Abstract
Although numerous long non-coding RNAs (lncRNAs) have been identified to be important in human cancer, their potential regulatory roles in epithelial tumorigenesis and tumor progression in ovarian cancer remain unclear. The purpose of the present study was to investigate lncRNAs that were differentially expressed (DE) in epithelial ovarian cancer and to explore their potential functions. The lncRNA profiles in five pairs of human epithelial ovarian cancer tissues and their adjacent normal tissues were described using microarrays. The results of the microarray analysis revealed that 672 upregulated and 549 downregulated (fold-change ≥2.0) lncRNAs were DE between the cancerous and normal tissues. Reverse transcription-quantitative polymerase chain reaction was used to validate the microarray results using four upregulated (RP11-1C1.7, XLOC_003286, growth arrest-specific 5 and ZNF295-AS1) and four downregulated (protein tyrosine kinase 7, maternally expressed gene 3, AC079776.2 and ribosomal protein lateral stalk subunit P0 pseudogene 2) lncRNAs. Furthermore, gene ontology and pathway analyses were used to carry out functional analyses of the candidate genes of DE lncRNAs. The results identified lncRNAs with significantly altered expression profiles in human epithelial ovarian cancer cells compared with those in adjacent normal cells. These data offer new insights into the occurrence and development of epithelial ovarian cancer, and these lncRNAs may provide novel molecular biomarkers for further research on epithelial ovarian cancer.
Keywords: epithelial ovarian cancer, long non-coding RNAs, microarray, gene ontology, RT-qPCR
Introduction
Ovarian cancer is the most common cause of mortality from gynecological tumors in women worldwide (1). The 5-year survival rate for patients with advanced ovarian cancer has been reported to be ~30% (2). The incidence of ovarian cancer in Asian countries is considerably lower than that in developed countries, but the difference is reducing (3). In China, the estimated incidence of ovarian cancer during 1999–2010 was 7.91 per 100,000 people (4). Epithelial ovarian cancer accounts for nearly 90% of all ovarian tumors (5). The high mortality of epithelial ovarian cancer is attributed to late-stage diagnosis in >70% of the patients (6). Constant damage and repair of ovarian surface epithelial cells, use of gonadotropin-releasing hormone and steroid hormones, inflammation, genetic factors, and environmental factors have been previously shown to be associated with epithelial ovarian cancer (7–9); however, the exact molecular mechanisms of its occurrence and development remain to be fully identified.
For more than half a century, the concept of gene was limited to the messenger RNA (mRNA) coding region of the genome. With progress in life science research in the post-genome era, numerous studies have demonstrated the involvement of non-coding RNAs (ncRNAs) at various levels in the cell, including transcription, and post-transcriptional regulation of nuclear internal and external signal communication (10). In addition, these RNAs have been demonstrated to be closely associated with the pathological processes of numerous serious diseases (11). Long ncRNAs (lncRNAs) are non-coding RNAs >200 nt in length. Accumulating evidence indicates that lncRNAs serve an important role in various biological processes such as genomic imprinting, transcription activation and inhibition, chromosome recombination, intranuclear transportation, and organ development (12,13). Certain studies have indicated that aberrant regulation of lncRNAs is associated with various types of human cancer (14). Furthermore, lncRNAs are often used as a potential biomarker in the diagnosis and prognosis of tumors (15). Although a few lncRNAs have been implicated in the progression of epithelial ovarian cancer, the functions of the majority of lncRNAs remain to be investigated.
Therefore, the present study used an lncRNA microarray to identify lncRNAs that are differentially expressed (DE) in epithelial ovarian cancer. The microarray results were verified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for specific DE lncRNAs. The present data may provide a molecular basis for understanding the pathogenesis of epithelial ovarian cancer.
Materials and methods
Tissue collection
For tissue collection, five patients with epithelial ovarian cancer were recruited between May and July 2014 at the Department of Gynecology, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University (Nanjing, China). The patients were pathologically confirmed as having epithelial ovarian cancer. Epithelial ovarian cancer tissues and surrounding normal tissues were collected following surgery, snap frozen in liquid nitrogen, and stored at −80°C. Written informed consent was obtained from all patients and the study was approved by the ethics committee of Nanjing Medical University.
