Abstract
Breast cancer is the most common type of malignant tumor in females, and metastasis is the most common cause of breast cancer-associated mortality. Previous studies have identified that abnormal expression of microRNAs is commonly observed in human cancer and may be crucial for cancer metastasis. In the present study, microRNA-452 (miR-452) was investigated for its ability to act as a tumor suppressor in breast cancer. miR-452 expression was quantified in breast cancer tissue samples and cell lines with reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Transwell migration and invasion assays were used to investigate the effect of miR-452 on the migration and invasion capabilities of breast cancer cells. Potential target genes of miR-452 were identified with miRanda and TargetScan. A luciferase reporter assay was performed to validate RAB11A as a putative target of miR-452, and was corroborated by RT-qPCR and western blot analyses. Finally, small interfering RNA (siRNA) was used to knockdown RAB11A expression and confirm whether miR-452 inhibited breast cancer cell migration and invasion via the negative regulation of RAB11A. The results revealed that miR-452 was downregulated in breast cancer tissues and cell lines, and that its downregulation may be associated with breast cancer metastasis, as miR-452 expression inhibited the migration and invasion capacities of breast cancer cells. RT-qPCR and western blot analyses indicated that miR-452 negatively regulated the expression of RAB11A mRNA and protein. The luciferase reporter assay revealed that miR-452 specifically bound to the 3′-untranslated region of RAB11A. Furthermore, inhibition of RAB11A with siRNA inhibited breast cancer cell migration and invasion. In conclusion, the present study has demonstrated that miR-452 may act as a tumor suppressor gene via inhibition of cell migration and invasion by targeting RAB11A in breast cancer.
Keywords: breast cancer, microRNA-452, metastasis, RAB11A
Introduction
Breast cancer is the most common type of malignancy in women and the second most prevalent cause of tumor-associated mortality worldwide. It is estimated that there were 234,190 new cases and 40,730 mortalities due to breast cancer in 2015 (1–3). A number of factors have been verified to contribute to the carcinogenesis of breast cancer, including genetic alterations, epigenetic alterations and environmental factors (4,5). The progression of breast cancer is characterized by aggressive local invasion, early metastasis, and a low sensitivity to chemotherapy (6,7). Metastasis is the complication that may occur the most often during cancer and is the most prevalent cause of breast cancer-associated mortality (8). Subsequent to surgery and adjuvant treatment, 30–75% of breast cancer patients will develop metastatic disease. Patients with metastatic breast cancer have a median survival time of ~2 years following the diagnosis of metastasis (9). The invasion-metastasis cascade of breast cancer comprises multiple steps, including local invasion, entry into the circulation, arrival at distant secondary sites, extravasation and colonization in distant organs (10–12). Therefore, further elucidation of the molecular signaling cascades that control the metastasis of breast cancer is urgent, and may provide new strategies to prevent breast cancer metastasis.
MicroRNAs (miRNAs) are a group of small, non-coding and endogenous RNAs of ~22 nucleotides that negatively regulate gene expression in eukaryotes (13). miRNAs exert their functions by binding to the 3′-untranslated region (3′-UTR) of the targeted mRNAs, promoting mRNA degradation, inhibiting mRNA translation, and affecting transcription (14,15). It has been previously demonstrated that miRNAs control a wide range of biological functions, participating in the processes of cell differentiation, proliferation, apoptosis, cell cycle, angiogenesis, metabolism, invasion and metastasis (16–18). miRNAs are differentially expressed in a variety of tissues and cells; therefore, they are important biomarkers and therapeutic targets for numerous types of disease (19). Increasing evidence supports the hypothesis that the alteration of miRNA expression may serve a pivotal role in carcinogenesis and progression in numerous human cancer types, including breast cancer (20–22). In breast cancer, miRNAs may function as oncogenes (which are expressed at higher levels in cancer and decrease the expression of tumor suppressor genes) or as tumor suppressors (which are downregulated in cancer and decrease the expression of oncogenes) (23). Therefore, miRNAs may be novel therapeutic targets for the treatment of breast cancer.
miR-452 has been reported to be abnormally expressed in numerous types of human cancer (24–26). However, no specific studies have been performed to reveal the expression and biological roles underlying miR-452 in breast cancer. The aim of the present study was to investigate the expression level, biological roles and its underlying mechanism in breast cancer. In the present study, significantly lower levels of miR-452 were identified in breast cancer tissues compared with paired normal breast tissues. In addition, the miR-452 expression level was lower in breast cancer tissues from patients with metastasis compared with the breast cancer tissue from patients without metastasis. miR-452 was also downregulated in all examined breast cancer lines. Induction of miR-452 expression suppressed migration and invasion abilities in the cell lines. Further analysis revealed that miR-452 decreased breast cancer cell metastasis by directly targeting RAB11A.
