Abstract
Prevalent mutations in the mitogen-activated protein kinase 1 (MAPK1)/extracellular signal-regulated kinase 2 (ERK2) pathway have been identified in cervical squamous cell carcinoma in a large-scale genome sequencing effort. Furthermore, mutations in the rat sarcoma viral oncogene homolog (RAS)/Raf/Mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway have also been revealed to have important roles in the pathogenesis of human cancer. However, whether the potential hotspot mutations in ERK2 and other components of the RAS/RAF/MEK/ERK signaling pathway also exist in Chinese patients with cervical carcinoma remains to be elucidated. In the present study, a total of 260 patients with cervical carcinoma of distinct subtypes were analyzed for the presence of potential hotspot mutations in the RAS/RAF/MEK/ERK signaling pathway. No ERK2 mutations were detected in these samples; however, Kirsten RAS (KRAS) p.G12D (c.35G>A) mutation was identified in 2/26 (7.7%) cervical adenocarcinoma cases, including 1/20 cervical mucinous adenocarcinoma and 1/6 cervical endometrioid carcinoma cases. In addition, no mutations in the ERK1, neuroblastoma RAS, Harvey RAS or B-Raf proto-oncogene serine/threonine kinase genes were detected in the present study. These results indicated that ethnic differences may be a primary reason for the discrepancy in ERK2 mutation frequencies between the current study and previous studies. Furthermore, mutation in the KRAS gene, but not other genes in the RAS/RAF/MEK/ERK signaling pathway, may have an active role in the pathogenesis of cervical carcinoma.
Keywords: mitogen-activated protein kinase 1, rat sarcoma viral oncogene homolog/Raf/mitogen-activated protein kinase activated kinase/mitogen-activated protein kinase 1 signaling pathway, mutation, cervical carcinoma
Introduction
Cervical carcinoma is the second most frequent type of gynecological malignancy (1). Despite the availability of Pap smear-based screening decreasing the incidence and mortality of cervical carcinoma, it remains a primary cause of cancer-associated mortality in females globally (2). It is well established that human papillomavirus infection has an important role in the development of cervical carcinoma, in addition to genetic alterations that have also been demonstrated to be required for the initiation and progression of this malignancy (3). Although there have been developments in the understanding of the molecular etiology of cervical carcinoma, the underlying mechanistic details remain to be elucidated, emphasizing the requirement for identifying novel molecular genetic alterations.
The RAS/RAF/MEK/ERK cascade is an important signaling pathway that regulates diverse cellular functions, including cell proliferation, survival, differentiation and migration (4). A number of previous studies have demonstrated that mutations in the kinases of the RAS/RAF/MEK/ERK transduction pathway are frequently observed in human cancer, including cervical carcinoma (5–8). Of these, V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS p.G12, p.G13 and p.Q61), neuroblastoma RAS (NRAS; p.G12, p.G13 and p.Q61), Harvey RAS (HRAS; p.G12, p.G13 and p.Q61) and B-Raf proto-oncogene serine/threonine kinase (BRAF; p.V600E) mutations were among the most typically observed and were considered to be hotspot mutations in these genes (6,9,10).
Furthermore, a large-scale exome- and RNA-sequencing study has identified frequent ERK2 mutations (7.6%, 6/79) in cervical squamous cell carcinoma; the hotspot mutation is ERK2 p.E322K (11). By contrast, no ERK2 mutations were detected in 24 cases of cervical adenocarcinoma (11). As homologous residues of paralogous genes are mutated frequently in human cancer, it was investigated whether cervical carcinoma also contained mutations in the ERK1 p.E339 residue, the homologous residue of ERK2 p.E322 that is often mutated in cervical squamous cell carcinoma (12,13).
Presently, the detailed mutational status of the RAS/RAF/MEK/ERK signaling pathway in Chinese patients with cervical carcinoma remains to be investigated. The current study recruited and analyzed tumor tissue samples from a cohort of 260 Chinese patients with distinct subtypes of cervical carcinoma for the presence of the aforementioned potential mutations in the KRAS (p.G12, p.G13 and p.Q61), NRAS (p.G12, p.G13 and p.Q61), HRAS (p.G12, p.G13 and p.Q61), BRAF (p.V600E), ERK2 (p.E322) and ERK1 (p.E339) genes in the RAS/RAF/MEK/ERK signaling pathway.