RNA extraction
Total RNA was extracted from five pairs of epithelial ovarian cancer and adjacent normal tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol, and quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). The RNA integrity of each sample was assessed using standard denaturing gel electrophoresis, as previously described (16).
Microarray and data analysis
Microarray analysis was performed by Kangchen Biotech Co., Ltd. (Shanghai, China). Arraystar Human LncRNA Microarray V3.0 (Arraystar Inc., Rockville, MD, USA) is designed for the global profiling of human lncRNAs and protein-coding transcripts. This software is capable of detecting ~30,586 lncRNAs and 26,109 coding transcripts (17). Briefly, mRNA was purified from total RNA upon removal of ribosomal RNA using the mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre, Madison, WI, USA). Then, each sample was amplified and transcribed into fluorescent complementary RNA (cRNA) along the entire length of the transcripts without 3′-bias using the Quick Amp Labeling kit, One-Color (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol. The labeled cRNAs were purified using the RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA). The concentration and specific activity of the labeled cRNAs (pmol cyanine 3/µg cRNA) were measured by the NanoDrop ND-1000. First, 1 µg of each labeled cRNA was fragmented by adding 5 µl of 10X blocking agent and 1 µl of 25X fragmentation buffer (both Agilent Technologies, Inc.). The mixture was then heated at 60°C for 30 min, and subsequently, 25 µl of 2X hybridization buffer (GE Healthcare Life Sciences, Little Chalfont, UK) was added to dilute the labeled cRNA. For microarray analysis, 50 µl of the hybridization solution was dispensed into the gasket slide and assembled to the lncRNA expression microarray slide. The slides were incubated for 17 h at 65°C in a Microarray Hybridization Oven (Agilent Technologies, Inc.). The hybridized arrays were washed with Gene Expression Wash Buffer (Agilent Technologies, Inc.) and scanned with using the G2505C Microarray Scanner System (Agilent Technologies, Inc.). Feature Extraction software version 11.0.1.1 (Agilent Technologies, Inc.) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies, Inc.).
Gene ontology (GO) and pathway analyses
GO and pathway analyses were used to determine the roles of DE mRNAs in biological pathways or GO terms. Differentially regulated mRNAs were uploaded into the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov/), which utilized GO terms to identify the molecular function represented in the gene profile. Pathway analysis was carried out based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/).
RT-qPCR validation
Total RNA was reverse transcribed into complementary DNA (cDNA) using the AMV Reverse Transcriptase (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocol. RT-qPCR was performed using an Applied Biosystems 7300 Real-Time PCR Sequence Detection System (Thermo Fisher Scientific, Inc.). RT-qPCR was conducted using 1 µl of cDNA, 12.5 µl of 2X SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.), 10.5 µl of diethyl pyrocarbonate-treated water, and 0.5 µl of 10 µM forward and reverse primers, in a total volume of 25 µl. The following specific primers were used for PCR: RP11-1C1.7 forward, 5′-CTCAGGCTTGGCTCAGACAC-3′ and reverse, 5′-GCAAACAGCCTTGGAGAAGC-3′; XLOC_003286 forward, 5′-AAGGGATCTGGTCTTCAACA-3′ and reverse, 5′-TTCCACCATGTAATGGGTCC-3′; growth arrest specific 5 (GAS5) forward, 5′-TGAAGTCCTAAAGAGCAAGCC-3′ and reverse, 5′-ACCAGGAGCAGAACCATTAAG-3′; ZNF295-AS1 forward, 5′-CCCAGGAGGGAGGTGATACT-3′ and reverse, 5′-TGGGTAGCTTGTGAACCACC-3′; protein tyrosine kinase 7 (PTK7) forward, 5′-GGAAGCCACACTTCACCTAGCAG-3′ and reverse, 5′-CTGCCACAGTGAGCTGGACATGG-3′; maternally expressed gene 3 (MEG3) forward, 5′-GCTCTACTCCGTGGAAGCAC-3′ and reverse, 5′-CAAACCAGGAAGGAGACGAG-3′; AC079776.2, forward, 5′-GCCGATGGTAGAGAAGACCG-3′ and reverse, 5′-GGGGCTCAGAAGCCATCTTT-3′; and ribosomal protein lateral stalk subunit P0 pseudogene 2 (RPLP0P2) forward, 5′-AAAAACGATCAACGAACCTT-3′ and reverse, 5′-AATCGTCTCTGCTTTTCTTG-3′. The PCR conditions were as follows: Denaturation at 95°C for 10 min, followed by 40 cycles of amplification and quantification at 95°C for 15 sec and 60°C for 1 min. GAPDH (forward, 5′-CCGGGAAACTGTGGCGTGATGG-3′ and reverse, 5′-AGGTGGAGGTATGGGTGTCGCTGTT-3′) was used as the internal control. The experiments were performed in triplicate. The relative fold-change was calculated using the 2−ΔΔCq method (18).