Patients and methods
Patients and tissue samples
A total of 61 pairs of breast cancer tissues and adjacent normal tissues were obtained from 3201 Hospital (Hanzhong, Shaanxi, China). All patients included in the study had not received chemotherapy or any other treatment prior to the surgery. All tissues were snap frozen in liquid nitrogen immediately following surgery, and stored in a −80°C freezer until they were used. The present study was approved by the Ethics Committee of San Er Ling Yi Hospital. Written informed consent was also obtained from all patients enrolled in the present study.
Cell culture
Human breast cancer cells (MCF-7, BT-474, MDA-MB-453, MDA-MB-231, SKBR3) and HEK293T cells were purchased from American Type Culture Collection (Manassas, VA, USA). A normal mammary epithelial cell line (MCF-10A) was purchased from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cell lines were cultured in RPMI-1640 medium (MCF-7, BT-474 and SKBR3; Gibco; Thermo Fisher Scientific Inc., Waltham, MA, USA) or Dulbecco's modified Eagle's medium (DMEM; MDA-MB-453, MDA-MB-231, HEK293T and MCF-10A) with 10% (v/v) fetal bovine serum (FBS) and penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
Oligonucleotide transfection
The miR-452 mimic, negative control mimic (miR-NC), RAB11A siRNA and negative control siRNA (siR-NC) were all obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cells were transfected with miR-452 mimics (50 pmol/ml), miR-NC (50 pmol/ml), RAB11A siRNA (50 pmol/ml), siR-NC (50 pmol/ml), using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific Inc.), following the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tissues and cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The relative expression level of miR-452 were determined using a SYBR PrimeScript miRNA RT-PCR kit (Takara Bio, Inc., Otsu, Japan), and normalized to U6. The reaction system contained 12.5 µl 2X One Step SYBR® RT-PCR Buffer 4, 1.5 µl TaKaRa Ex Taq™ HS Mix, 0.5 µl PrimeScript™ PLUS RTase Mix, 1 µl forward primer and 1 µl reverse primer, 2 µl cDNA and 6.5 µl double distilled water. The cycling conditions were as follows: 42°C for 5 min; 95°C for 10 sec; and 40 cycles of 95°C for 5 sec, 55°C for 30 sec and 70°C for 30 sec. For detection of mRNA expression, total RNA was quantified and cDNA was generated by RT using M-MLV Reverse Transcriptase (Promega Corporation, Madison, WI USA). RT-qPCR was performed using SYBR Green Real-time PCR Master Mix (Toyobo Co., Ltd., Osaka, Japan) on an Applied Biosystems 7500 Sequence Detection system (Thermo Fisher Scientific, Inc.), with GADPH as an internal control. The reaction system contained 10 µl SYBR Green PCR master mix, 2 µl forward primer and 2 µl reverse primer, 2 µl cDNA and double distilled water. The thermocycling conditions were as follows: 95°C for 10 min, then 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primer sequences used were as follows: miR-452 forward, 5′-GCGAACTGTTTGCAGAGG-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; U6 forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; RAB11A forward, 5′-ATCTTCTCCTCGCTTCTGG-3′ and reverse, 5′-GCCTGCTGGCTGGTTATCA-3′; and GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG-3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′. Each sample was analyzed in triplicate, and data were analyzed using the 2−ΔΔCq method (27).
Transwell migration assay
Transwell chambers (8.0-µm pores; BD Biosciences, San Jose, CA, USA) were used to evaluate the migration abilities of cells. At 48 h after transfection, as previously described, 5×104 cells were suspended in FBS-free DMEM medium and added to the upper chamber, and 500 µl medium with 20% FBS was placed into the lower compartment. After incubating for 24 h at 37°C with 5% CO2, the non-migrated cells in the upper chamber were removed with a cotton swab. The migrated cells were fixed with 100% methanol, stained with 0.5% crystal violet and washed with PBS (Gibco; Thermo Fisher Scientific, Inc.). The membranes were observed under a light microscope (Olympus Corporation, Tokyo, Japan) and five fields were randomly selected to represent each membrane.
Transwell invasion assay
Transwell chambers pre-coated with Matrigel (BD Biosciences) were used to evaluate the invasion abilities of cells. Cells and medium were inserted into the chambers as described for the Transwell migration assay. After incubating for 48 h at 37°C with 5% CO2, the cells remaining in the upper chamber were removed with a cotton swab. The invading cells were fixed and analyzed as described in the Transwell migration assay section.
