Abstract
While earlier studies of Apaf-1 holo-apoptosome architecture revealed the spectacular heptameric wheel-like structure formed by Apaf-1, the central CARD disk responsible for caspase-9 recruitment remained incompletely resolved. In a recent issue of Structure, Su et al. (2017) describe a crystal structure of the complex between Apaf-1 CARD and caspase-9 CARD. Together with two recent cryo-EM structures, this work brings us closer to a full view of the holo-apoptosome.
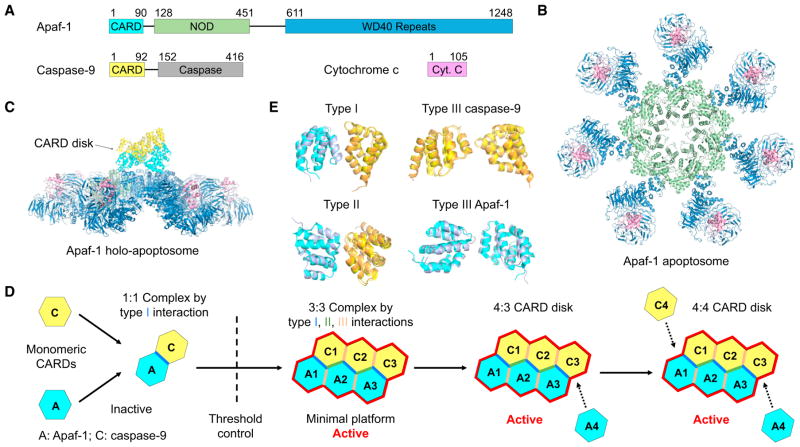
The human apoptosome is the central platform for caspase-9 activation in response to mitochondrial damage and cytochrome c release in the intrinsic apoptosis pathway, which is critically important for embryonic development and the maintenance of cellular homeostasis. The human holo-apoptosome contains Apaf-1, cytochrome c, and caspase-9 (Figure 1A), and previous cryo-EM structures have established that the nucleotide-binding oligomerization domain (NOD) of Apaf-1 mediates heptameric oligomerization, while its tryptophan-aspartic acid (WD40) domain interacts with cytochrome c (Yuan and Akey, 2013) (Figure 1B). However, understanding how its caspase recruitment domain (CARD) brings in caspase-9 via CARD/ CARD interactions to form the holo-apoptosome remained unclear, even in the context of what we have learned on helical oligomerization of the death domain (DD)-fold family, of which CARD is a member (Ferrao and Wu, 2012).
Figure 1. CARD Disk in the Apaf-1 Apoptosome.
(A) Domain architectures of Apaf-1, caspase-9, and cytochrome c.
(B) Top view of the cryo-EM structure of the Apaf-1 apoptosome (PDB ID: 3JBT).
(C) Side view of the Apaf-1 holo-apoptosome showing the 4:4 Apaf-1:caspase-9 CARD disk above the heptameric wheel-like structure (PDB ID: 5JUY).
(D) A schematic of structurally captured steps in CARD disk assembly and activation of Apaf-1 holo-apoptosome.
(E) Superimposed type I, II, and III (for both Apaf-1:Apaf-1 and caspase-9:caspase-9) interactions between those in the crystal structure (PDB ID: 5WVC, Apaf-1 in slate and caspase-9 in orange) and the cryo-EM structure (PDB ID: 5JUY, Apaf-1 in cyan and caspase-9 in yellow).
The helical oligomerization typically involves three types of asymmetric interactions, type I, II, and III, which cooperate to generate homo- and hetero-oligomers from relatively small assemblies to open-ended filaments. The first crystal structure of the Apaf-1CARD/caspase-9CARD complex revealed a 1:1 asymmetric complex that uses type I interface (Qin et al., 1999). However, cryo-EM structure of the holo-apoptosome revealed an oligomeric CARD disk above the heptameric apoptosome ring (reviewed in Yuan and Akey, 2013), and the molar ratio between Apaf-1 and caspase-9 was estimated to be around 7:4 (Hu et al., 2014). Moreover, another crystal structure of the Apaf-1CARD/caspase-9CARD complex presented a trimeric assembly containing 2 Apaf-1CARD and 1 caspase-9CARD, assembled using all three types of interfaces (Hu et al., 2014). Extensive mutagenesis further confirmed that all type I, II, and III interfaces are crucial for caspase-9 activation by the apoptosome (Hu et al., 2014). Nonetheless, the 2:1 Apaf-1CARD/caspase-9CARD complex structure is still too small to explain the size of the CARD disk, leaving the nature of the CARD assembly unresolved.