Materials and methods
Ethics statement
The present study was approved by the Institutional Review Board of Jiangxi Provincial Maternal and Child Health Hospital (Nanchang, China), and informed consent was obtained from each patient prior to analysis. The current study was conducted according to the Declaration of Helsinki.
Patients and tissue samples
A set of 456 available formalin-fixed, paraffin-embedded (FFPE) cervical cancer tissues were prepared from samples obtained via surgical removal from July 2008 to August 2013, stored at room temperature, were obtained from the archives of the Department of Pathology at Jiangxi Provincial Maternal and Child Health Hospital. All of these samples were reviewed by two experienced pathologists who were blinded to the patient characteristics. Only those >40% of cancerous cells, as determined via staining with 0.5% haematoxylin for 5 min, followed by 1% eosin for 10 min and then analyzed with a light microscope under magnification, ×20-x200 (BX53T-32P01; Olympus Corporation, Tokyo, Japan) after dehydration, were included in the present study. According to these criteria, 260/456 tissue samples were further selected for the present study, including 207 cases of squamous cell carcinoma, 27 of adenosquamous carcinoma and 26 of adenocarcinoma. For the 26 patients with adenocarcinoma, 20 were mucinous and 6 were endometrioid (Table I).
Table I.
Mutation frequencies of the genes in the RAS/RAF/MEK/ERK signaling pathway in 260 Chinese patients with distinct subtypes of cervical carcinoma.
| Gene and mutation codon | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | No. | KRAS p.G12 | KRAS p.G13 | KRAS p.Q61 | NRAS p.G12 | NRAS p.G13 | NRAS p.Q61 | HRAS p.G12 | HRAS p.G13 | HRAS p.Q61 | BRAF p.V600 | ERK1 p.E339 | ERK2 p.E322 |
| Squamous cell | 207 | 0/207 | 0/207 | 0/207 | 0/207 | 0/207 | 0/205 | 0/206 | 0/206 | 0/204 | 0/204 | 0/203 | 0/201 |
| Adenosquamous | 27 | 0/27 | 0/27 | 0/27 | 0/27 | 0/27 | 0/27 | 0/27 | 0/27 | 0/26 | 0/27 | 0/25 | 0/26 |
| Adenocarcinoma | 26 | 2/26 | 0/26 | 0/26 | 0/26 | 0/26 | 0/26 | 0/26 | 0/26 | 0/25 | 0/26 | 0/25 | 0/25 |
| Mucinous | 20 | 1/20 | 0/20 | 0/20 | 0/20 | 0/20 | 0/20 | 0/20 | 0/20 | 0/19 | 0/20 | 0/19 | 0/19 |
| Endometrioid | 6 | 1/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
RAS, rat sarcoma viral oncogene homolog; MEK, mitogen-activated protein kinase activated kinase; ERK, mitogen-activated protein kinase; KRAS, Kirsten RAS; NRAS, neuroblastoma RAS; HRAS, Harvey RAS; BRAF, B-Raf proto-oncogene serine/threonine kinase; bp, base pairs.
DNA isolation and polymerase chain reaction (PCR) amplification
The genomic DNA of FFPE tissue samples was isolated using a commercially available DNA isolation kit (cat. no., R6954-02, Omega Bio-Tek, Inc., Norcross, GA, USA). The genomic regions spanning the potential mutation hotspots in the KRAS, NRAS, HRAS, BRAF, ERK1 and ERK2 genes were amplified using PCR with their respective primer pairs (Table II). For the PCR reaction, a total of 300 ng genomic DNA was used as a template for the nine amplicons in a final volume of 25 µl, amplified with LA Taq DNA Polymerase (Takara Biotechnology Co., Ltd, Dalian, China). The PCR was run with the following program: 94°C for 3 min, 35 cycles of 94°C for 30 sec, 52–60°C for 30 sec and 72°C for 20 sec, with a final 72°C extension for 10 min (Table II). All PCR reactions were performed in a Thermal Cycler 2720 (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Table II.