Western blotting
Cells were harvested, and lysed with radioimmunoprecipitation assay lysis buffer. Bulk protein concentration was determined with a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were electrophoresed on an 10% SDS-PAGE gel and transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% skimmed milk powder in Tris-buffered saline (TBS) for 1 h, then probed with the primary antibodies (Abcam, Cambridge, UK) at 4°C overnight. The primary antibodies were a monoclonal mouse anti-human RAB11A antibody (ab170134; dilution, 1:1,000) and a monoclonal mouse anti-human GADPH antibody (ab9484; dilution, 1:1,000). The membranes were washed with TBS containing 0.1% Tween-20, and incubated with a corresponding horseradish peroxidase-conjugated secondary antibody (ab6789; 1:3,000; Abcam) for 1 h at 37°C. Subsequent to washing, the membranes were visualized using a Pierce Enhanced Chemiluminescence Detection System (Thermo Fisher Scientific, Inc.). The densitometry was analyzed by Quantity One software (version 4.6.2; Bio-Rad Laboratories, Inc., Hercules, CA, USA). For bioinformatic analysis, the potential target genes of miR-452 were analyzed using miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/).
Luciferase reporter assay
Plasmids containing the wild-type RAB11A 3′-UTR (PmirGLO-RAB11A-3′-UTR-WT) or a mutant RAB11A 3′-UTR (PmirGLO-RAB11A-3′-UTR-Mut; Fig. 1) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). HEK293T cells were co-transfected with PmirGLO-RAB11A-3′-UTR-WT or PmirGLO-RAB11A-3′-UTR-Mut together with a miR-452 mimic or miR-NC. At 48 h after transfection, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega Corporation), following the manufacturer's protocol. Firefly luciferase activities were normalized to Renilla luciferase activities.
Figure 1.

miR-452 mimic sequence alongside RAB11A-3′-UTR-WT and RAB11A-3′-UTR-Mut mRNA sequences, including the miR-452-binding site of the WT sequence, and the equivalent area from the Mut sequence. miR-452, microRNA-452; 3′-UTR, 3′-untranslated region; WT, wild type; Mut, mutant.
Statistical analysis
Each assay was repeated three times. Data are presented as the mean ± standard deviation. Statistical analysis was performed using two-tailed Student's t-test or one-way analysis of variance using SPSS software (version 16; SPSS, Inc., Chicago, IL, USA). SNK was used to compare between two groups in multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-452 are downregulated in breast cancer tissues
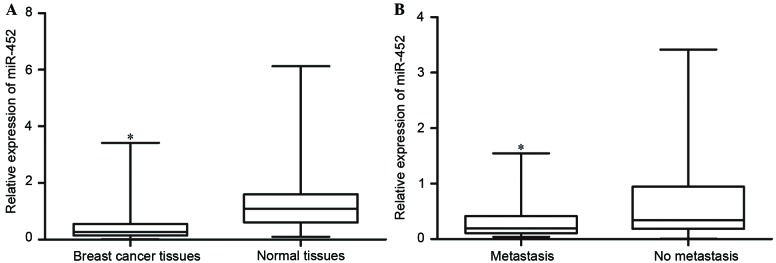
Initially, the expression levels of miR-452 in breast cancer tissues and matched adjacent normal tissues were quantified with RT-qPCR. The results revealed that miR-452 expression levels in breast cancer tissues were significantly lower compared with those in the matched normal tissues (P<0.05; Fig. 2A).
Figure 2.
Expression levels of miR-452 in breast cancer tissues assessed by reverse transcription-quantitative polymerase chain reaction. (A) miR-452 was downregulated in breast cancer tissue, compared with paired adjacent normal tissue. *P<0.05 vs. normal tissues. (B) miR-452 levels in breast cancer with metastasis were significantly lower than in those without metastasis. Data are presented as the mean ± standard deviation. *P<0.05 vs. no metastasis. miR-452, microRNA-452.
miR-452 expression levels were additionally compared between breast cancer tissues in patients with and without metastatic breast cancer. It was identified that the levels of miR-452 in breast cancer with metastasis were significantly lower than those in breast cancer without metastasis (P<0.05; Fig. 2B). These results indicated that miR-452 was downregulated in breast cancer tissues, and may be associated with breast cancer metastasis.
Expression levels of miR-452 are decreased in breast cancer cell lines
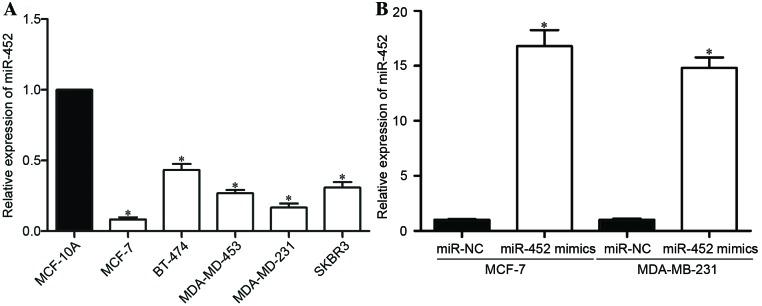
miR-452 expression levels were quantified for a number of breast cancer cell lines. The miR-452 expression levels were revealed to be significantly downregulated in all the assessed breast cancer cell lines (MCF-7, BT-474, MDA-MB-453, MDA-MB-231, SKBR3) compared with that in MCF-10A, a normal human breast epithelial cell line (P<0.05; Fig. 3A).
