Abstract
The present report describes a unique infantile acute lymphoblastic leukemia (ALL) case with cryptic mixed-lineage leukemia (MLL) rearrangements with 11q23 chromosomal translocation. MLL break-apart signals were identified by fluorescence in situ hybridization, and transcriptome sequencing revealed MLL-myeloid/lymphoid or mixed-lineage leukemia; translocated To, 10 (MLLT10)/AF10 fusion transcripts. Analysis also revealed a previously unreported MLLT10/AF10-homeobox protein Mohawk (MKX) transcript, where the 5′ portion of MLLT10/AF10 at 10p12.31 was fused out-of-frame with the 3′ portion of MKX at 10p12.1, which is closely located to MLLT10/AF10. Furthermore, the reciprocal 3′-MLL gene segment was fused in-frame to AT-rich interaction domain (ARID)5B at 10q21. Previously, common allelic variants in ARID5B, which are directly associated with hematopoietic differentiation and development, have been repeatedly and significantly associated with childhood ALL. The heterozygous genotype in ARID5B (RefSNP: rs10821936) increased the risk for leukemia with MLL-rearrangement. In particular, single nucleotide polymorphisms of ARID5B conferred increased risk for MLL-MLLT3/AF9. Based on these findings, the authors propose that while the presence of reciprocal MLL alleles has been detected in this patient, different pathological disease mechanisms may be at play due to individual recombination events.
Keywords: acute lymphoblastic leukemia, molecular biology, transcriptome sequencing, ARID5B, MLL-MLLT10/AF10
Introduction
The incidence of translocations in the mixed-lineage leukemia (MLL) gene at chromosome band 11q23 is high in acute lymphoblastic leukemia (ALL) and acute myeloid leukemia in infants (1,2). MLL forms rearrangements with >60 translocation partner genes (3,4). The most common rearrangements are AFF1/AF4, MLLT1/ENL, and MLLT3/AF9, and the less common are ELL, myeloid/lymphoid or mixed-lineage leukemia; translocated To, 10 (MLLT10)/AF10 and MLLT4/AF6 (3). However, in a significant fraction of patients with leukemia and MLL rearrangements, such alterations are absent (3). To address the pathobiology of leukemia with MLL rearrangements and determine potential involvement of fusion genes, the authors of the present report previously performed transcriptome sequencing in an infant with ALL and MLL rearrangement. The infant was negative for MLL-AF4, MLL-ENL, MLL-ELL and MLL-AF9 fusion transcripts, and the presence of a MLL-MLLT10/AF10 fusion transcript was detected (5).
In the present report, a new case of infantile ALL with MLL-MLLT10/AF10 and 10;11 rearrangements was presented. A chromosomal mechanism leading to MLL-MLLT10/AF10 fusion and alternative splicing of an MLL-exon-8-MLLT10 fusion genes, resulting in two different isoforms, was described. In addition, it was determined that MLLT10/AF10-homeobox protein Mohawk (MKX) resulted from an inversion (10p12.1;10p12.31). Furthermore, transcriptome sequencing revealed a separate chromosomal translocation leading to a previously unreported AT-rich interaction domain (ARID)5B-MLL-positive 10;11 rearrangement in this patient. ARID5B polymorphisms are important determinants of childhood ALL susceptibility, and treatment outcomes and contribute to racial disparities in ALL (6). Taken together, these results support the hypothesis of the authors that precise control of MLL and MLLT10/AF10 fusion transcripts is crucial in leukemogenesis.
