Abstract
This study examined the association of CD44V6 expression in ovarian cancer. We recruited 38 patients with ovarian cancer, 23 with benign ovarian tumor, and 20 with normal ovaries using RT-PCR and western block analysis. Compared with normal ovaries, the expression of CD44V6 mRNA was significantly elevated in benign ovarian tumor and ovarian cancer. At the protein level, we found no significant differences in CD44V6 expression between normal ovarian tissue and benign ovarian tumor. However, the expression of CD44V6 in ovarian cancer was significantly elevated compared to normal ovaries and benign ovarian tumor. These results were supported by ELISA and western blot analysis. Immunohistochemistry showed that CD44V6 protein in ovarian cancer cells accumulated at high levels on the membrane of ovarian cancer cells. CD44V6 expression is closely associated with the tumorous transformation of ovarian tissue, suggesting that CD44V6 can promote the occurrence and progression of ovarian cancer.
Keywords: CD44V6, ovarian cancer, RT-PCR, enzyme-linked immunosorbent assay, correlation
Introduction
As one of the most common gynecological diseases, the incidence rate of ovarian cancer has exceeded both endometrial and cervical cancer, becoming one of the gynecological diseases with high mortality rate (1). Although research on ovarian cancer has achieved some progress, definite diagnosis of ovarian cancer typically occurs at advanced stages. Reasons for the late diagnosis include the deep location in the pelvic cavity of ovarian cancer (2) and the absence of obvious symptoms in early stages, which hampers the subsequent treatment (3). Therefore, early detection and treatment are critical goals for the recovery of ovarian cancer patients (4). In recent years, the development of molecular biological techniques found that the CD44 gene in chromosome 11 plays an importance regulatory role in lymphocyte homing, movement, activation (5,6), signal transduction, and adhesion among cells (7,8). The CD44 gene contains 20 highly conserved exons including constitutive exons and alternatively spliced exons in the V-area. The structural domain encoded by the alternative exons in the V-area has a close correlation with the incidence and deterioration of many malignant tumors (9). For example, the positive expression of CD44V is closely associated with the poor prognosis of colon and breast cancer (10). Moreover, the deficiency of CD44V has a close relationship with the poor prognosis of bladder cancer (10), whereas CD44V6 is involved in peritoneal metastasis, and high expression of CD44V6 can severely disturb postoperative recovery.
In the present study, we studied the expression of CD44V6 in normal ovarian tissue, benign ovarian tissue, and ovarian cancer. The results provide new experimental findings to assist in the diagnosis and treatment of ovarian cancer.
Patients and methods
Patient data
We recruited 38 patients, admitted to the Hubei Cancer Hospital (Hubei, China), who received gynecological surgery for ovarian cancer from February 2014 to February, 2015. This experimental group had an average age of 45.4 years. We also recruited 23 patients with benign ovarian tumor with an average age of 43.4 years and 20 with normal ovarian tissue with an average age of 44.3 years. The patients with ovarian cancer were diagnosed according to the Ovarian Cancer International Federation of Gynecology and Obstetrics. Exclusion criteria for the study were: Patients with other related gynecological diseases such as cervical and endometrial cancer. This study was approved by the Ethics Committee of Hubei Cancer Hospital. Signed written informed consents were obtained from all participants before the study.
RNA extraction from ovarian cancer cells
Ovarian samples were kept in liquid nitrogen at °C until needed, then 0.1 g, was placed on ice, to which 0.6 ml of RNA Plus was added (Takara Bio, Dalian, China). Samples were ground quickly in precooled mortar and homogenates were transferred to 1.5 ml tube, washed with 0.3 ml of RNA Plus, and transferred to a centrifuge tube, after which 200 µl of chloroform was added, agitated for 15 sec and incubated on ice for 15 min. We centrifuged samples at 10,000 × g for 15 min at 4°C. The supernatant was transferred to a tube without RNase, and equivalent isopropanol was added, mixed, and incubated on ice for 10 min, then centrifuged again at 1,000 × g for 15 min at 4°C. The supernatant was removed and 750 µl of 75% ethanol was added, with gentle mixing and centrifugation at 10,000 × g for 10 min at 4°C. The supernatant was then removed. RNase-free water was added and the total RNA was measured.