Figure 3.
Expression levels of miR-452 in breast cancer cell lines detected by RT-qPCR. (A) The relative expression of miR-452 was downregulated in breast cancer cell lines compared with the normal breast epithelial cell line, MCF-10A. (B) The transfection efficiency of an miR-452 mimic was assessed by comparing the detected miR-452 levels of mimic-transfected cells, with miR-NC-transfected cells, using RT-qPCR. Data are presented as the mean ± standard deviation. *P<0.05 vs. MCF-10A or miR-NC. miR-452, microRNA-452; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR-NC, negative control microRNA.
For further studies, MCF-7 and MDA-MB-231 cells were selected, as these had the lowest miR-452 expression level. An miR-452 mimic was introduced into MCF-7 and MDA-MB-231 cells to increase the miR-452 expression level, and miR-NC was used as a control. The RT-qPCR results demonstrated that miR-452 levels were significantly higher in MCF-7 and MDA-MB-231 cells following transfection with the miR-452 mimic than with the miR-NC (P<0.05; Fig. 3B).
miR-452 inhibits the migration and invasion capacities of breast cancer cells
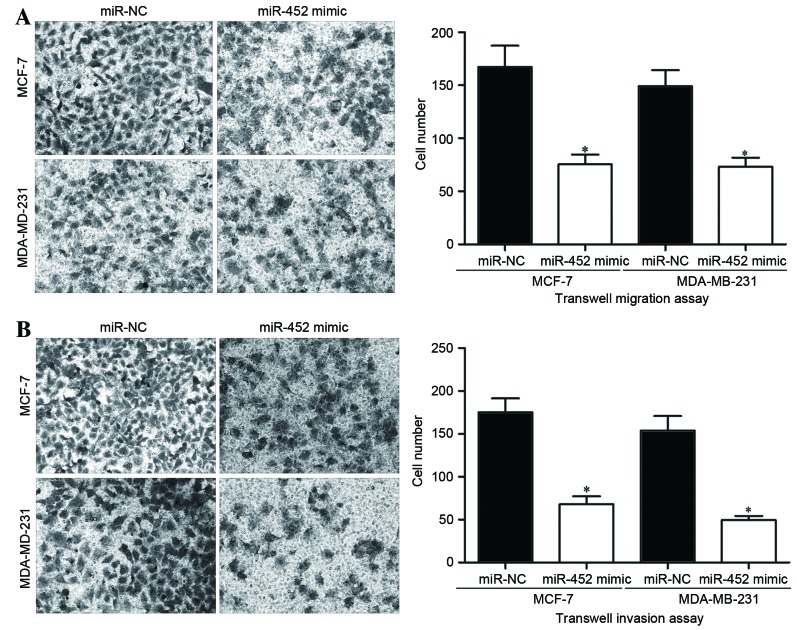
As the expression level of miR-452 had been associated with metastatic breast cancer in previous results, Transwell migration and invasion assays were performed to assess the effect of miR-452 on the migration and invasion capacities of breast cancer cells. The Transwell migration assay results demonstrated that miR-452 overexpression significantly inhibited the migration capacities of MCF-7 and MDA-MB-231 cells (P<0.05; Fig. 4A). Similarly, the invasion capacities of MCF-7 and MDA-MB-231 cells were also significantly inhibited, as demonstrated by the Transwell invasion assays (P<0.05; Fig. 4B). These results suggested that miR-452 may be associated with the regulation of metastasis in breast cancer cells.
Figure 4.
miR-452 inhibits the migration and invasion abilities of breast cancer cells. (A) Transwell migration and (B) Transwell invasion assays were performed to investigate the effects of miR-452 on MCF-7 and MDA-MB-231 breast cancer cells, revealing reduced migration and invasion capabilities in miR-452 mimic-transfected cells. Data are presented as the mean ± standard deviation. *P<0.05 vs. corresponding miR-NC group. miR-452, microRNA-452; miR-NC, negative control microRNA.
miR-452 directly targets RAB11A in breast cancer cells
To explore the molecular mechanisms by which miR-452 may inhibit the migration and invasion of breast cancer cells, bioinformatics analysis was performed with two miRNA algorithms: miRanda (www.microrna.org) and TargetScan (www.targetscan.org). The analysis revealed that RAB11A was a likely target gene of miR-452 (Fig. 1). To confirm whether miR-452 negatively regulated RAB11A expression, the expression of RAB11A at the mRNA and protein levels was measured by RT-qPCR and western blot analyses. RAB11A expression was significantly reduced at the mRNA (P<0.05; Fig. 5A) and protein (P<0.05; Fig. 5B) levels in the MCF-7 and MDA-MB-231 cells transfected with an miR-452 mimic, compared with the cells transfected with miR-NC.
Figure 5.