Case report
Patient characteristics
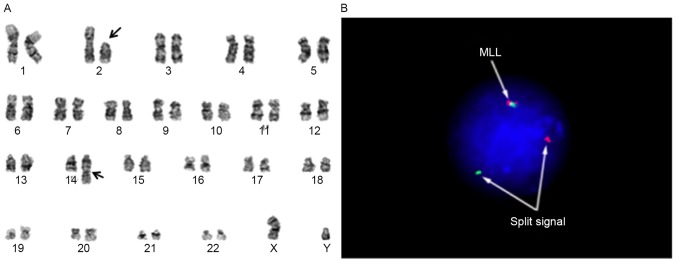
A 2-month-old Japanese male infant was admitted to Tokyo University Hospital (Tokyo, Japan) in January 2008. Laboratory tests demonstrated a leukocyte count of 5.44×1010/l (normal range, 4.6×109-18.9×109/l) with 88% blasts, hemoglobin of 9.0 g/dl (normal range, 9.5–13.7 g/dl), and platelet count of 3.9×1010/l (normal range, 25×1010-82×1010/l). Leukemic cells were cytogenetically characterized as 46, XY, t(2;14)(p11.2;q32), add(11)(q23) (Fig. 1A) and were found to express cluster of differentiation (CD)10 and CD19 by bone marrow biopsy. Analysis with fluorescence in situ hybridization using the MLL break-apart probe for the determination of add(11)(q23) revealed the typical split signal (Fig. 1B). Based on the above data, the diagnosis was established as infantile B-precursor ALL with MLL rearrangement. The patient achieved complete remission with chemotherapy and received stem cell transplantation. Treatment was well tolerated, and he has been in complete remission for 7 years.
Figure 1.
Cytogenetic analysis suggested the evidence for 11q23 rearrangement. (A) G-banded karyogram from bone marrow cells at diagnosis showed to be 46, XY, t(2;14)(p11.2;q32), add(11)(q23) in 14 of 20 bone marrow cells. The arrow indicates the breakpoint at 11q23. (B) Fluorescence in situ hybridization analysis with MLL probe (Vysis) on interphase nuclei of bone marrow cells at diagnosis. A 11q23 split-signal type was observed: One green signal and one orange signal (divided arrows). A normal signal pattern for the MLL probe (green and red fusion signals) was also observed in the bone marrow cells (arrows). MLL, mixed-lineage leukemia.
The present study was approved by the Gene Analysis Research Ethics Committee at the University of Tokyo (Tokyo, Japan). Informed consent was obtained from the guardian of the patient.
Paired-end RNA sequencing and identification of fusion genes
High-quality RNA with an RNA integrity number >6.0 from the patient was used to prepare RNA sequencing libraries, according to the TruSeq® RNA (Illumina, San Diego, CA, USA) protocol, which were then sequenced on an Illumina HiSeq 2000 device. An in-house pipeline, Genomon-fusion, was used to identify fusion transcripts. All candidate gene fusions that were >2 paired reads were confirmed by reverse transcription-polymerase chain reaction (RT-PCR) and Sanger sequencing.
RT-PCR and Sanger sequencing
Total RNA (4 µg) was reverse transcribed to cDNA in a total volume of 33 µl with random primers using the Ready-To-Go You-Prime First-Strand beads (Pharmacia Biotech; GE Healthcare, Chicago, IL, USA). RT-PCR and Sanger sequencing were performed as previously described (4). In brief, 1 µl cDNA was used as a template in RT-PCR and the reaction was performed for 35 cycles in a GeneAmp PCR System 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), with denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 1 min and a final cycle of 72°C for 7 min. RT-PCR experiment was repeated three times. MLL-MLLT10/AF10 was amplified using the following forward (F) and reverse (R) primers: MLL F1, 5′-CCTGAGGACTGTGGTGTTTGTAC-3′ and MLLT10/AF10 R1, 5′-CCTGACTGAGAGAAGATCCAGAT-3′. ARID5B-MLL was amplified using the following forward and reverse primers: ARID5B F1, 5′-TCGATGCTGAAACGCATCCA-3′ and MLL R1, 5′-CACTGCTCTCTTTGCTGTCT-3′. MLLT10/AF10-MKX was amplified using the following forward and reverse primers: MLLT10/AF10 F1, 5′-ATGGAAGTTTACAGAGCCTCAG-3′ and MKX R1, 5′-TTCGTTCATGTGGGTTCTTGG-3′. Nucleotide sequences of PCR products and, if necessary, subcloned PCR products were analyzed as described previously (4).
Detection of fusion transcripts
To identify high-confidence fusion transcripts, an in-house pipeline for RNA sequencing data analysis, Genomon-fusion, was used for analysis of RNA sequencing data from the patient bone marrow cells. A total of 49 fusion transcripts supported by discordant read pairs as well as one perfectly matched junction-spanning read, with the other end of the read-pair mapping to either of the fusion gene partners, were identified.
