wash, lane 2: negative control experiment with no UV irradiation, lane 3: negative control experiment with no UV irradiation and no Gdn-HCl wash, lane 4: PNA input as a control. The values (%) represent crosslinking yields.20 (b) Photoaffinity labeling of probe 2 or probe 5 at 100 nM with PNA, ECA, or RCA (100 nM) in PBS.
To demonstrate the generality of the multivalent carbohydrate photoaffinity probes to capture other lactose binding proteins, we next employed probes 2 and 5 for PAL reactions with ECA and RCA under the identical reaction conditions as that for PNA (Fig. 2b). Both benzophenone and aryl azide probes crosslinked ECA with similar yields (7% for 2 and 4) as in the case with PNA, while benzophenone probe was more reactive with RCA (2: 71%, 5: 37%).21 RCA was crosslinked more efficiently by probe 2 and 5 compared to PNA, which is consistent with the higher affinity of RCA toward d-lactose. On the other hand, ECA was crosslinked with markedly lower yields despite its higher binding affinity toward d-lactose than PNA. These data may be due to the differences in the geometry of carbohydrate binding sites on the proteins. Both PNA and RCA are tetrameric proteins having a pair of two carbohydrate binding site facing the same direction with ∼6 nm apart,22 potentially allowing two Lac ligands on a probe to bind at the same time and crosslink more efficiently. ECA, in contrast, is a dimeric protein with the carbohydrate binding site at the opposite faces,23 which likely binds single Lac ligand on a probe thus crosslinking less efficiently.
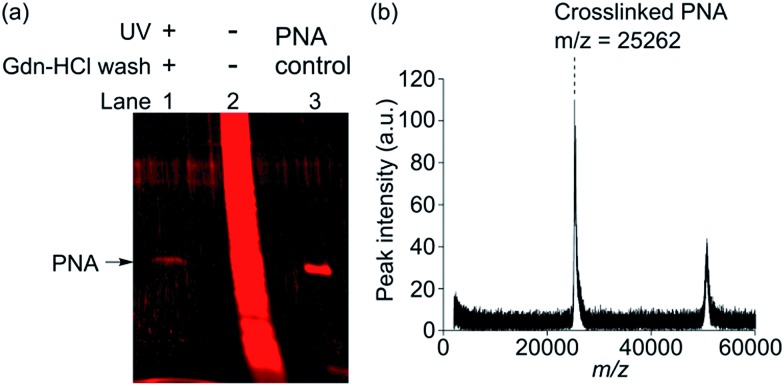
In order to test if the AuNP-based multivalent photoaffinity probes can efficiently crosslink and isolate a low-affinity carbohydrate-binding protein in a complex protein mixture, we reacted probe 2 and PNA mixed with HeLa cell lysate proteins at a ratio of 1 : 100 (w/w). Probe 2 was used at 50 nM, which approximately corresponds to the K d value, to minimize ligand-independent photocrosslinking of nonspecific proteins. The probe-crosslinked proteins were enriched by three repeated cycles of centrifugation and stringent washing using 3 M Gdn-HCl buffer. Subsequently, they were cleaved from AuNPs and were analysed by SDS-PAGE.24 As shown in Fig. 3a, a protein band corresponding to PNA was observed only under the reaction condition involving UV irradiation (Fig. 3a, lane 1). In a stark contrast, when probe 2 and a protein mixture was treated under the negative control condition involving no UV irradiation and no Gdn-HCl washing, a multitude of nonspecific protein bands was obtained (Fig. 3a, lane 2), which made it difficult to identify PNA as a specific binding protein.
Fig. 3. Photoaffinity labeling of PNA (1 μg, 50 nM) mixed in cell lysate (100 μg, 0.5 mg mL–1) using probe 2 (50 nM). (a) SDS-PAGE analysis showing protein band(s) eluted from probe 2. Lane 1: crosslinked-PNA enriched by Gdn-HCl wash, lane 2: negative control experiment with no UV irradiation and no Gdn-HCl wash, lane 3: PNA input as a control. (b) MALDI-TOF MS analysis showing the m/z peaks corresponding to PNA captured by probe 2.