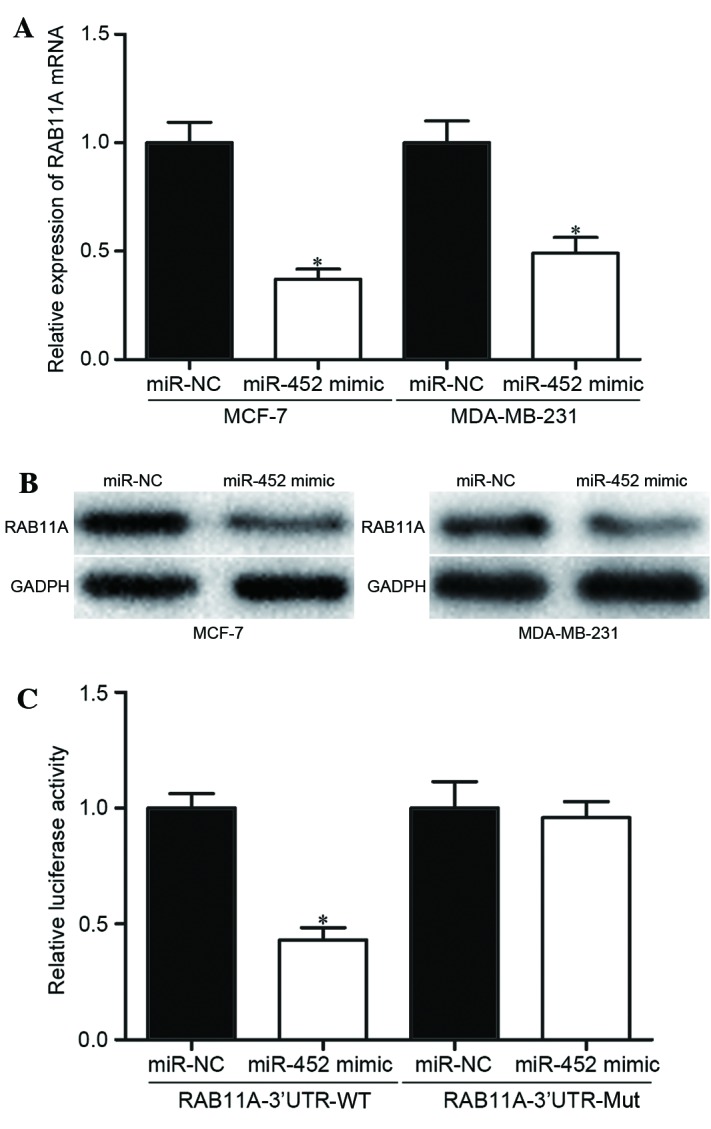
miR-452 directly targets RAB11A in breast cancer cells. The relative RAB11A (A) mRNA and (B) protein expression levels were significantly reduced in the miR-452 mimic-transfected breast cancer cells, compared with miR-NC-transfected cells. (C) A luciferase reporter assay was performed on HEK293T cells co-transfected with an miR-452 mimic or miR-NC, and PmirGLO-RAB11A-3′-UTR-WT or PmirGLO-RAB11A-3′-UTR-Mut, revealing that miR-452 decreased activity only in cells transfected with the WT RAB11A 3′-UTR. Data are presented as the mean ± standard deviation. *P<0.05 vs. corresponding miR-NC group. miR-452, microRNA-452; miR-NC, microRNA negative control; 3′-UTR, 3′-untranslated region; WT, wild type; Mut, mutant.
Luciferase reporter assays were performed to explore whether miR-452 regulated RAB11A expression directly through binding to the 3′-UTR of RAB11A mRNA. The luciferase activity significantly decreased following co-transfection with PmirGLO-RAB11A-3′-UTR-WT and an miR-452 mimic, compared with co-transfection with PmirGLO-RAB11A-3′-UTR-Mut and the miR-452 mimic (P<0.05; Fig. 5C), indicating that miR-452 specifically binds to the 3′-UTR of RAB11A mRNA. Taken together, the results demonstrated that miR-452 negatively regulates RAB11A expression directly through binding the 3′-UTR of RAB11A mRNA.