Statistical analysis
The lncRNAs and mRNAs that exhibited significantly different expression levels between the two groups were identified through P-value/false discovery rate filtering. DE lncRNAs and mRNAs were identified by fold-change filtering and Student's t test. All data were expressed as means ± standard deviation. Statistical analysis was performed using SPSS 10.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
DE lncRNAs and mRNAs
A total of 1221 lncRNAs were significantly DE between the tumor and control groups (fold-change ≥2.0), among which, 672 were upregulated and 549 were downregulated. Among the DE mRNAs between the two groups, 525 were upregulated and 418 were downregulated. Partial results for the DE lncRNAs and mRNAs are listed in Tables I and II, respectively.
Table I.
Screening of differentially expressed lncRNAs (tumor vs. normal).
| Regulation | lncRNA | Fold-change | Chromosomal localization | RNA length, bp |
|---|---|---|---|---|
| Up | RP5-857K21.3 | 91.6369032 | Chr1 | 437 |
| Up | uc001zjx.1 | 64.7598797 | Chr15 | 641 |
| Up | DQ573539 | 39.8247748 | Chr9 | 1,713 |
| Up | RP11-1C1.7 | 38.8887511 | Chr5 | 483 |
| Up | XLOC_004134 | 25.2266495 | Chr4 | 261 |
| Up | RP11-872J21.3 | 21.4447620 | Chr14 | 1,512 |
| Up | LOC338817 | 18.7987392 | Chr12 | 3,684 |
| Up | CDKN2B-AS1 | 15.7325039 | Chr9 | 1,067 |
| Up | HLA-DRB6 | 15.1244408 | Chr6 | 715 |
| Up | UCA1 | 12.9894370 | Chr19 | 1,413 |
| Up | BX004987.5 | 11.7750069 | Chr1 | 736 |
| Up | FOLH1B | 10.8238534 | Chr11 | 2,163 |
| Up | ZNF295-AS1 | 9.3852619 | Chr21 | 1,073 |
| Up | AK054990 | 9.1453539 | Chr2 | 2,070 |
| Up | AP001615.9 | 8.1669081 | Chr21 | 461 |
| Up | GAS5 | 7.8179616 | Chr1 | 822 |
| Up | LINC00152 | 7.0158480 | Chr2 | 455 |
| Up | XLOC_003286 | 6.5502125 | Chr3 | 409 |
| Up | DPY19L2P2 | 4.4375165 | Chr7 | 3,433 |
| Up | AL833634 | 2.2275523 | Chr11 | 1,885 |
| Down | CTD-2536I1.1 | 58.1029053 | Chr15 | 614 |
| Down | BC071789 | 46.6526362 | Chr3 | 2,730 |
| Down | RP11-548O1.3 | 41.2599738 | Chr3 | 483 |
| Down | MEG3 | 35.0543457 | Chr14 | 1,351 |
| Down | RP11-471J12.1 | 30.7697326 | Chr4 | 892 |
| Down | LEMD1-AS1 | 24.3438594 | Chr1 | 2,781 |
| Down | CLCN6 | 20.5708229 | Chr1 | 5,697 |
| Down | AL132709.5 | 19.7389918 | Chr14 | 644 |
| Down | XLOC_010463 | 17.3764962 | Chr13 | 9,590 |
| Down | CACNA1G-AS1 | 15.5318244 | Chr17 | 1,450 |
| Down | AC079776.2 | 12.6763061 | Chr2 | 400 |
| Down | RP11-998D10.2 | 10.6026574 | Chr14 | 548 |
| Down | LOC253044 | 7.5169687 | Chr15 | 1,735 |
| Down | PVT1 | 4.8097586 | Chr8 | 654 |
| Down | AX747026 | 4.3736710 | Chr1 | 2,133 |
| Down | OPA1-AS1 | 3.4889195 | Chr3 | 513 |
| Down | PTK7 | 3.1639252 | Chr6 | 4,040 |
| Down | RP11-799B12.4 | 2.5604262 | Chr18 | 735 |
| Down | RPLP0P2 | 2.4997850 | Chr11 | 573 |
| Down | HOTAIR | 2.1863176 | Chr12 | 2,370 |
lncRNA, long non-coding RNA; Chr, chromosome.