In separate experiments, probe 2 and PNA were subjected to the same PAL reaction and the Gdn-HCl washing conditions and then were directly analysed by MALDI-TOF MS (Fig. 3b). Only two distinctive mass peaks were observed (m/z = 25 262, 50 663), which approximately corresponded to PNA (calculated M w = 25 189 Da, observed m/z = 25 246, Fig. S9†) and its dimer. Taken together, we successfully showed that probe 2 enabled highly selective crosslinking and efficient isolation of PNA. Similar results were also obtained by using probe 5 (Fig. S8 and S9†). Given that PNA binds lactose only weakly (K d = 400 μM),12 highly selective photoaffinity labeling by probe 2 and 5 is remarkable. The ligand-independent photocrosslinking of nonspecific proteins typically becomes significant in a PAL experiment at a probe concentration of >1 μM.11 We therefore reasoned that high selectivity was achieved because the reactions could be conducted at low probe concentration in the nM range by virtue of the multivalent effect. For unambiguous detection of PNA as a specific binding protein, the application of the stringent washing condition by using a protein denaturing buffer was critical in efficiently removing nonspecifically bound proteins. Use of such a washing condition was possible due to the covalent nature of the probe-protein crosslinking. These results clearly demonstrated the advantage of performing PAL over the conventional affinity pull-down method for the efficient isolation of specific binding proteins.
Conclusions
In conclusion, novel multivalent carbohydrate photoaffinity probes were developed based on AuNPs for a streamlined approach, which addresses the issues of low efficiency and specificity associated with directly capturing carbohydrate-binding proteins. The use of the AuNP scaffolds provided unique advantages on two levels. First, the probe design can be optimized rapidly due to the ease of preparing the AuNP-based photoaffinity probes with various combinations and ratios of a ligand and a photoreactive group. Secondly, the physicochemical properties of AuNPs provided a basis for a novel bifunctional chemical tool, which serves both as a photoaffinity probe in solution and a solid affinity reagent which can be readily separated by centrifugation. We demonstrated that the optimized probes facilitated highly selective PAL and straightforward isolation of a low-affinity carbohydrate binding protein in cell lysate. To our knowledge, these results represent the first example of a streamlined photoaffinity labeling approach, where crosslinking, enrichment and isolation of binding proteins were performed using a single probe. Notably, the covalent crosslinking of a binding protein by PAL as opposed to a noncovalent affinity capture was found to be more effective in isolating a specific binding protein due to an efficient removal of nonspecifically bound proteins. Our results therefore showed that the AuNP-based multivalent photoaffinity probes would be useful for the discovery of carbohydrate-binding proteins.
Acknowledgments
We thank Professor Keiichi Noguchi for the technical assistance with LC-MS analysis. MALDI-TOF MS was made available by a grant from the Low-Carbon Research Network Japan (LCnet). This work was funded by the Human Frontier Science Program Young Investigator Grant and Grant-in-Aid for Scientific Research C by JSPS.