To focus on fusion transcripts identified in infantile leukemia, from the initial list of 49 fusion transcripts, two in-frame fusion transcripts (MLL-MLLT10/AF10 and ARID5B-MLL) and one out-of-frame fusion transcript (MLLT10/AF10-MKX) that were supported by >10 junction-spanning reads were identified. The ARID5B-MLL fusion transcript has not been previously reported. No fusion events involving genes on chromosomes 2 and 14 were detected.
Validation of fusion transcripts
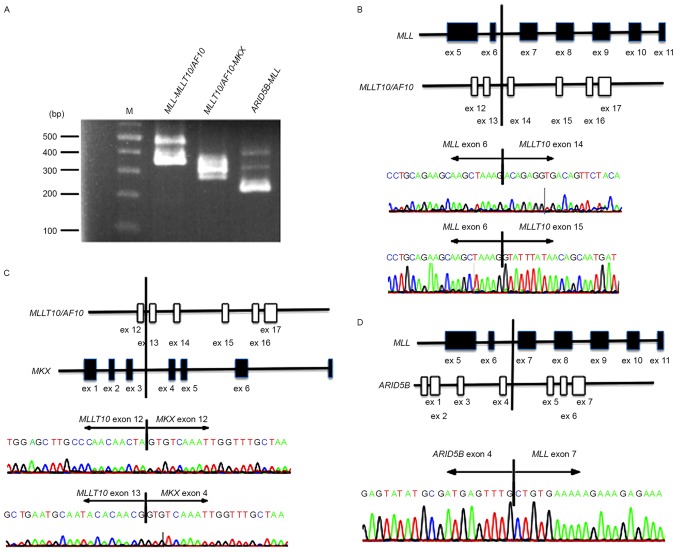
To evaluate the authenticity and validity of the gene fusion transcripts, Sanger sequencing of RT-PCR amplification products spanning fusion junction points was performed to validate all transcripts detected in the patient bone marrow cells. Although the RT step is able to potentially produce artifactual fusion transcripts, all three fusion transcripts, MLL-MLLT10/AF10, ARID5B-MLL, and MLLT10/AF10-MKX, were detected by Sanger sequencing (Fig. 2). Specifically, all fusion transcripts showing >10 junction-spanning reads were identifiable by Sanger sequencing.
Figure 2.
Validation of fusions. (A) Identification of MLL-MLLT10/AF10, MLLT10/AF10-MKX and ARID5B-MLL fusion transcripts in bone marrow cells from the patient using RT-PCR. Marker represents a 1 kb DNA ladder. M, size marker. (B-D) Schematic representation of reverse transcription-polymerase chain reaction products on der(10) and der(11) resulted in MLL-M14LLT10/AF10, MLLT10/AF10-MKX and ARID5B-MLL fusion genes. Sanger sequencing chromatograms showing the reading frame at the breakpoint and putative translation of the fusion protein in the patient's bone marrow cells. ARID5B, AT-rich interaction domain 5B; MLL, mixed-lineage leukemia; MLLT10, myeloid/lymphoid or mixed-lineage leukemia; translocated To, 10.
Discussion
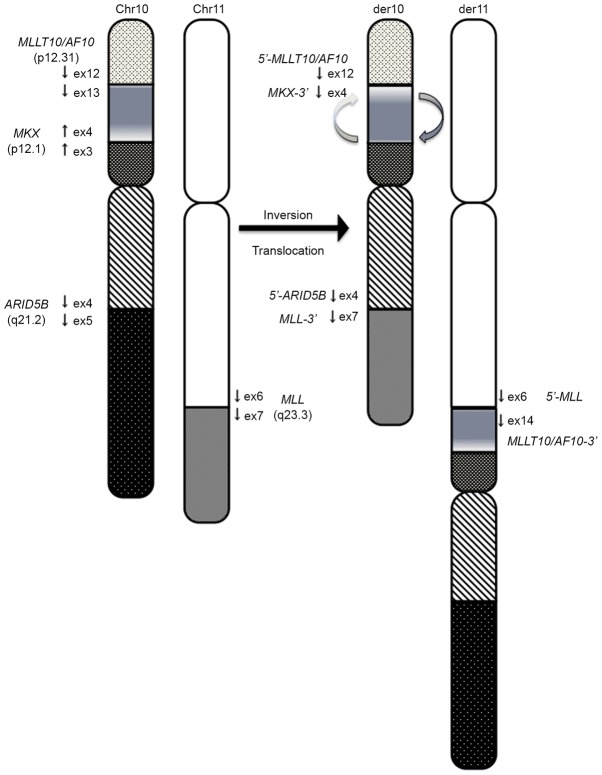
The authors herein presented a new case with inv(10)(p12.1;p12.31) and t(10;11)(q21;q23). In this case, the MLL-MLLT10/AF10 fusion resulted from several rearrangements, including inversion followed by translocation. This patient exhibited complex rearrangements, which included inversion of 10p12.1-p12.31 followed by translocation of 11q23 to 10p12 that was associated with the generation of MLL-MLLT10 (Fig. 3). The proximal inversion breakpoint at 10p was heterogeneous and contained a region within 10p12.1-p12.31. MLLT10/AF10 has been reported to be involved in a complex rearrangement that resulted in an inversion of either 10p or 11q followed by translocation of 10p;11q or insertion of the inverted segment into MLLT10/AF10 or MLL (7,8). MLLT10/AF10 was also one of the 20 genes involved in complex rearrangements, with three- or four-way translocations resulting in >2 fusion alleles (8). While the 3′ portions of the translocation partner genes were regularly fused to the 5′-portion of MLL, the genes involved in complex rearrangements, including MLLT10/AF10, were fused to the 3′ portion of MLL (8). CALM-MLLT10/AF10 and reciprocal MLLT10/AF10-CALM transcripts have been detected in patients with various hematological malignancies (9). Silliman et al (10) emphasized that truncated MLLT10/AF10 protein in normal cells and truncated MLLT10/AF10-CALM protein in leukemic cells retain a cysteine-rich motif and leukemia-associated protein (LAP) (11) and would function as dominant-negative inhibitors of full-length AF10 or associated proteins (12). There is a high likelihood that the rearrangements in the patient in the present report also involved a complex mechanism, given that the 3′ portion of MKX was fused to the 5′ portion of MLLT10/AF10. MLLT10/AF10-MKX proteins in leukemic