Primers for the mutational analysis of the genes in the RAS/RAF/MEK/ERK signaling pathway.
| Gene | Potential mutation codon | Amplicon Bp | Sample detected n=260 | Annealing °C | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|---|---|
| KRAS | p.G12-G13 | 162 | 260 | 52 | CTGCTGAAAATGACTGAATA | ATGGTCCTGCACCAGTAATA |
| KRAS | p.Q61 | 132 | 260 | 52 | CCAGACTGTGTTTCTCCCTTC | AAAGAAAGCCCTCCCCAGT |
| NRAS | p.G12-G13 | 118 | 260 | 52 | GACTGAGTACAAACTGGTGGTGG | GGGCCTCACCTCTATGGTG |
| NRAS | p.Q61 | 201 | 258 | 52 | CAAGTGGTTATAGATGGTGAAACC | AAGATCATCCTTTCAGAGAAAATAAT |
| HRAS | p.G12-G13 | 139 | 259 | 52 | CAGGAGACCCTGTAGGAGGA | TCGTCCACAAAATGGTTCTG |
| HRAS | p.Q61 | 194 | 255 | 52 | TCCTGCAGGATTCCTACCGG | GGTTCACCTGTACTGGTGGA |
| BRAF | p.V600 | 228 | 257 | 52 | TGCTTGCTCTGATAGGAAAATG | AGCATCTCAGGGCCAAAAAT |
| ERK1 | p.E339 | 246 | 253 | 60 | CTGCTCTCACTACTGCAAAACC | TGGCAGCAGGTATATCTCAGG |
| ERK2 | p.E322 | 231 | 252 | 55 | CTGACTCCTGCCCTTCCATA | GGGTGGTAGAGACAGCAAGG |
RAS, rat sarcoma viral oncogene homolog; MEK, mitogen-activated protein kinase activated kinase; ERK, mitogen-activated protein kinase; KRAS, Kirsten RAS; NRAS, neuroblastoma RAS; HRAS, Harvey RAS; BRAF, B-Raf proto-oncogene serine/threonine kinase; bp, base pairs.
Protein sequence homology analysis
Human ERK2 (NP_002736) and ERK1 (NP_002737) protein sequences were obtained from GenBank database (www.ncbi.nlm.nih.gov/genbank/), and the DNASTAR® Lasergene version 7.2 software (DNASTAR, Inc., Madison, WI, and USA) was used to analyze the protein sequence homology.
Direct sequencing of the RAS/RAF/MEK/ERK pathway mutations
The PCR products were subsequently purified using a TIANquick Midi Purification kit (cat. no., DP204-02, Tiangen Biotech Co., Ltd., Beijing, China), according to the manufacturer's protocol. Subsequently, the purified PCR products were analyzed by Sanger sequencing with an ABI Prism 3730 DNA sequencer (Applied Biosystems; Thermo Fisher Scientific, Inc.) as described previously (14). All identified mutations were confirmed with an independent PCR and bidirectional sequencing; the potential somatic mutations were confirmed by sequencing of the paired adjacent control tissues.
Results
Absence of ERK2 and ERK1 mutation
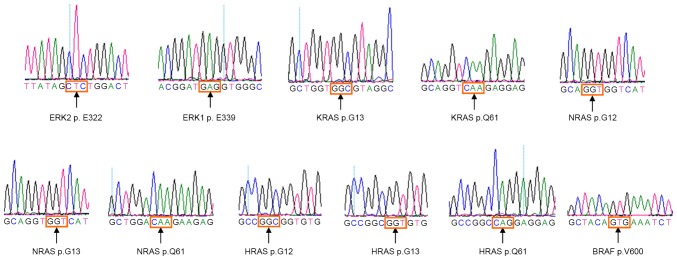
The results of sequence homology analysis demonstrated that ERK2 p.E322 and ERK1 p.E339 residues were the homolog residues (Fig. 1). The 260 patients involved included 207 cases of squamous cell carcinoma with a median age of 44 years (range, 27–67), 27 of adenosquamous carcinoma with a median age of 41 years (range, 23–58) and 26 of adenocarcinoma with a median age of 43 years (range, 31–59). There were no ERK2 p.E322 mutations detected in the 260 tumor samples, nor any ERK1 p.E339 mutations in these tissue samples (Table I; Fig. 2).
Figure 1.