Fluorogenic quantitative PCR
The procedure was conducted according to Takara fluorogenic quantitative PCR instructions, with some modifications (11). The primers for fluorogenic quantitative PCR were synthesized by Suzhou Jinweizhi Biotechnology (Suzhou, China) and the sequences are shown in Table I.
Table I.
Fluorogenic quantitative PCR primers.
| Primers | Sequences |
|---|---|
| CD44V6-F | GTCGATGCTAGCTAGCCGTAGCATG |
| CD44V6-R | CGAGCTAGTCGTAGTCGATCGATCG |
| GAPDH-F | GTCGATGGCTAGTCGTAGCATCGAT |
| GAPDH-R | TGCTAGCTGGCATGCCCGATCGATC |
F, forward; R, reverse.
ELISA
We conducted enzyme-linked immunosorbent assay (ELISA) following the instructions of the ELISA kit (Takara Company) with minor modifications (12). The standard protein sample for ELISA was diluted with assay buffer at 1:50 to create standard curve. After dilution with phosphate-buffered saline (PBS) (pH 7.2) at 1:100, the samples were mixed with 100 µl of sample solution and 50 µl of detection solution, and incubated for 2 h at room temperature. Then, TMB chromogenic substrate was added and light absorption was measured at 495 nm. CD44V6 concentration was calculated according to the standard curve. The primary mouse monoclonal CD44V6 antibody (dilution, 1:500; cat. no. ab78960) and secondary goat anti-rabbit (HRP) IgG antibody (dilution, 1:2000; cat. no. ab6721) were from Abcam (Cambridge, MA, USA).
Western blot analysis
Total protein in the sample was extracted with the protein extraction kit from Roche according to the manufacturer's recommendations. Then, we proceeded with standard western blot analysis procedures.
Immunohistochemistry
Immunohistochemistry SP-linking method was applied in this study. Normal ovarian tissue, benign ovarian tumor, and ovarian cancer samples were fixed in 10% formaldehyde, embedded in paraffin, and sectioned at 4 µm. The sections were fixed on a glass slide and placed in an oven for 2 h at 60°C. Then, we de-paraffinized with xylene, re-dehydrated in ethyl alcohol gradients, and hydrated by washing in PBS 5 times, heated and boiled for 2 min in a pressure cooker. After cooling, the sections were placed in PBS for 30 min at room temperature. Then, we added 50 µl of peroxidase blocker, incubated at 37°C for 10 min, and washed in PBS 5 times. PBS solution was removed, and 45 µl of pre-immune serum was added and incubated for 10 min at room temperature. Then, primary antibody was added and incubated at room temperature for 2 h (or overnight at 4°C), and washed 5 times in PBS; 50 µl of streptomycin-HRP was added and incubated for 2 h at room temperature, and washed 5 times in PBS; and 100 µl of coloration solution was added and observed under the microscope. After 10 min, followed washing with distilled water and re-dyeing by hematoxylin for 5 min, washing again, dehydration in ethyl alcohol gradient, air drying, and then the slides were sealed.
Quantification of stained slides
Positive immunohistochemical staining is the accumulation of yellow particles in membrane or cytoplasm, which was judged as positive. The standards for immunohistochemistry showed (9) that membrane stain <10% or negative tumor cells after stained were determined as negative. Only the stained membrane or tumor cell with membrane stain >10% was identified as positive. The results were judged through the KI index, which was the numbers of positive cells in each field.
Statistical analysis
SPSS 20.0 software (Chicago, IL, USA) was used to process and analyze the data. CD44V6 protein expression in different tissue samples was detected by χ2. The result of immunohistochemistry KI index was tested by t-test.
Results
CD44V6 mRNA expression
To determine the role of CD44V6 in ovarian cancer, we examined CD44V6 mRNA levels in ovarian cancer, and benign and normal ovaries. Compared with normal ovaries, MEG-A3 mRNA content in benign ovarian tumor and ovarian cancer were higher. CD44V6 mRNA expression in benign ovarian tumor was 4.5 times higher than in normal ovarian tissues. Moreover, the expression of CD44V6 mRNA in ovarian cancer was 13.6-fold higher than that in the benign ovarian tumor. These results demonstrated that CD44V6 mRNA expression strongly associates with ovarian tumors, with the highest values identifying ovarian cancer (Fig. 1).