Table II.
Screening of differentially expressed mRNAs (tumor vs. normal).
| Regulation | mRNA | Fold-change | Chromosomal localization | RNA length, bp |
|---|---|---|---|---|
| Up | GAL | 112.8379148 | Chr11 | 778 |
| Up | LAMC2 | 94.8845376 | Chr1 | 5,623 |
| Up | CCNA1 | 80.0032110 | Chr13 | 1,841 |
| Up | MUC1 | 62.3494142 | Chr1 | 878 |
| Up | WDR69 | 54.5549954 | Chr2 | 1,669 |
| Up | ENKUR | 47.3980040 | Chr10 | 3,382 |
| Up | STOML3 | 32.8593469 | Chr13 | 1,936 |
| Up | KIAA0101 | 27.1783242 | Chr15 | 1,345 |
| Up | CCNB2 | 20.5538621 | Chr15 | 1,566 |
| Up | SLC1A3 | 18.5310330 | Chr5 | 3,670 |
| Up | SAA2 | 16.4134886 | Chr11 | 594 |
| Up | FGF18 | 14.3701010 | Chr5 | 1,999 |
| Up | UBE2C | 12.9501868 | Chr20 | 520 |
| Up | NAA16 | 9.5211755 | Chr13 | 1,833 |
| Up | KCNIP4 | 7.5399784 | Chr4 | 2,371 |
| Up | SLITRK6 | 6.5738521 | Chr13 | 4,199 |
| Up | CEP44 | 4.4520673 | Chr4 | 3,290 |
| Up | C20orf201 | 3.2842057 | Chr20 | 868 |
| Up | DHCR7 | 2.3597491 | Chr11 | 2,665 |
| Up | RNLS | 2.0603828 | Chr10 | 2,420 |
| Down | ITM2A | 110.4209953 | ChrX | 1,719 |
| Down | ZBTB16 | 82.7721198 | Chr11 | 2,417 |
| Down | CPXM1 | 80.2367909 | Chr20 | 2,409 |
| Down | GATA4 | 69.2038646 | Chr8 | 3,419 |
| Down | APOD | 54.0064083 | Chr3 | 1,130 |
| Down | DCN | 48.0233786 | Chr12 | 1,336 |
| Down | GNG11 | 37.6068614 | Chr7 | 964 |
| Down | DHRS2 | 32.5328881 | Chr14 | 1,709 |
| Down | ACADL | 28.4221424 | Chr2 | 2,565 |
| Down | LCE1C | 24.4961395 | Chr1 | 695 |
| Down | MATN2 | 18.1700061 | Chr8 | 4065 |
| Down | LCE2C | 16.5275274 | Chr1 | 614 |
| Down | PPP1R14A | 10.4409979 | Chr19 | 782 |
| Down | OSR2 | 8.4374819 | Chr8 | 1,907 |
| Down | AKT3 | 6.6042407 | Chr1 | 7,091 |
| Down | IL28RA | 5.1905507 | Chr1 | 4,432 |
| Down | PIK3IP1 | 3.6901547 | Chr22 | 2,478 |
| Down | SULF1 | 3.4824364 | Chr8 | 5,716 |
| Down | DCAF4L2 | 2.8399696 | Chr8 | 3,339 |
| Down | MARK3 | 2.6464566 | Chr14 | 3,519 |
mRNA, messenger RNA; Chr, chromosome.