cells retain a cysteine-rich motif and LAP. MKX is a member of the three-amino acid loop extension superclass of homeobox genes and is associated with cryptorchidism (13). The DNA recognition motif for MKX is largely dependent on a core set of amino acids in helix III, and residues at the N terminus of the homeodomain that make direct contact with the DNA (14,15). However residues at the N terminus were not maintained in the MLLT10/AF10-MKX fusion transcript due to the presence of an out-of frame fusion transcript, and MKX may be biologically inactivated. These results suggested that MLLT10/AF10-MKX may have a similar role in leukemogenesis as MLLT10/AF10-CALM.
Figure 3.
Proposed chromosomal mechanism, which leads to MLL-MLLT10/AF10, ARID5B-MLL and MLLT10/AF10-MKX rearrangements in the patient. Paracentric inversion of 10p12.31-p12.1 with a breakpoint in the MLLT10/AF10 gene, followed by an additional break 3′ of MLL prior to translocation of the 11q segment into ARID5B located at 10q21. ARID5B, AT-rich interaction domain 5B; MLL, mixed-lineage leukemia; MLLT10, myeloid/lymphoid or mixed-lineage leukemia; translocated To, 10.
In the present study, ARID5B-MLL fusion was caused by the presence of a translocation between derivative chromosomes 10q21 and 11q23. ARID5B gene variants were consistently shown to increase the risk of childhood ALL in various populations (16–18). The ARID5B gene encodes a member of the AT-rich interaction domain (ARID) family of DNA-binding proteins. The encoded protein forms a histone H3K9me2 demethylase complex together with PHD finger protein 2 to regulate the transcription of target genes involved in adipogenesis and liver development (19). Although the function of ARID5B in lympho-hematopoiesis has not been well studied, it may be involved in epigenetic regulation of gene expression in hematopoietic stem cells and early lymphoid progenitors, similar to other AT-rich DNA-binding proteins (20–23). Interestingly, a single nucleotide polymorphism in ARID5B (RefSNP: rs10821936) was demonstrated to increase the risk of MLL-MLLT3/AF9 (17). This finding suggests that genetic susceptibility may be associated with the differences regarding MLL breakpoints and partner genes. Although the role of truncated isoforms of ARID5B in tumorigenesis remains unknown, epigenetic dysregulation may occur in early B progenitor cells in ALL with ARID5B-MLL fusion. Two recent studies clearly demonstrated that reciprocal MLL fusion proteins may have an important role in cancer development (24,25). During follow-up analyses, a large collection of reciprocal MLL fusions was identified, and ~15% of these were in-frame fusions that may be readily expressed as reciprocal fusion proteins (8). All other characterized reciprocal MLL alleles represented out-of-frame fusions with either a chromosomal locus or a reciprocal translocation partner gene (8). However, even these events allowed the transcription and expression of a 5′-truncated MLL protein, termed MLL* (26). This truncated version of MLL has no ability to bind Menin1, lens epithelium-derived growth factor or MYB, but it is able to carry out all enzymatic functions necessary to execute H4K16 acetylation events by associating with MOF or H3K4 methylation via the SET domain complex (27). Together with these results, the findings of the present report suggested that reciprocal MLL fusion proteins, including ARID5B-MLL, may inhibit early B-cell development and have oncogenic functions. Additional functional studies are required to understand the role of these fusion genes in the development of MLL-associated infantile leukemia.