Inhibition of RAB11A protein levels by RAB11A siRNA inhibits breast cancer cell migration and invasion
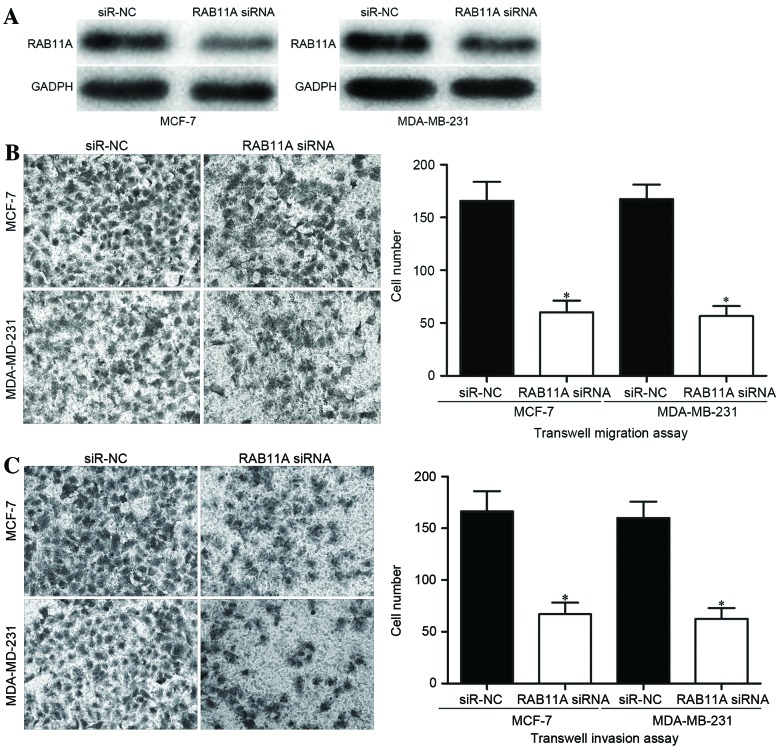
To explore whether miR-452 inhibited breast cancer cell migration and invasion through negative regulation of RAB11A, siRNA was used to knockdown the RAB11A expression level in breast cancer cells. The migration and invasion abilities of these cells were then evaluated. Following transfection, western blot analysis measured the expression of RAB11A protein. The results confirmed that RAB11A was significantly downregulated in MCF-7 and MDA-MB-231 cells subsequent to transfection with RAB11A siRNA, compared with transfection with an siR-NC (P<0.05; Fig. 6A). Transwell migration and invasion assays revealed that the numbers of migrating (P<0.05; Fig. 6B) and invading (P<0.05; Fig. 6C) cells were significantly decreased in the MCF-7 and MDA-MB-231 cells transfected with RAB11A siRNA compared with in the cells transfected with siR-NC. These results demonstrated that miR-452 may decrease the migration and invasion capacities of breast cancer cells via the downregulation of RAB11A.
Figure 6.
Inhibition of RAB11A inhibits migration and invasion of MCF-7 and MDA-MB-231 breast cancer cells. (A) Compared with siR-NC, RAB11A siRNA decreased RAB11A protein expression in breast cancer cells. (B) Transwell migration and (C) Transwell invasion assays were performed, revealing that RAB11A knockdown decreased breast cancer cell migration and invasion capacities. Data are presented as the mean ± standard deviation. *P<0.05 vs. corresponding siR-NC group. siRNA, small interfering RNA; siR-NC, negative control siRNA.
Discussion
Breast cancer is the most common type of malignant tumor in women. The main therapy options for patients with breast cancer are surgery, radiotherapy and chemotherapy (28). However, patients with metastatic breast cancer do not respond well to these treatments. Metastasis is the predominant cause of breast cancer-associated mortality (29). Thus, it is important to explore the prevention of metastasis in breast cancer. Studies have recently identified that miRNAs may inhibit (30,31) or enhance the metastasis of breast cancer cells (32) or prostate cancer cells (33). The pivotal role of miRNAs in metastasis via the modulation of various target genes has promoted intensive research into miRNAs as a novel perspective on the metastatic process.
In the present study, miR-452 expression levels in breast cancer tissues and matched adjacent normal tissues were quantified with RT-qPCR. The result indicated that miR-452 was significantly downregulated in breast cancer tissues compared with that in matched adjacent normal tissues, and the level of miR-452 in breast cancer with metastasis was significantly lower than in breast cancer without metastasis. In addition, the expression levels of miR-452 were lower in all examined breast cancer cell lines than in the human breast epithelial cell line MCF-10A. Similar results have been reported for non-small cell lung cancer (24), prostate cancer (25) and glioma (26). These findings suggest that the low expression of miR-452 may be a common event in human cancer, and may be associated with metastasis of breast cancer.
To investigate the effect of miR-452 on metastasis of breast cancer, an miR-452 mimic was transfected into breast cancer cells to enhance its expression. The restoration of miR-452 expression inhibited the migration and invasion capacities of breast cancer cells, as determined by Transwell migration and invasion assays, respectively. In non-small cell lung cancer, overexpression of miR-452 has been shown to decrease the invasion ability of cancer cells (24). Furthermore, Liu et al (26) reported that downregulation of miR-452 serves an important function in glioma carcinogenesis. Collectively, these findings indicate that miR-452 may act as a tumor suppressor, and may be a potential therapeutic target in these types of cancer.
However, the expression level and function of miR-452 is tissue-specific. miR-452 was identified as upregulated in various types of human cancer, including hepatocellular carcinoma (34), esophageal cancer (22) and urothelial carcinoma (23). miR-452 enhanced proliferation, migration and invasion, induced cell cycle G1 to S transition, and inhibited apoptosis in hepatocellular carcinoma cells. The difference between cancer types may be explained by the distinct context of various tissue microenvironments, and the ‘imperfect complementarity’ of the interactions between miRNAs and target mRNAs (24).