Footnotes
References
- Varki A. and Lowe J. B., in Essential of Glycobiology, ed. A. Varki, R. D. Cumming, J. D. Esko, H. H. Freeze, P. Stanley, C. R. Bertozzi, G. W. Hart and M. E. Etzler, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2nd edn, 2009, ch. 6. [PubMed] [Google Scholar]
- Paulson J. C., Blixt O., Collins B. E. Nat. Chem. Biol. 2006;2:238. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- (a) Ziegler S., Pries V., Hedberg C., Waldmann H. Angew. Chem., Int. Ed. Engl. 2013;52:2744. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]; (b) Kawatani M., Osada H. MedChemComm. 2014;5:277. [Google Scholar]
- (a) Lauc G., Lee R. T., Dumič J., Lee Y. C. Glycobiology. 2000;10:357. doi: 10.1093/glycob/10.4.357. [DOI] [PubMed] [Google Scholar]; (b) Ballell L., Scherpenzeel M. V., Buchalova K., Liskamp R. M. J., Pieters R. J. Org. Biomol. Chem. 2006;4:4387. doi: 10.1039/b611050a. [DOI] [PubMed] [Google Scholar]; (c) Park S., Shin I. Org. Lett. 2007;9:619. doi: 10.1021/ol062889f. [DOI] [PubMed] [Google Scholar]; (d) Chang T.-C., Lai C.-H., Chien C.-W., Liang C.-F., Adak A. K., Chuang Y.-J., Chen Y.-C., Lin C.-C. Bioconjugate Chem. 2013;24:1895. doi: 10.1021/bc400306g. [DOI] [PubMed] [Google Scholar]; (e) Wibowo A., Peters E. C., Hsieh-Wilson L. C. J. Am. Chem. Soc. 2014;136:9528. doi: 10.1021/ja502482a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Lee Y. C., Lee R. T. Acc. Chem. Res. 1995;28:321. [Google Scholar]; (b) Mammen M., Choi S.-K., Whitesides G. M. Angew. Chem., Int. Ed. Engl. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; (c) Gestwicki J. E., Cairo C. W., Strong L. E., Oetjen K. A., Kiessling L. L. J. Am. Chem. Soc. 2002;124:14922. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]; (d) Fasting C., Schalley C. A., Weber M., Seitz O., Hecht S., Koksch B., Dernedde J., Graf C., Knapp E.-W., Haag R. Angew. Chem., Int. Ed. Engl. 2012;51:10472. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]; (e) Pieters R. J. Org. Biomol. Chem. 2009;7:2013. doi: 10.1039/b901828j. [DOI] [PubMed] [Google Scholar]
- (a) Kotzyba-Hibert F., Kapfer I., Goeldner M. Angew. Chem., Int. Ed. Engl. 1995;34:1296. [Google Scholar]; (b) Yu S., Wands A. M., Kohler J. J. J. Carbohydr. Chem. 2012;31:325. doi: 10.1080/07328303.2012.676118. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sakurai K. Asian J. Org. Chem. 2015;4:116. [Google Scholar]
- (a) Marradi M., Chiodo F., Garcia I., Penades S. Chem. Soc. Rev. 2013;42:4728. doi: 10.1039/c2cs35420a. [DOI] [PubMed] [Google Scholar]; (b) Adak A. K., Lin H.-J., Lin C.-C. Org. Biomol. Chem. 2014;12:5563. doi: 10.1039/c4ob00827h. [DOI] [PubMed] [Google Scholar]
- (a) Lin C.-C., Yeh Y.-C., Yang C.-Y., Chen C.-L., Chen G.-F., Chen C.-C., Wu Y.-C. J. Am. Chem. Soc. 2002;124:3508. doi: 10.1021/ja0200903. [DOI] [PubMed] [Google Scholar]; (b) Hernaiz J., de la Fuente J. M., Barrientos A. G., Penades S. Angew. Chem., Int. Ed. Engl. 2002;41:1554. doi: 10.1002/1521-3773(20020503)41:9<1554::aid-anie1554>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Chen S.-H., Chien Y.-Y., Chang Y.-W., Liao H.-K., Chang C.-Y., Jan M.-D., Wang K.-T., Lin C.-C. ChemBioChem. 2005;6:1169. doi: 10.1002/cbic.200500023. [DOI] [PubMed] [Google Scholar]
- Love J. C., Estroff L. A., Kriebel J. K., Nuzzo R. G., Whitesides G. M. Chem. Rev. 2005;105:1103. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- Sakurai K., Ozawa S., Yamada R., Yasui T., Mizuno S. ChemBioChem. 2014;15:1399. doi: 10.1002/cbic.201402051. [DOI] [PubMed] [Google Scholar]