Acknowledgements
The authors of the present report would like to thank Ms. Masayo Matsumura and Ms. Yin Yi, Department of Pediatrics, University of Tokyo (Tokyo, Japan), or special technical assistance. The present study was supported by KAKENHI of Japan Society of Promotion of Science (grant nos. 25253095 and 26293242), Research on Measures for Intractable Diseases, Health and Labor Sciences Research Grants, the Ministry of Health, Labor and Welfare, Research on Health Sciences Focusing on Drug Innovation, the Japan Health Sciences Foundation, Core Research for Evolutional Science and Technology, Japan Science and Technology Agency and the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT) (grant no. 886695).
Glossary
Abbreviations
- ALL
acute lymphoblastic leukemia
- RT-PCR
reverse transcription-polymerase chain reaction
- FISH
fluorescence in situ hybridization
References
- 1.Felix CA. Secondary leukemias induced by topoisomerase targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/S0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Chen A, Yan XM, Huang G. Disordered epigenetic regulation in MLL-related leukemia. Int J Hematol. 2012;96:428–437. doi: 10.1007/s12185-012-1180-0. [DOI] [PubMed] [Google Scholar]
- 3.Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 4.Hiwatari M, Taki T, Taketani T, Taniwaki M, Sugita K, Okuya M, Eguchi M, Ida K, Hayashi Y. Fusion of an AF4-related gene, LAF4, to MLL in childhood acute lymphoblastic leukemia with t(2;11)(q11;q23) Oncogene. 2003;22:2851–2855. doi: 10.1038/sj.onc.1206389. [DOI] [PubMed] [Google Scholar]
- 5.Chaplin T, Ayton P, Bernard OA, Saha V, Valle V Della, Hillion J, Gregorini A, Lillington D, Berger R, Young BD. A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 6.Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, Neale G, Scheet P, Burchard EG, Torgerson DG, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30:751–757. doi: 10.1200/JCO.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Limbergen H, Poppe B, Janssens A, De Bock R, De Paepe A, Noens L, Speleman F. Molecular cytogenetic analysis of 10;11 rearrangements in acute myeloid leukemia. Leukemia. 2002;16:344–351. doi: 10.1038/sj.leu.2402397. [DOI] [PubMed] [Google Scholar]
- 8.Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, de Oliveira M Pombo, Renneville A, Villarese P, Macintyre E, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27:2165–2176. doi: 10.1038/leu.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashihara E, Nakamura S, Inaba T, Taki T, Hayashi Y, Shimazaki C. A novel AF10-CALM fusion transcript in gamma/delta-T cell type lymphoblastic lymphoma. Am J Hematol. 2007;82:859–860. doi: 10.1002/ajh.21021. [DOI] [PubMed] [Google Scholar]
- 10.Silliman CC, McGavran L, Wei Q, Miller LA, Li S, Hunger SP. Alternative splicing in wild-type AF10 and CALM cDNAs and in AF10-CALM and CALM-AF10 fusion cDNAs produced by the t(10;11)(p13-14;q14-q21) suggests a potential role for truncated AF10 polypeptides. Leukemia. 1998;12:1404–1410. doi: 10.1038/sj.leu.2401109. [DOI] [PubMed] [Google Scholar]