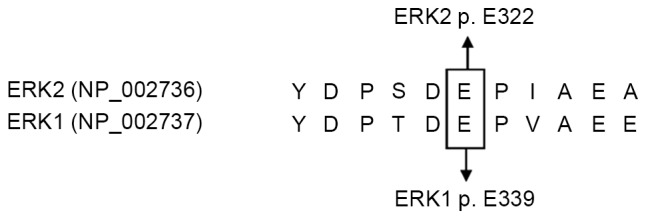
Protein sequence homology analysis of ERK2 and ERK1. Human ERK2 (NP_002736) and ERK1 (NP_002737) protein sequences were obtained from the GenBank database, and the amino acid sequences were aligned with DNASTAR® Lasergene version 7.2 software, according to the software manuals. The 317th-327th amino acids of ERK2 and 334th-344th amino acids from ERK1 are displayed here. ERK, mitogen-activated protein kinase.
Figure 2.
Representative sequencing electropherograms of the potential mutation site in ERK2 p.E322, ERK1 p.E339, KRAS (p.G13 and p.Q61), NRAS (p.G12, p.G13 and p.Q61), HRAS (p.G12, p.G13 and p.Q61) and BRAF (p.V600E); the arrows refer to the wild type sequence with potential hotspot mutations. ERK, mitogen-activated protein kinase; KRAS, Kirsten rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS; HRAS, Harvey RAS; BRAF, B-Raf proto-oncogene serine/threonine kinase.
Mutations in other genes of the RAS/RAF/MEK/ERK pathway
The 260 patients were also screened for potential hotspot mutations in the KRAS, NRAS, HRAS and BRAF genes in the RAS/RAF/MEK/ERK pathway (Table II). In total, the KRAS p.G12D (c.35G>A) mutation was identified in 2/26 (7.7%) cervical adenocarcinoma cases, including 1/20 (5.0%) cases of cervical mucinous carcinoma (CCC-199) and 1/6 (16.7%) patients with cervical endometrioid carcinoma (CCC-190; Table I; Fig. 3), who were 43 and 31 years, respectively; however, these mutations were not detected in the remaining tissue samples. Additionally, no mutations were detected in the KRAS (p.G13 and p.Q61), NRAS (p.G12, p.G13 and p.Q61), HRAS (p.G12, p.G13 and p.Q61) and BRAF (p.V600E) genes in the remaining tissue samples (Table I; Fig. 2).
Figure 3.
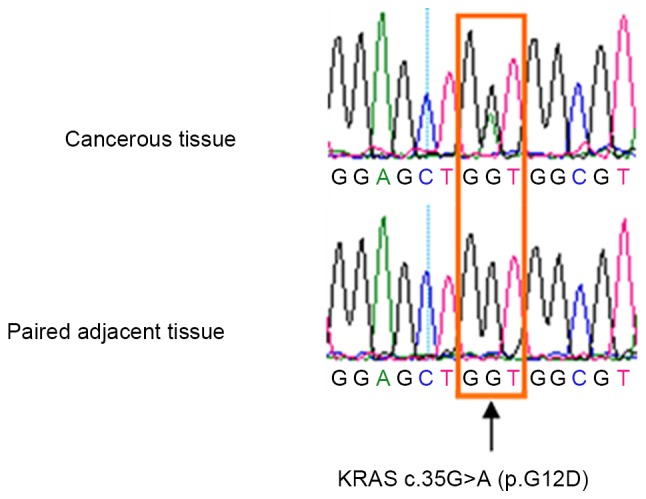
Representative sequencing electropherogram of KRAS p.G12D (c.35G>A) mutation; the arrow refers to the location of the mutation. KRAS, Kirsten rat sarcoma viral oncogene homolog.
Discussion
Using whole-exome sequencing and validation analyses, Ojesina et al (11) reported existing ERK2 p E322K mutations (5.1%, 4/79) in cervical squamous cell carcinoma. However, the results of the current study were contrary to those of Ojesina et al (11) as there were no ERK2 p.E322 mutations detected in this cohort of 260 Chinese patients with distinct subtypes of cervical carcinoma (11).