Figure 1.
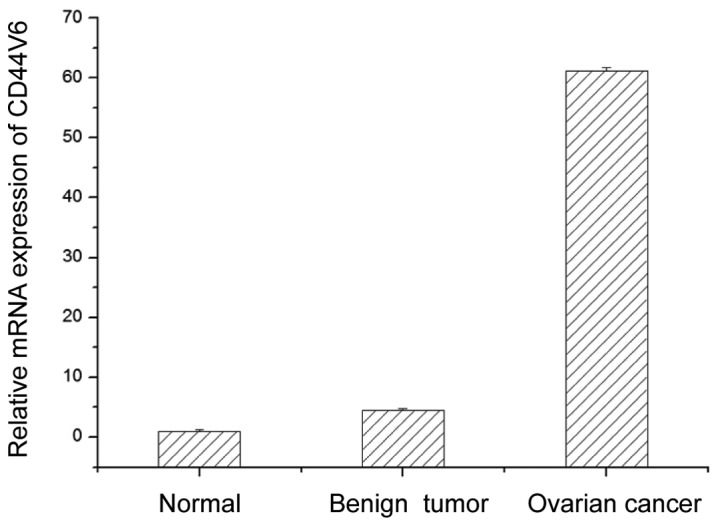
CD44V6 mRNA expression in normal ovarian tissue, benign ovarian tumor and ovarian cancer.
CD44V6 protein expression by western blot analysis
After identifying strong upregulation of CD44V6 at the mRNA level, we examined the expression of CD44V6 protein by western blot analysis in normal ovarian tissue, benign ovarian tumor, and ovarian cancer. We found no significant difference in the expression of CD44V6 protein between normal ovarian tissue and benign ovarian tumors (Fig. 2). However, CD44V6 protein content in ovarian cancer tissue was significantly higher compared with normal ovarian tissue and benign ovarian tumors (Fig. 2), which is consistent with the upregulation of CD44V6 mRNA.
Figure 2.
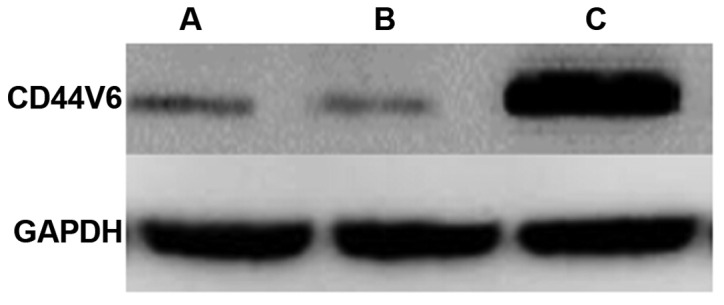
CD44V6 expression of in normal ovarian tissue, benign ovarian tumor and ovarian cancer. Lane A, normal ovarian tissue; lane B, benign ovarian tumor; lane C, ovarian cancer.
CD44V6 protein expression by ELISA
After identifying strong upregulation of CD44V6 protein in ovarian cancer by western blot analysis, we verified this result using a more quantitative technique. We found no significant difference in CD44V6 protein expression between normal ovarian tissue and benign ovarian tumors (Table II). However, CD44V6 expression was significantly higher in ovarian cancer tissue than in normal ovaries and in benign ovarian tumors (Table II). This further supports the association of CD44V6 protein with ovarian cancer.
Table II.
CD44V6 protein expression in normal ovarian tissue, benign ovarian tumor and ovarian cancer.
| Groups | CD44V6 protein (µg) | P-value |
|---|---|---|
| Normal ovarian tissue | 2.1±0.13 | |
| Benign ovarian tumor | 2.8±0.18 | >0.05 |
| Ovarian cancer | 16.2±0.21 | <0.05 |
P<0.05, the difference is statistically significant.