Validation of de lncRNAs
The results of the microarray analysis were confirmed by RT-qPCR of eight randomly selected lncRNAs. GAPDH was used as a normalization control. Of these randomly selected lncRNAs, four (RP11-1C1.7, XLOC_003286, GAS5 and ZNF295-AS1) were upregulated and the other four (PTK7, MEG3, AC079776.2 and RPLP0P2) were downregulated in epithelial ovarian cancer samples compared with their expression levels in adjacent normal tissues of the same individual. As the results of RT-qPCR and microarray analyses are consistent (Fig. 1), these data can be used with confidence in further research.
Figure 1.
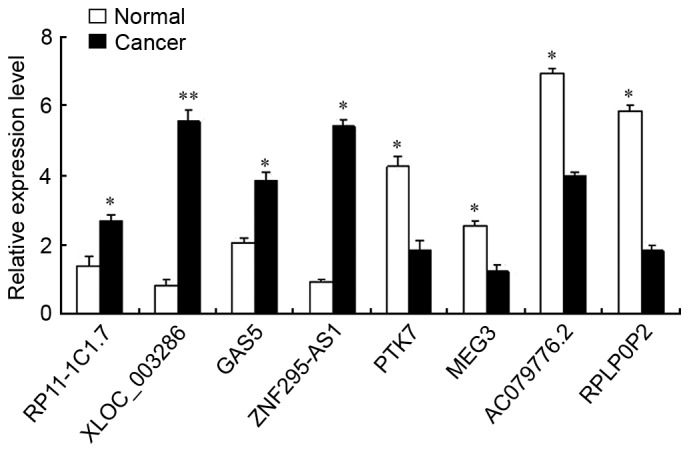
Validation of differentially expressed long non-coding RNAs using reverse transcription-quantitative polymerase chain reaction. *P<0.05, **P<0.01 vs. either normal or cancer.
Pathway analysis
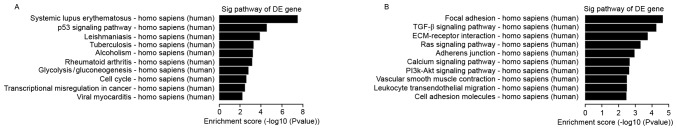
Pathway analysis is a functional method of mapping genes to KEGG pathways (19). Based on the KEGG database (http://www.genome.jp/kegg), KEGG pathway analysis was employed for DE mRNAs. Each P-value denoted the significance of the corresponding pathway, while the EASE Score, Fisher's P-value or hypergeometric P-value denoted the significance of the pathway correlated to the conditions. A low P-value indicated a marked significance of the pathway (P-value cut-off, 0.05). The bar plots in Fig. 2 show the top 10 enrichment scores [-log10 (P-value)] of the significant enrichment pathway. Fig. 2 presents the results of the KEGG pathway analysis for the upregulated and downregulated mRNAs.
Figure 2.
Pathway analysis. The bar plots show the top 10 enrichment scores [-log10 (P-value)] of the most significant enrichment pathways. (A) Upregulation in cancer vs. normal cells. (B) Downregulation in cancer vs. normal cells. TGF, transforming growth factor; ECM, extracellular matrix; PI3k, phosphoinositide 3 kinase; Sig, significant; DE, differentially expressed.
GO analysis
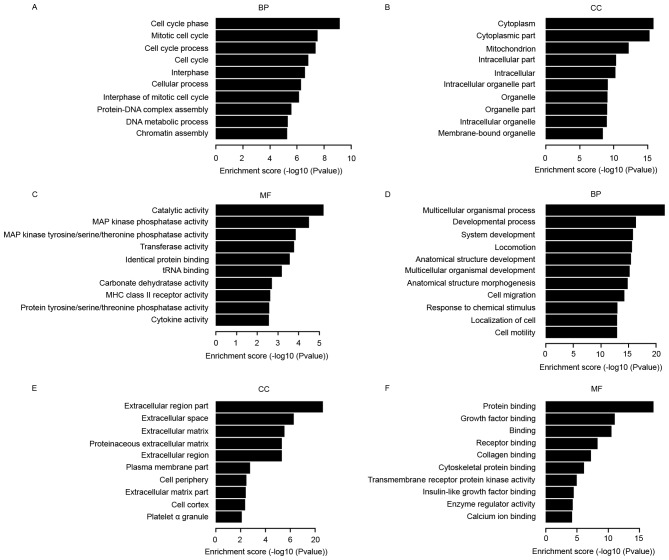
The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). The ontology covers three domains: Biological processes, cellular components and molecular function. Fisher's exact test is used to determine if there are any more overlaps between the DE gene list and the GO annotation list than what is expected by chance. The P-value denotes the significance of enrichment of GO terms in the DE genes. The lower the P-value, the more significant is the GO term (P≤0.05 is recommended) (20). The bar plots in Fig. 3 show the 10 most significant enrichment terms with the most number of DE genes.