In previous studies, the genes BMI1 proto-oncogene (24), insulin-like growth factor-1 receptor (35), lymphoid enhanced binding factor 1 (26), transcription factor 4 (26), and cyclin-dependent kinase inhibitor 1B (34) were demonstrated to be direct targets of miR-452. To understand the mechanisms by which miR-452 inhibited metastasis in breast cancer, predicted targets of miR-452 were identified using the TargetScan and miRanda search tools. The tools predicted that RAB11A was a target of miR-452. To assess the regulatory association between miR-452 and RAB11A, RT-qPCR and western blot analyses were adopted to detect RAB11A expression at the mRNA and protein level in breast cancer cells following transfection with an miR-452 mimic. The results revealed that RAB11A mRNA and protein levels were significantly reduced in miR-452-transfected breast cancer cells. Additionally, a luciferase reporter assay indicated that miR-452 binds directly to the 3′-UTR of the RAB11A mRNA. The inhibition of RAB11A protein expression by RAB11A siRNA inhibited breast cancer cell migration and invasion, thus further verifying that RAB11A is a direct, functional target gene of miR-452 in breast cancer.
RAB11A is a member of the Rab family, which also includes RAB11B and RAB25. Rab proteins are small (21–25 kDa), monomeric GTPases that constitute the largest branch of the Ras superfamily and are evolutionarily conserved between yeast and humans (36). It has been reported that Rab proteins exert an important function in various types of human cancer, including breast, colon, lung, ovarian and bladder cancer (37). RAB11A has previously been demonstrated to be involved in breast cancer carcinogenesis and progression (38–40). Wang et al (41) reported that RAB11A was a direct target of miR-320a in breast cancer, and contributed to the growth and invasion of cancer cells. All these findings support the results of the present study in implicating RAB11A as a direct functional target of miR-452 in breast cancer.
In conclusion, the present study revealed that miR-452 is a novel tumor suppressor gene in breast cancer. miR-452 was significantly downregulated in breast cancer, and its downregulation was also correlated with metastasis in patients with breast cancer. Forced miR-452 expression decreased breast cancer cell metastasis through the direct targeting of RAB11A. Therefore, these findings provide a potential prognostic target for breast cancer patients.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Xue J, Chen Z, Gu X, Zhang Y, Zhang W. MicroRNA-148a inhibits migration of breast cancer cells by targeting MMP-13. Tumour Biol. 2016;37:1581–1590. doi: 10.1007/s13277-015-3926-9. [DOI] [PubMed] [Google Scholar]
- 4.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann MW, Niederacher D, Schnurch HG, Gusterson BA, Bender HG. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med (Berl) 1997;75:429–439. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- 6.Gradishar WJ. Treatment of metastatic breast cancer. J Natl Compr Canc Netw. 2014;12(5 Suppl):S759–S761. doi: 10.6004/jnccn.2014.0184. [DOI] [PubMed] [Google Scholar]
- 7.Martin HL, Smith L, Tomlinson DC. Multidrug-resistant breast cancer: Current perspectives. Breast Cancer (Dove Med Press) 2014;6:1–13. doi: 10.2147/BCTT.S37638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi RU, Miyazaki H, Ochiya T. The roles of MicroRNAs in breast cancer. Cancers (Basel) 2015;7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamucci T, D'Ottavio AM, Magnolfi E, Barduagni M, Vaccaro A, Sperduti I, Moscetti L, Belli F, Meliffi L. Weekly epirubicin plus docetaxel as first-line treatment in metastatic breast cancer. Br J Cancer. 2007;97:1040–1045. doi: 10.1038/sj.bjc.6603982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 11.Wan L, Pantel K, Kang Y. Tumor metastasis: Moving new biological insights into the clinic. Nat Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Ouyang G. Periostin: A bridge between cancer stem cells and their metastatic niche. Cell Stem Cell. 2012;10:111–112. doi: 10.1016/j.stem.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Teng G, Papavasiliou FN. Shhh! Silencing by microRNA-155. Philos Trans R Soc Lond B Biol Sci. 2009;364:631–637. doi: 10.1098/rstb.2008.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. doi: 10.12688/f1000research.2-136.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gromak N. Intronic microRNAs: A crossroad in gene regulation. Biochem Soc Trans. 2012;40:759–761. doi: 10.1042/BST20120023. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 18.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla GC, Singh J, Barik S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Zhou Y, Pan H, Zhou J, Fan Y, Qu P. microRNA-99a inhibiting cell proliferation, migration and invasion by targeting fibroblast growth factor receptor 3 in bladder cancer. Oncol Lett. 2014;7:1219–1224. doi: 10.3892/ol.2014.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu D, Pan H, Zhou Y, Zhang Z, Qu P, Zhou J, Wang W. Upregulation of microRNA-204 inhibits cell proliferation, migration and invasion in human renal cell carcinoma cells by downregulating SOX4. Mol Med Rep. 2015;12:7059–7064. doi: 10.3892/mmr.2015.4259. [DOI] [PubMed] [Google Scholar]