The discrepancy in the mutation frequency of ERK2 between the present and previous study (11) requires consideration. Regarding the methods of this mutation screen, sensitive Sanger sequencing was adopted, whilst the previous study (11) detected the potential mutations using the more sensitive exome- and RNA-sequencing techniques; therefore, it is possible the current study may have missed potential mutations. However, it was noted that each tissue sample analyzed in the present study contained >40% of tumor cells; this is beyond the detection sensitivity of Sanger sequencing, which has a detection threshold of 6.6–20% of the mutated allele in a background of the wild-type allele (15–19). Furthermore, a high frequency of KRAS mutations were detected in the cervical adenocarcinoma tissues using Sanger sequencing, which is comparable to that observed by numerous previous studies wherein the analyzed patient tissue samples contained high proportions of cancerous cells (≥40% malignant cells), comparable to the proportion of malignant cells in the present study (11,20,21). Therefore, a possible explanation for the discrepancy in mutation frequencies is the difference in ethnic background, as the prior study (11) focused on European females whilst the current sample cohort was Chinese in origin, suggesting that the ERK2 mutations may be specific to certain populations.
In addition, neither the 27 cases of adenosquamous carcinoma, nor the 26 adenocarcinoma cases, were identified to have ERK2 mutations; these results are concordant with those of previous studies that demonstrated that ERK2 mutations occurred only in squamous cell carcinoma and not in adenocarcinoma (11,22).
A number of previous studies have demonstrated that the homologous residues of paralogous genes are frequently mutated in human cancer (12,13). However, the current study did not detect any mutations in ERK1 p.E339, the homologous residue of ERK2 p.E322, in the 260 tissue samples of distinct subtypes of cervical carcinoma, implicating that ERK1 p.E339 mutations may not be actively involved in the pathogenesis of these diseases.
Considering that numerous genes in the RAS/RAF/MEK/ERK signaling pathway are frequently mutated in human cancer (23,24), the current study screened cervical carcinoma samples for the potential hotspot mutations in the KRAS, NRAS, HRAS and BRAF genes. Amongst these, a KRAS p.G12D mutation was identified at a high frequency (7.7%, 2/26) in cervical adenocarcinoma, but was absent in the 207 cases of squamous cell carcinoma and in the 27 of adenosquamous carcinoma. The frequency of the KRAS mutation observed in the tissue samples analyzed in the present study was similar to that identified in numerous previous studies, where the frequency of the KRAS mutation ranged from 8.0–17.5% in cervical adenocarcinoma, to absent or rare in cervical squamous cell carcinoma (11,20,21). Taken together, these results further support the hypothesis that KRAS mutations are frequent and may be a driving factor for the development of cervical adenocarcinoma (20,21), but not for squamous cell carcinoma or adenosquamous carcinoma.
In addition, previous studies identified that the frequencies of NRAS, HRAS and BRAF hotspot mutations were absent or low in cervical squamous cell carcinoma or adenosquamous carcinoma (11,21,25,26). Similarly, the current study also did not identify the potential hotspot mutations in these genes in the 260 cases of cervical carcinoma with distinct subtypes. However, due to the limited sample sized analyzed here, it is hard to draw a definite conclusion that whether the mutations in NRAS, HRAS and BRAF would possess a role in the pathogenesis of cervical squamous cell carcinoma or adenosquamous carcinoma.
In conclusion, ERK2 hotspot mutations were not detected in the cervical squamous cell carcinoma cases of the present study cohort, and the ethnic variation compared with previous studies may be one of the primary reasons. In addition, mutations in KRAS, but not NRAS, HRAS, BRAF or ERK1, in the RAS/RAF/MEK/ERK signaling pathway were detected in cervical adenocarcinoma tissues with a higher frequency, indicating that the KRAS p.G12D mutation may serve an active role in the pathogenesis of cervical adenocarcinoma.