CD44V6 immunohistochemistry
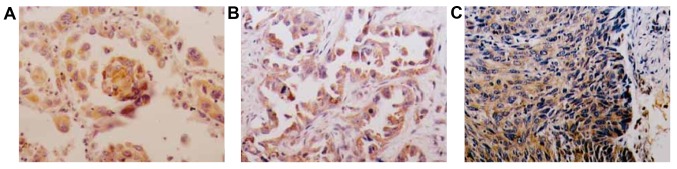
To examine the accumulation and distribution of CD44V6, we conducted immunohistochemistry on normal ovarian tissue, benign ovarian tumor, and ovarian cancer (Table III). We found that CD44V6 was low in normal ovarian tissue and slightly higher in benign ovarian tumor, although the difference was not significant (Fig. 3 and Table III). However, we found a large number of CD44V6-positive cells in ovarian cancer (Fig. 3 and Table III). These results are consistent with the results with ELISA and western blot analysis.
Table III.
CD44V6-positive cells in normal ovarian tissue, benign ovarian tumor, and ovarian cancer.
| Groups | Total no. of cells (n) | CD44V6-positive cells | CD44V6-positive cells | KI value (%) | P-value |
|---|---|---|---|---|---|
| Normal ovarian tissue | 400 | 35 | 365 | 8.75 | |
| Benign ovarian tumor | 400 | 51 | 349 | 12.75 | >0.05 |
| Ovarian cancer | 400 | 328 | 72 | 82.0 | <0.01 |
P<0.05, the difference is statistically significant.
Figure 3.
CD44V6 immunohistochemistry in normal ovarian tissue, benign ovarian tumor and ovarian cancer. (A) Normal ovarian tissue. (B) Benign ovarian tumor. (C) Ovarian cancer.
Discussion
CD44V6 is an adhesive molecule that plays an important role in normal signal transduction and intercellular material exchange (13–15). Overexpression of CD44V is associated with malignant cell changes (16). Overexpression of CD44V can promote the incidence and deterioration of a malignant tumor to some extent (17). CD44V6 can be involved in the process of cell invasion and the transformation of tumor cells (18). Further research showed that CD44V6 can promote the fusion between capillary endothelial cells and tumor cells, which enables tumor cells to avoid the immune system and promote the transformation of tumor cells (19). In recent years, with research on gynecological diseases such as endometrial cancer, it was proposed that CD44V could be involved in the incidence and progression of many gynecological diseases. For instance, CD44V expression in cervical cancer tissue is significantly higher than that of normal tissue (20).
The growing incidence of ovarian cancer has resulted in more studies and relevant molecular and cellular discoveries. In the present study, we found that CD44V6 is highly expressed in ovarian cancer, suggesting that a high expression of CD44V6 can promote the incidence and progression of ovarian cancer. Moreover, we found that CD44V6 mRNA levels are significantly higher in benign ovarian tumor compared to normal ovaries, but there was no difference at the protein level. These results suggest a close connection with the transformation of ovarian tissue, where CD44V6 mRNA is first upregulated in benign tumor and higher upregulation in malignant tumor results in elevated CD44V6. Thus, CD44V6 expression is strongly associated with the tumorous transformation of ovarian cells. Together with research in other cancers, these results suggest that CD44V6 is directly responsible for the transition from normal ovary to benign tumor to cancer. Understanding the role of CD44V6 in ovarian cancer can help in designing tests to improve early diagnosis and treatment of advanced-stage ovarian cancer.