Figure 3.
Gene ontology analysis. The bar plots show the 10 most significant enrichment terms with the most number of differentially expressed genes. The ontology covers three domains: BP, CC and MF. (A-C) Upregulation in cancer vs. normal cells. (D-F) Downregulation in cancer vs. normal cells. MAP, mitogen-activated protein; MHC, major histocompatibility complex; BP, biological processes; CC, cellular components; MF, molecular function.
Discussion
As increasing research has focused on the function of lncRNAs in epithelial ovarian cancer, an increasing number of lncRNAs have been identified. For example, Gao et al demonstrated that the lncRNA human ovarian cancer-specific transcript 2 promotes tumor cell migration, invasion and proliferation in epithelial ovarian cancer by modulating microRNA let-7b availability (21). lncRNA H19 expression was inhibited by histone H1.3, which contributes to the suppression of epithelial ovarian carcinogenesis (22). However, the genome-wide expression and the biological functional significance of lncRNAs in epithelial ovarian cancer remain unknown.
In the present study, microarray analysis was used to compare lncRNA expression in epithelial ovarian cancer cells and adjacent normal tissues, and 1221 DE lncRNAs (672 upregulated and 549 downregulated) were identified. These results were further confirmed via RT-qPCR for eight randomly selected lncRNAs.
A previous study has reported that Hox transcript antisense intergenic RNA (HOTAIR) is a 2.2-kb lncRNA located at the HOXC locus (23). It has been reported that suppression of HOTAIR expression in highly metastatic epithelial ovarian cancer cell lines significantly reduced cell invasion, and the HOTAIR expression levels were highly positively correlated with the International Federation of Gynecology and Obstetrics stage (24). The MEG3 gene is located in chromosome 14q32 (25), and is expressed in numerous normal tissues, but its expression level has been reported by various previous studies to be either downregulated or absent in a variety of tumor tissues, including ovarian cancer cells and epithelial ovarian cancer tissues (26–28). In the present study, HOTAIR was upregulated and MEG3 was downregulated in epithelial ovarian cancer vs. normal tissues. These results confirmed that HOTAIR and MEG3 serve a critical role in the occurrence, development and invasion of epithelial ovarian cancer.
GAS5 is encoded at chromosome 1q25, and was originally isolated from NIH-3T3 cells by subtractive hybridization (29). Several recent studies have shown that GAS5 is an lncRNA that functions as a tumor suppressor. For example, Cao et al noticed that patients with cervical cancer with reduced expression of GAS5 have significantly poorer overall survival than those with higher GAS5 expression (30). Shi et al reported that GAS5 expression was downregulated in non-small cell lung cancer tissues compared with that in noncancerous tissues, and was highly associated with tumor size and tumor-node-metastasis stage (31). However, in the present study, it was observed that the expression of GAS5 was upregulated in epithelial ovarian cancer compared with that in adjacent healthy tissues. The majority of scholars agree that glucocorticoids serve an important role in the regulation of ovarian epithelial function, and they are closely associated with the occurrence and development of ovarian cancer (32,33). In another study, glucocorticoids were demonstrated to significantly inhibit the proliferation of human ovarian cancer cells (34). Therefore, it can be hypothesized that, as a glucocorticoid receptor response element (GRE) analogue, GAS5 may be able to inhibit glucocorticoid production by competing with GRE to associate with the DNA-binding domain of the glucocorticoid receptor (35).