- 11.Saha V, Chaplin T, Gregorini A, Ayton P, Young BD. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins; Proc Natl Acad Sci USA; 1995; pp. 9737–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumon K, Kobayashi H, Maseki N, Sakashita A, Sakurai M, Tanizawa A, Imashuku S, Kaneko Y. Mixed-lineage leukemia with t(10;11)(p13;q21): An analysis of AF10-CALM and CALM-AF10 fusion mRNAs and clinical features. Genes Chromosomes Cancer. 1999;25:33–39. doi: 10.1002/(SICI)1098-2264(199905)25:1<33::AID-GCC5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Mroczkowski HJ, Arnold G, Schneck FX, Rajkovic A, Yatsenko SA. Interstitial 10p11.23-p12.1 microdeletions associated with developmental delay, craniofacial abnormalities, and cryptorchidism. Am J Med Genet A 164A. 2014:1–2626. doi: 10.1002/ajmg.a.36627. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DM, George R, Noyes MB, Rowton M, Liu W, Jiang R, Wolfe SA, Wilson-Rawls J, Rawls A. Characterization of the DNA-binding properties of the Mohawk homeobox transcription factor. J Biol Chem. 2012;287:35351–35359. doi: 10.1074/jbc.M112.399386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emerenciano M, Barbosa TC, Lopes BA, Blunck CB, Faro A, Andrade C, Meyer C, Marschalek R, Pombo-de-Oliveira MS, Brazilian Collaborative Study Group of Infant Acute Leukemia ARID5B polymorphism confers an increased risk to acquire specific MLL rearrangements in early childhood leukemia. BMC Cancer. 2014;14:127. doi: 10.1186/1471-2407-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, Sheridan E, Gast A, Kinsey SE, Lightfoot T, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115:1765–1767. doi: 10.1182/blood-2009-09-241513. [DOI] [PubMed] [Google Scholar]
- 19.Patsialou A, Wilsker D, Moran E. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33:66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb CF, Bryant J, Popowski M, Allred L, Kim D, Harriss J, Schmidt C, Miner CA, Rose K, Cheng HL, et al. The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Mol Cell Biol. 2011;31:1041–1053. doi: 10.1128/MCB.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Will B, Vogler TO, Bartholdy B, Garrett-Bakelman F, Mayer J, Barreyro L, Pandolfi A, Todorova TI, Okoye-Okafor UC, Stanley RF, et al. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat Immunol. 2013;14:437–445. doi: 10.1038/ni.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, Kouro T, Iida R, Kokame K, Miyata T, et al. The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity. 2013;38:1105–1115. doi: 10.1016/j.immuni.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokota T, Kanakura Y. Role of tissue-specific AT-rich DNA sequence-binding proteins in lymphocyte differentiation. Int J Hematol. 2014;100:238–245. doi: 10.1007/s12185-014-1602-2. [DOI] [PubMed] [Google Scholar]
- 24.Bursen A, Schwabe K, Rüster B, Henschler R, Ruthardt M, Dingermann T, Marschalek R. The AF4.MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115:3570–3579. doi: 10.1182/blood-2009-06-229542. [DOI] [PubMed] [Google Scholar]
- 25.Emerenciano M, Kowarz E, Karl K, de Almeida Lopes B, Scholz B, Bracharz S, Meyer C, Pombo-de-Oliveira MS, Marschalek R. Functional analysis of the two reciprocal fusion genes MLL-NEBL and NEBL-MLL reveal their oncogenic potential. Cancer Lett. 2013;332:30–34. doi: 10.1016/j.canlet.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 26.Kowarz E, Burmeister T, Lo Nigro L, Jansen MW, Delabesse E, Klingebiel T, Dingermann T, Meyer C, Marschalek R. Complex MLL rearrangements in t(4;11) leukemia patients with absent AF4.MLL fusion allele. Leukemia. 2007;21:1232–1238. doi: 10.1038/sj.leu.2404686. [DOI] [PubMed] [Google Scholar]
- 27.Scharf S, Zech J, Bursen A, Schraets D, Oliver PL, Kliem S, Pfitzner E, Gillert E, Dingermann T, Marschalek R. Transcription linked to recombination: A gene-internal promoter coincides with the recombination hot spot II of the human MLL gene. Oncogene. 2007;26:1361–1371. doi: 10.1038/sj.onc.1209948. [DOI] [PubMed] [Google Scholar]