References
- 1.Zhang X, Ng WL, Wang P, Tian L, Werner E, Wang H, Doetsch P, Wang Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012;72:4707–4713. doi: 10.1158/0008-5472.CAN-12-0639. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 2.Ouyang YB, Lu Y, Yue S, Giffard RG. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion. 2012;12:213–219. doi: 10.1016/j.mito.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb PM, Purdie DM, Grover S, Jordan S, Dick ML, Green AC. Symptoms and diagnosis of borderline, early and advanced epithelial ovarian cancer. Gynecol Oncol. 2004;92:232–239. doi: 10.1016/j.ygyno.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Singer CF, Tea MK, Pristauz G, Hubalek M, Rappaport C, Riedl C, Helbich T. Guideline for the prevention and early detection of breast and ovarian cancer in high risk patients, particularly in women from HBOC (hereditary breast and ovarian cancer) families. Wien Klin Wochenschr. 2012;124:334–339. doi: 10.1007/s00508-012-0173-6. (In German) [DOI] [PubMed] [Google Scholar]
- 5.Yu P, Zhou L, Ke W, Li K. Clinical significance of pAKT and CD44v6 overexpression with breast cancer. J Cancer Res Clin Oncol. 2010;136:1283–1292. doi: 10.1007/s00432-010-0779-x. [DOI] [PubMed] [Google Scholar]
- 6.Afify A, Purnell P, Nguyen L. Role of CD44s and CD44v6 on human breast cancer cell adhesion, migration, and invasion. Exp Mol Pathol. 2009;86:95–100. doi: 10.1016/j.yexmp.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Liang J, Jiang D, Griffith J, Yu S, Fan J, Zhao X, Bucala R, Noble PW. CD44 is a negative regulator of acute pulmonary inflammation and lipopolysaccharide-TLR signaling in mouse macrophages. J Immunol. 2007;178:2469–2475. doi: 10.4049/jimmunol.178.4.2469. [DOI] [PubMed] [Google Scholar]
- 8.Matsumura Y, Hanbury D, Smith J, Tarin D. Non-invasive detection of malignancy by identification of unusual CD44 gene activity in exfoliated cancer cells. BMJ. 1994;308:619–624. doi: 10.1136/bmj.308.6929.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Zha XM, Wang R, Li XD, Xu B, Xu YJ, Yin YM. Regulation of CD44 expression by tumor necrosis factor-α and its potential role in breast cancer cell migration. Biomed Pharmacother. 2012;66:144–150. doi: 10.1016/j.biopha.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 10.Nagel S, Hirschmann P, Dirnhofer S, Günthert U, Tzankov A. Coexpression of CD44 variant isoforms and receptor for hyaluronic acid-mediated motility (RHAMM, CD168) is an International Prognostic Index and C-MYC gene status-independent predictor of poor outcome in diffuse large B-cell lymphomas. Exp Hematol. 2010;38:38–45. doi: 10.1016/j.exphem.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Zhou Z, Di W, Li N. Correlation of CD44v6 expression with ovarian cancer progression and recurrence. BMC Cancer. 2013;13:182. doi: 10.1186/1471-2407-13-182. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Zhao S, He JL, Qiu ZX, Chen NY, Luo Z, Chen BJ, Li WM. Prognostic value of CD44 variant exon 6 expression in non-small cell lung cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:6761–6766. doi: 10.7314/APJCP.2014.15.16.6761. [DOI] [PubMed] [Google Scholar]
- 13.Jung T, Gross W, Zöller M. CD44v6 coordinates tumor matrix-triggered motility and apoptosis resistance. J Biol Chem. 2011;286:15862–15874. doi: 10.1074/jbc.M110.208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Xiao L, Luo CH, Zhou H, Zeng L, Zhong J, Tang Y, Zhao XH, Zhao M, Zhang Y. CD44v6 promotes β-catenin and TGF-β expression, inducing aggression in ovarian cancer cells. Mol Med Rep. 2015;11:3505–3510. doi: 10.3892/mmr.2015.3145. [DOI] [PubMed] [Google Scholar]
- 15.Misra S, Heldin P, Hascall VC, Karamanos NK, Skandalis SS, Markwald RR, Ghatak S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heider KH, Kuthan H, Stehle G, Munzert G. CD44v6: a target for antibody-based cancer therapy. Cancer Immunol Immunother. 2004;53:567–579. doi: 10.1007/s00262-003-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marhaba R, Zöller M. CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol. 2004;35:211–231. doi: 10.1023/B:HIJO.0000032354.94213.69. [DOI] [PubMed] [Google Scholar]
- 18.Spiegelberg D, Kuku G, Selvaraju R, Nestor M. Characterization of CD44 variant expression in head and neck squamous cell carcinomas. Tumour Biol. 2014;35:2053–2062. doi: 10.1007/s13277-013-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 20.Rassouli FB, Matin MM, Bahrami AR, Ghaffarzadegan K, Cheshomi H, Lari S, Memar B, Kan MS. Evaluating stem and cancerous biomarkers in CD15+CD44+ KYSE30 cells. Tumour Biol. 2013;34:2909–2920. doi: 10.1007/s13277-013-0853-5. [DOI] [PubMed] [Google Scholar]