To understand the function of the targets of DE lncRNAs, GO terms and KEGG pathway annotation were applied in the present study to the target gene pool. The GO analysis revealed that the DE genes were associated with mitogen-activated protein kinase phosphatase activity, major histocompatibility complex class II receptor activity and DNA metabolic processes, which is consistent with previous research (36–38). Previous studies have demonstrated that signaling pathways, including the Ras, p53 and transforming growth factor-β signaling pathways, serve a critical role in the regulation of pathophysiological processes in ovarian cancer (39–41). In addition to these signaling pathways, the present study also demonstrated that focal adhesion, extracellular matrix-receptor interaction, cell adhesion molecules, cell cycle, transcriptional misregulation in cancer and other signaling pathways were involved in the pathogenesis of epithelial ovarian cancer.
In summary, the present study identified lncRNAs that were aberrantly expressed in epithelial ovarian cancer compared with their expression in matched normal tissue. Further studies are required to reveal the possible biological functions and mechanism of these lncRNAs.
Acknowledgements
The present study was supported by the Science and Technology Development Foundation of Nanjing Medical University (grant no. 2014NJMUZD050). The authors thank Kangchen Biotech Co., Ltd. (Shanghai, China) for their technical assistance.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Lowe KA, Chia VM, Taylor A, O'Malley C, Kelsh M, Mohamed M, Mowat FS, Goff B. An international assessment of ovarian cancer incidence and mortality. Gynecol Oncol. 2013;130:107–114. doi: 10.1016/j.ygyno.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Chiang YC, Chen CA, Chiang CJ, Hsu TH, Lin MC, You SL, Cheng WF, Lai MS. Trends in incidence and survival outcome of epithelial ovarian cancer: 30-year national population-based registry in Taiwan. J Gynecol Oncol. 2013;24:342–351. doi: 10.3802/jgo.2013.24.4.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Liu SZ, Zheng RS, Zhang F, Chen WQ, Sun XB. Time Trends of Ovarian Cancer Incidence in China. Asian Pac J Cancer Prev. 2014;15:191–193. doi: 10.7314/APJCP.2014.15.1.191. [DOI] [PubMed] [Google Scholar]
- 5.Lawrie TA, Bryant A, Cameron A, Gray E, Morrison J. Pegylated liposomal doxorubicin for relapsed epithelial ovarian cancer. Cochrane Database Syst Rev: CD006910. 2013 doi: 10.1002/14651858.CD010482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadducci A, Cosio S, Zola P, Landoni F, Maggino T, Sartori E. Surveillance procedures for patients treated for epithelial ovarian cancer: A review of the literature. Int J Gynecol Cancer. 2007;17:21–31. doi: 10.1111/j.1525-1438.2007.00826.x. [DOI] [PubMed] [Google Scholar]
- 7.Adams SV, Quraishi SM, Shafer MM, Passarelli MN, Freney EP, Chlebowski RT, Luo J, Meliker JR, Mu L, Neuhouser ML, Newcomb PA. Dietary cadmium exposure and risk of breast, endometrial and ovarian cancer in the Women's Health Initiative. Environ Health Perspect. 2014;122:594–600. doi: 10.1289/ehp.1307054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schock H, Surcel HM, Zeleniuch-Jacquotte A, Grankvist K, Lakso HÅ, Fortner RT, Kaaks R, Pukkala E, Lehtinen M, Toniolo P, Lundin E. Early pregnancy sex steroids and maternal risk of epithelial ovarian cancer. Endocr Relat Cancer. 2014;21:831–844. doi: 10.1530/ERC-14-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haruta S, Furukawa N, Yoshizawa Y, Tsunemi T, Nagai A, Kawaguchi R, Tanase Y, Yoshida S, Kobayashi H. Molecular genetics and epidemiology of epithelial ovarian cancer. Oncol Rep. 2011;26:1347–1356. doi: 10.3892/or.2011.1456. [DOI] [PubMed] [Google Scholar]
- 10.Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: Regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 12.Hung T, Chang HY. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: Where do we go from here? Int J Mol Sci. 2015;16:1395–1405. doi: 10.3390/ijms16011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–357. doi: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 16.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Long W, Li Q, Zhou Q, Wang Y, Wang H, Zhou B, Li J. Distinct expression profiles of lncRNAs between regressive and mature scars. Cell Physiol Biochem. 2015;35:663–675. doi: 10.1159/000369727. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M. Molecular network analysis of diseases and drugs in KEGG. Methods Mol Biol. 2013;939:263–275. doi: 10.1007/978-1-62703-107-3_17. [DOI] [PubMed] [Google Scholar]