In a recent issue of Structure, the Lin group presented the crystal structure of a 3:3 Apaf-1CARD/caspase-9CARD complex (Su et al., 2017). Almost concurrently, the Akey (Cheng et al., 2016) and Shi (Li et al., 2017) groups reported two independently determined cryo-EM structures of the CARD disk in the context of the holo-apoptosome (Figure 1C), which were refined without imposing the seven-fold symmetry and reached sufficient resolution to visualize α helices in the CARDs.
Collectively, these three studies reveal a novel helical assembly of the CARD disk involving a core 3:3 arrangement with a final 4:3 or 4:4 Apaf-1CARD/cas-pase-9CARD complex and implicate a unique assembly pathway for the formation of the CARD disk (Figure 1D). First, Apaf-1CARD does not oligomerize in the absence of caspase-9CARD, as evidenced by the presence of CARD disk density only in the holo-apoptosome (Yuan and Akey, 2013). The crystal structure and the associated biochemical data clearly established that in solution, Apaf-1CARD and caspase-9CARD initially form a 1:1 complex, which at higher concentrations is further oligomerized into a 3:3 complex (Su et al., 2017) (Figure 1D). The 1:1 Apaf-1CARD/caspase-9CARD complex, but not the 3:3 oligomer, is stable at high salt (Qin et al., 1999; Su et al., 2017), justifying the lack of a higher-order structure in the initial crystal obtained under 1.75 M phosphate despite the high concentration used in crystallization (Qin et al., 1999). Second, given the ability of many DD-fold proteins to form open-ended, heterogeneous oligomeric mixtures, it is surprising that the Apaf-1CARD/caspase-9CARD complex stops oligomerizing at 3:3. The Lin group argued that this is because the complex assembles from the stable 1:1 Apaf-1CARD/caspase-9CARD “protomer,” and the helical rise per protomer is too short, so that the 4th protomer would clash with the first protomer. The 3:3 complex then forms the “open jump ring” or “split ring” architecture in the words of the authors (Su et al., 2017). A similar size restriction mechanism dubbed “lock washer” was seen in the tetrameric structure of the tandem CARD of RIG-I (Yin et al., 2015). Third, the 3:3 complex essentially represents the core of the 4:3 and 4:4 Apaf-1CARD/caspase-9CARD CARD disk in the context of the holo-apoptosome (Cheng et al., 2016; Li et al., 2017). The 4th Apaf-1CARD is not assembled as an Apaf-1CARD/caspase-9CARD protomer, but is brought into the CARD disk as a low affinity subunit at the high Apaf-1 concentration in the apoptosome (Figure 1D). Linkers connecting 4 Apaf-1 CARDs in the CARD disk to their cognate Apaf-1 molecules are partially visible (Cheng et al., 2016), further demonstrating the correctness of the composition. Similarly, the 4th caspase-9CARD is observed only in some holo-apoptsome cryo-EM maps, and must also represent a low affinity subunit, which is mediated by a type II interaction with Apaf-1 and type I and III interactions with other caspase-9CARD subunits in the CARD disk (Figure 1D).
Although apoptosomes are found from invertebrates to mammals in response to death signals, structural studies of the Dark apoptosome in the fly D. melanogaster and the CED-4 apoptosome in the nematode C. elegans revealed a double octameric wheel-like structure and an octameric dome-like structure, respectively (Yuan and Akey, 2013; Pang et al., 2015). While eight versus seven subunits in these apoptosomes may represent a certain degree of divergence at their respective NODs, the CARD disks may be much more divergent among these complexes. In the Dark apoptosome with the caspase Dronc, the CARD disk contains eight pairs of heterodimeric DarkCARD/DroncCARD complexes assembled via the type II interface located between the two octameric wheels (Pang et al., 2015). In the CED-4 apoptosome, a two-layered, 4-fold symmetric octameric CARD disk is formed via type II interfaces between the top and bottom layers (Yuan and Akey, 2013), but the mode of recruitment of CED-3 is currently unknown. Despite the variable structural architecture, the universal presence of the type II interaction in Dark and CED-4, as well as the observed type II interaction in the PelleDD/TubeDD complex from D. melanogaster, led the Lin group to propose that the type II interface may have appeared first in the evolution of the DD-fold family (Su et al., 2017); only in higher organisms do true helical complexes using all three types of interactions become more prevalent, as seen in almost every signaling pathway in mammalian innate immunity (Yin et al., 2015).