Acknowledgements
The authors would like to thank all participants involved in the current study. The present study was supported by grants from the Natural Science Foundation of Jiangxi Province (grant no. 20142BAB215003, 20161ACB21021) and the Sailing Project of Jiangxi Province (for ZY).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Luyten A, Scherbring S, Reinecke-Lüthge A, Braun BE, Pietralla M, Theiler K, Petry KU. Risk-adapted primary HPV cervical cancer screening project in Wolfsburg, Germany-experience over 3 years. J Clin Virol. 2009;46(Suppl 3):S5–S10. doi: 10.1016/S1386-6532(09)70294-X. [DOI] [PubMed] [Google Scholar]
- 3.Moody CA, Laimins LA. Human papillomavirus oncoproteins: Pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 4.Knight T, Irving JA. Ras/Raf/MEK/ERK pathway activation in childhood acute lymphoblastic leukemia and its therapeutic targeting. Front Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaans VM, Trietsch MD, Crobach S, Stelloo E, Kremer D, Osse EM, Haar NT, van Eijk R, Muller S, van Wezel T, et al. Designing a high-throughput somatic mutation profiling panel specifically for gynaecological cancers. PLoS One. 2014;9:e93451. doi: 10.1371/journal.pone.0093451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotoula V, Sozopoulos E, Litsiou H, Fanourakis G, Koletsa T, Voutsinas G, Tseleni-Balafouta S, Mitsiades CS, Wellmann A, Mitsiades N. Mutational analysis of the BRAF RAS and EGFR genes in human adrenocortical carcinomas. Endocr Relat Cancer. 2009;16:565–572. doi: 10.1677/ERC-08-0101. [DOI] [PubMed] [Google Scholar]
- 8.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang S, Atkins MB. Which drug and when, for patients with BRAF-mutant melanoma? Lancet Oncol. 2013;14:e60–e69. doi: 10.1016/S1470-2045(12)70539-9. [DOI] [PubMed] [Google Scholar]
- 11.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Y, Zeng Y, Zhang DF, Zou SH, Cheng YF, Yao YG. IDH1 and IDH2 mutations are frequent in Chinese patients with acute myeloid leukemia but rare in other types of hematological disorders. Biochem Biophys Res Commun. 2010;402:378–383. doi: 10.1016/j.bbrc.2010.10.038. [DOI] [PubMed] [Google Scholar]
- 14.Platt AR, Woodhall RW, George AL., Jr Improved DNA sequencing quality and efficiency using an optimized fast cycle sequencing protocol. Biotechniques. 2007;43:58–62. doi: 10.2144/000112499. [DOI] [PubMed] [Google Scholar]
- 15.Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, Tietze L, Schildhaus HU, Büttner R, Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monzon FA, Ogino S, Hammond ME, Halling KC, Bloom KJ, Nikiforova MN. The role of KRAS mutation testing in the management of patients with metastatic colorectal cancer. Arch Pathol Lab Med. 2009;133:1600–1606. doi: 10.5858/133.10.1600. [DOI] [PubMed] [Google Scholar]
- 17.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: Diagnostic and clinical implications. J Mol Diagn. 2010;12:425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao YG, Ogasawara Y, Kajigaya S, Molldrem JJ, Falcão RP, Pintão MC, McCoy JP, Jr, Rizzatti EG, Young NS. Mitochondrial DNA sequence variation in single cells from leukemia patients. Blood. 2007;109:756–762. doi: 10.1182/blood-2006-01-011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao YG, Kajigaya S, Young NS. Mitochondrial DNA mutations in single human blood cells. Mutat Res. 2015;779:68–77. doi: 10.1016/j.mrfmmm.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang S, Kim HS, Seo SS, Park SY, Sidransky D, Dong SM. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol. 2007;105:662–666. doi: 10.1016/j.ygyno.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 21.Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, MacConaill LE, Shoni M, Wagle N, Jones RT, et al. Oncogenic mutations in cervical cancer: Genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013;119:3776–3783. doi: 10.1002/cncr.28288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung TK, Van Hummelen P, Chan PK, Cheung TH, Yim SF, Yu MY, Ducar MD, Thorner AR, MacConaill LE, Doran G, et al. Genomic aberrations in cervical adenocarcinomas in Hong Kong Chinese women. Int J Cancer. 2015;137:776–783. doi: 10.1002/ijc.29456. [DOI] [PubMed] [Google Scholar]
- 23.Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 24.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Montalto G, Cervello M, Nicoletti F, Fagone P, Malaponte G, Mazzarino MC, et al. Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget. 2012;3:954–987. doi: 10.18632/oncotarget.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pappa KI, Choleza M, Markaki S, Giannikaki E, Kyroudi A, Vlachos G, Voulgaris Z, Anagnou NP. Consistent absence of BRAF mutations in cervical and endometrial cancer despite KRAS mutation status. Gynecol Oncol. 2006;100:596–600. doi: 10.1016/j.ygyno.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 26.Iida K, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A, Yeasmin S, Otsuki Y, Kobayashi H, Nakayama S, Miyazaki K. EGFR gene amplification is related to adverse clinical outcomes in cervical squamous cell carcinoma, making the EGFR pathway a novel therapeutic targe. Br J Cancer. 2011;105:420–427. doi: 10.1038/bjc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]