- 20.Gu S, Li G, Zhang X, Yan J, Gao J, An X, Liu Y, Su P. Aberrant expression of long noncoding RNAs in chronic thromboembolic pulmonary hypertension. Mol Med Rep. 2015;11:2631–2643. doi: 10.3892/mmr.2014.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, Liu Y, Ou J, Wang D, Yao L, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 22.Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, McDonald JF, Bouhassira EE, Fan Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 27.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng X, Li J, Yang L, Chen Z, Zhao Q, Tan L, Zhou Y, Li J. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. 2014;32:277–285. doi: 10.3892/or.2014.3208. [DOI] [PubMed] [Google Scholar]
- 29.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 30.Cao S, Liu W, Li F, Zhao W, Qin C. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol. 2014;7:6776–6783. [PMC free article] [PubMed] [Google Scholar]
- 31.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54:E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 32.Melhem A, Yamada SD, Fleming GF, Delgado B, Brickley DR, Wu W, Kocherginsky M, Conzen SD. Administration of glucocorticoids to ovarian cancer patients is associated with expression of the anti-apoptotic genes SGK1 and MKP1/DUSP1 in ovarian tissues. Clin Cancer Res. 2009;15:3196–3204. doi: 10.1158/1078-0432.CCR-08-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson RE, Fegan KS, Ren X, Hillier SG, Duncan WC. Glucocorticoid regulation of SLIT/ROBO tumour suppressor genes in the ovarian surface epithelium and ovarian cancer cells. PLoS One. 2011;6:e27792. doi: 10.1371/journal.pone.0027792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu MJ, Fang GE, Liu YJ, Song LN. Effects of glucocorticoid on proliferation, differentiation and glucocorticoid receptor expression in human ovarian carcinoma cell line 3AO. Acta Pharmacol Sin. 2002;23:819–823. [PubMed] [Google Scholar]
- 35.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beauchamp MC, Yasmeen A, Knafo A, Gotlieb WH. Targeting insulin and insulin-like growth factor pathways in epithelial ovarian cancer. J Oncol. 2010;2010:257058. doi: 10.1155/2010/257058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dadmarz RD, Ordoubadi A, Mixon A, Thompson CO, Barracchini KC, Hijazi YM, Steller MA, Rosenberg SA, Schwartzentruber DJ. Tumor-infiltrating lymphocytes from human ovarian cancer patients recognize autologous tumor in an MHC class II-restricted fashion. Cancer J Sci Am. 1996;2:263–272. [PubMed] [Google Scholar]
- 38.Beesley J, Jordan SJ, Spurdle AB, Song H, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, McGuire V, Whittemore AS, et al. Association between single-nucleotide polymorphisms in hormone metabolism and DNA repair genes and epithelial ovarian cancer: results from two Australian studies and an additional validation set. Cancer Epidemiol Biomarkers Prev. 2007;16:2557–2565. doi: 10.1158/1055-9965.EPI-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mei FC, Young TW, Liu J, Cheng X. RAS-mediated epigenetic inactivation of OPCML in oncogenic transformation of human ovarian surface epithelial cells. FASEB J. 2006;20:497–499. doi: 10.1096/fj.05-4586fje. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Zhang Y, Gao Y, Cui Y, Liu H, Li M, Tian Y. Downregulation of HNF1 homeobox B is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;32:979–988. doi: 10.3892/or.2014.3297. [DOI] [PubMed] [Google Scholar]
- 41.Ji M, Shi H, Xie Y, Zhao Z, Li S, Chang C, Cheng X, Li Y. Ubiquitin specific protease 22 promotes cell proliferation and tumor growth of epithelial ovarian cancer through synergy with transforming growth factor β1. Oncol Rep. 2015;33:133–140. doi: 10.3892/or.2014.3580. [DOI] [PubMed] [Google Scholar]