In the holo-apoptosome, recruitment of caspase-9 may occur before oligomerization in the CARD disk, which presumably brings the caspase domain into proximity for their dimerization and activation. As pointed out by the Lin group, this scenario is quite different from most of the mammalian DD-fold complexes, in which upon stimulation, the upstream DD-fold protein can often form an oligomeric platform before recruitment of the downstream DD-fold protein (Su et al., 2017). In fact, unlike caspase-9, many of the downstream DD-folds can oligomerize on their own, for example, in the filaments of caspase-1CARD (Lu et al., 2016), and auto-inhibition of these proteins may be required to keep spontaneous activation at bay.
Although the 3:3 crystal structure represents the core of the 4:3 or 4:4 CARD disk from the Apaf-1 holo-apoptosome, small differences do exist in the exact molecular orientations that relate the pairs of CARDs in the type I, II, and III interactions (Figure 1E). This situation is reminiscent of the filament structure of the GFP-fused pyrin domain (PYD, a member of the DD-fold family) of AIM2, in which the GFP fusion may have tweaked the interfaces, leading to a different symmetry of the filament, yet maintaining its apparent stability and the ability to interact with ASCPYD (Lu et al., 2015). This structural plasticity may be intrinsic to DD-fold assemblies and may facilitate gene-duplication-mediated evolution from one primordial interaction pair to other interactions.
Structural elucidation of the CARD disk of the holo-apoptosome also raises additional questions. For example, it remains controversial why Apaf-1 does not use all seven CARDs to bind caspase-9CARD, and where the remaining Apaf-1 CARDs localize. The Shi group found an additional 2:1 Apaf-1CARD/ caspase-9CARD complex that binds at the periphery of the central heptameric hub (Li et al., 2017), while the Akey group found density at a similar location on the central hub, which was better fit by the catalytic domain of caspase-9, with the caveat that this density was at lower resolution (Cheng et al., 2016; Yuan and Akey, 2013). It is tempting to speculate that evolution of these systems has taken a path of least resistance, where assemblies that worked sufficiently well were selected although they may not have been the most efficient. A reductionist might reason that despite all the different architectures in apoptosomes and other molecular machines assembled by DD-folds, the oligomeric scaffolds all increase the local protein concentration to promote activation. In sum, these studies suggest that DD-folds may use diverse biophysical mechanisms to achieve their biological functions. It is therefore our job to dive deeper into DD-folds and unravel not only additional CARD tricks in apoptosomes, but also processes used by other versatile molecular machines of the DD-fold family.
References
- Cheng TC, Hong C, Akey IV, Yuan S, Akey CW. Elife. 2016;5:5. doi: 10.7554/eLife.17755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrao R, Wu H. Curr Opin Struct Biol. 2012;22:241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Wu D, Chen W, Yan Z, Yan C, He T, Liang Q, Shi Y. Proc Natl Acad Sci USA. 2014;111:16254–16261. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou M, Hu Q, Bai XC, Huang W, Scheres SH, Shi Y. Proc Natl Acad Sci USA. 2017;114:1542–1547. doi: 10.1073/pnas.1620626114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, Wu H. Cell Discov. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Li Y, Schmidt FI, Yin Q, Chen S, Fu TM, Tong AB, Ploegh HL, Mao Y, Wu H. Nat Struct Mol Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Bai XC, Yan C, Hao Q, Chen Z, Wang JW, Scheres SH, Shi Y. Genes Dev. 2015;29:277–287. doi: 10.1101/gad.255877.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- Su TW, Yang CY, Kao WP, Kuo BJ, Lin SM, Lin JY, Lo YC, Lin SC. Structure. 2017;25:407–420. doi: 10.1016/j.str.2016.12.019. [DOI] [PubMed] [Google Scholar]
- Yin Q, Fu TM, Li J, Wu H. Immunology. 2015;33:393–416. doi: 10.1146/annurev-immunol-032414-112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Akey CW. Structure. 2013;21:501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]