- Cagnoni A. J., Varela O., Uhrig M. L., Kovensky J. Eur. J. Org. Chem. 2013;5:972. [Google Scholar]
- Svensson C., Teneberg S., Nilsson C. L., Kjellberg A., Schwarz F. P., Sharon N., Krengel U. J. Mol. Biol. 2002;321:69. doi: 10.1016/s0022-2836(02)00554-5. [DOI] [PubMed] [Google Scholar]
- Sharma S., BharadwaJ S., Surolia A., Podder S. K. Biochem. J. 1998;333:539. doi: 10.1042/bj3330539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Garcia B., Salomé M., Lemelle L., Bridot J.-L., Gillet P., Perriat P., Roux S., Tillement O. Chem. Commun. 2005;3:369. doi: 10.1039/b411231h. [DOI] [PubMed] [Google Scholar]; (b) Karamanska R., Mukhopadhyay B., Russell D. A., Field R. A. Chem. Commun. 2005;26:3334. doi: 10.1039/b503843j. [DOI] [PubMed] [Google Scholar]
- Brown L. O., Hutchison J. E. J. Am. Chem. Soc. 1997;119:12384. [Google Scholar]
- (a) Maxwell D. J., Taylor J. R., Nie S. J. Am. Chem. Soc. 2002;124:9606. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]; (b) Hwu J. R., Lin Y. S., Josephrajan T., Hsu M.-H., Cheng F.-Y., Yeh C.-S., Su W.-C., Shieh D.-B. J. Am. Chem. Soc. 2009;131:66. doi: 10.1021/ja804947u. [DOI] [PubMed] [Google Scholar]; (c) Thygesen M. B., Sauer J., Jensen K. J. Chem.–Eur. J. 2009;15:1649. doi: 10.1002/chem.200801521. [DOI] [PubMed] [Google Scholar]
- (a) Tassa C., Duffner J. L., Lewis T. A., Weissleder R., Schreiber S. L., Koehler A. N., Shaw S. Y. Bioconjugate Chem. 2010;21:14. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gestwicki J. E., Strong L. E., Cairo C. W., Boehm F. J., Kiessling L. L. Chem. Biol. 2002;9:163. doi: 10.1016/s1074-5521(02)00102-3. [DOI] [PubMed] [Google Scholar]; (c) Dam T. K., Gerken T. A., Cavada B. S., Nascimento K. S., Moura T. R., Brewer C. F. J. Biol. Chem. 2007;282:28256. doi: 10.1074/jbc.M704677200. [DOI] [PubMed] [Google Scholar]
- Mammen M., Choi S.-K., Whitesides G. M. Angew. Chem., Int. Ed. Engl. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Crosslinking yields were calculated as follows: [fluorescent intensity of the eluted protein]/[fluorescent intensity of the reacted protein] × 100 (%)
- It's been known that benzophenone group tends to efficiently react with methionine, glycine and proline residues. One of the possible reasons probe 2 displayed higher crosslinking efficiency toward RCA than probe 5 may be that benzophenone group happened to be positioned close to these particular amino acid residues when RCA bound to probe 2 See: Deseke E., Nakatani Y., Ourisson G., Eur. J. Org. Chem., 1998, 2 , 243 Wittelsberger A., Thomasa B. E., Mierkeb D. F., Rosenblatt M., FEBS Lett., 2006, 580 , 1872, Wiegand M., Lindhorst T. K., Eur. J. Org. Chem., 2006. , 4841 . [Google Scholar]
- (a) Olsen K. W., Miller R. L. FEBS Lett. 1982;145:303. [Google Scholar]; (b) Sweeney W. C., Tonevitsky A. G., Temiakov D. E., Agapov I. I., Saward S., Palmer R. A. Proteins: Struct., Funct., Genet. 1997;28:586. doi: 10.1002/(sici)1097-0134(199708)28:4<586::aid-prot12>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Svensson C., Teneberg S., Nilsson C. L., Kjellberg A., Schwarz F. P., Sharon N., Krengel U. J. Mol. Biol. 2002;321:69. doi: 10.1016/s0022-2836(02)00554-5. [DOI] [PubMed] [Google Scholar]
- While the lipoic acid conjugated functionalities on probe 1–7 can be cleaved by thiolate-exchange reaction, they were apparently stable for several hours in cell lysate solution, which contains various reducing agents such as glutathione. Also see: Li X., Guo J., Asong J., Wolfert M. A., Boons G.-J., J. Am. Chem. Soc., 2011, 133 , 11147 .21678979 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.