- 22.Wang CZ, Yuan P, Li Y. MiR-126 regulated breast cancer cell invasion by targeting ADAM9. Int J Clin Exp Pathol. 2015;8:6547–6553. [PMC free article] [PubMed] [Google Scholar]
- 23.van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015;17:21. doi: 10.1186/s13058-015-0526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He Z, Xia Y, Pan C, Ma T, Liu B, Wang J, Chen L, Chen Y. Up-Regulation of MiR-452 inhibits metastasis of non-small cell lung cancer by regulating BMI1. Cell Physiol Biochem. 2015;37:387–398. doi: 10.1159/000430362. [DOI] [PubMed] [Google Scholar]
- 25.Kristensen H, Haldrup C, Strand S, Mundbjerg K, Mortensen MM, Thorsen K, Ostenfeld MS, Wild PJ, Arsov C, Goering W, et al. Hypermethylation of the GABRE~miR-452~miR-224 promoter in prostate cancer predicts biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2014;20:2169–2181. doi: 10.1158/1078-0432.CCR-13-2642. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Chen K, Wu J, Shi L, Hu B, Cheng S, Li M, Song L. Downregulation of miR-452 promotes stem-like traits and tumorigenicity of gliomas. Clin Cancer Res. 2013;19:3429–3438. doi: 10.1158/1078-0432.CCR-12-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Wei C, Luo Q, Sun X, Li D, Song H, Li X, Song J, Hua K, Fang L. MicroRNA-497 induces cell apoptosis by negatively regulating Bcl-2 protein expression at the posttranscriptional level in human breast cancer. Int J Clin Exp Pathol. 2015;8:7729–7739. [PMC free article] [PubMed] [Google Scholar]
- 29.Hong W, Dong E. The past, present and future of breast cancer research in China. Cancer Lett. 2014;351:1–5. doi: 10.1016/j.canlet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Bockhorn J, Prat A, Chang YF, Liu X, Huang S, Shang M, Nwachukwu C, Gomez-Vega MJ, Harrell JC, Olopade OI, et al. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Cancer Res. 2014;74:7406–7417. doi: 10.1158/0008-5472.CAN-14-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erturk E, Cecener G, Polatkan V, Gokgoz S, Egeli U, Tunca B, Tezcan G, Demirdogen E, Ak S, Tasdelen I. Evaluation of genetic variations in miRNA-binding sites of BRCA1 and BRCA2 genes as risk factors for the development of early-onset and/or familial breast cancer. Asian Pac J Cancer Prev. 2014;15:8319–8324. doi: 10.7314/APJCP.2014.15.19.8319. [DOI] [PubMed] [Google Scholar]
- 32.Sun EH, Zhou Q, Liu KS, Wei W, Wang CM, Liu XF, Lu C, Ma DY. Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray. Eur Rev Med Pharmacol Sci. 2014;18:2783–2788. [PubMed] [Google Scholar]
- 33.Cai ZK, Chen Q, Chen YB, Gu M, Zheng DC, Zhou J, Wang Z. microRNA-155 promotes the proliferation of prostate cancer cells by targeting annexin 7. Mol Med Rep. 2015;11:533–538. doi: 10.3892/mmr.2014.2744. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Q, Sheng Q, Jiang C, Shu J, Chen J, Nie Z, Lv Z, Zhang Y. MicroRNA-452 promotes tumorigenesis in hepatocellular carcinoma by targeting cyclin-dependent kinase inhibitor 1B. Mol Cell Biochem. 2014;389:187–195. doi: 10.1007/s11010-013-1940-z. [DOI] [PubMed] [Google Scholar]
- 35.Hu Q, Gong JP, Li J, Zhong SL, Chen WX, Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH, Tang JH. Down-regulation of miRNA-452 is associated with adriamycin-resistance in breast cancer cells. Asian Pac J Cancer Prev. 2014;15:5137–5142. doi: 10.7314/APJCP.2014.15.13.5137. [DOI] [PubMed] [Google Scholar]
- 36.Bhuin T, Roy JK. Rab11 in disease progression. Int J Mol Cell Med. 2015;4:1–8. [PMC free article] [PubMed] [Google Scholar]
- 37.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: An emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 38.Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rougé C, Lasorsa L, Letessier A, Ginestier C, Monville F, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 39.Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC. Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol. 2008;183:143–155. doi: 10.1083/jcb.200804140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B, Yang Z, Wang H, Cao Z, Zhao Y, Gong C, Ma L, Wang X, Hu X, Chen S. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am J Cancer Res. 2015;5:2719–2729. doi: 10.1158/1538-7445.AM2015-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]