Abstract
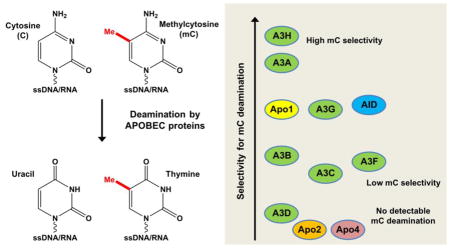
APOBECs are a family of cytidine deaminases involved in various important biological processes such as antibody diversification/maturation, restriction of viral infection, and generation of somatic mutations. Catalytically active APOBEC proteins execute their biological functions mostly through deaminating cytosine (C) to uracil on ssDNA/RNA. Activation-induced cytidine deaminase (AID), one of the APOBEC members, was reported to deaminate methylated cytosine (mC) on DNA and this mC deamination was proposed to be involved in demethylation of mC for epigenetic regulation. The mC deamination activity is later demonstrated for APOBEC3A (A3A), and more recently for APOBEC3B (A3B) and APOBEC3H (A3H). Despite extensive studies on APOBEC proteins, questions regarding whether the rest of APOBEC members have any mC deaminase activity and what are the relative deaminase activities for each APOBEC member remain unclear. Here we performed a family-wide analysis of deaminase activities on C and mC by using purified recombinant proteins for eleven known human APOBEC proteins under similar conditions. Our comprehensive analyses revealed each APOBEC has unique deaminase activity and selectivity for mC. A3A and A3H showed distinctively high deaminase activities on C and mC with relatively high selectivity for mC, whereas six other APOBEC members showed relatively low deaminase activity and selectivity for mC. Our mutational analysis showed that loop-1 of A3A is responsible for its high deaminase activity and selectivity for mC. These findings extend our understanding of APOBEC family proteins that have important roles in diverse biological functions as well as in genetic mutations.
Graphical abstract
Introduction
The human genome encodes eleven members in the APOBEC (“apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like”) family. APOBEC proteins share a conserved motif of the cytidine deaminase active site [1, 2], and have a typical 3D-fold composed of five beta-stranded core surrounded with six helices [3–12]. The catalytically active APOBECs induce mutations on ssDNA or RNA by editing cytosine (C) to uracil. APOBEC proteins play important roles in both acquired and innate immune systems [13–21]. For example, activation-induced cytidine deaminase (AID) induces somatic hypermutations by deaminating cytosines in the immunoglobulin locus of the maturing B cells and trigger antibody class-switch recombination, a critical process for antibody diversification and maturation for humoral immunity [22–26]. Genetic defects in AID cause Type-2 Hyper-IgM Syndrome (HIGM2), a disease related to immunodeficiency [27]. APOBEC1 (Apo1) edits the mRNA encoding apolipoprotein B (ApoB) to introduce an early stop codon and produce a truncated ApoB for lipid transport [28]. The APOBEC family has seven APOBEC3 (A3) subfamily members (A3A–H) that are involved in controlling intrinsic retroelements in the genome and restricting external viral infection and replication [14–21]. The viruses currently known to be restricted by A3 proteins include human immunodeficiency virus (HIV), hepatitis B virus (HBV), and human papillomavirus (HPV) [13, 29–39]. Unlike other APOBEC members, APOBEC2 (Apo2) and APOBEC4 (Apo4) appear to exhibit no deaminase activity or mutagenic activity in yeast and bacteria based mutator assays or in vitro tests [3, 40, 41], and their functions remain to be clarified.
In addition to the canonical deamination of cytosine, four APOBEC members, including AID, and A3A, and more recently A3B, and A3H have been demonstrated to deaminate methylated cytosine (mC) [42–51]. AID is the first APOBEC shown to deaminate mC to thymine [42, 43]. Multiple studies suggest that AID activity on mC deamination may be a part of DNA demethylation pathway in epigenetic regulation for stem cell maintenance and cell reprogramming [52, 53], even though such role of AID in demethylation of mC in genomic DNA remains controversial [45, 54]. Nonetheless, subsequent reports over recent years show that three other APOBECs, A3A, A3B and A3H deaminate mC to varying degrees [46–51]. In contrast, another APOBEC protein, A3G, has undetectable mC deamination activity [46, 47], and it is currently not well defined whether the rest of the APOBEC members have any mC deamination activity.
In order to clearly define the activity of mC deamination and to provide a comparative view of the C and mC deaminase activities for the entire APOBEC family members, we purified the recombinant proteins of eleven known APOBEC members and examined their deaminase activities on both C and mC. We found that nine APOBECs are active in deaminating C, and eight of nine active APOBECs showed detectable mC deaminase activity under our experimental conditions. The activities of A3A and A3H are distinctively high in both C and mC deamination, with over 3–4 orders of magnitude higher than the rest of the APOBEC members under similar assay conditions. When the relative mC deaminase activity (or mC selectivity factor) was considered, A3A, A3H, and AID have mC selectivity factor of over 10, with A3A and A3H reaching as high as 29.8 and 53.0, respectively. Through mutational analysis of A3A, we identified loop-1 region of A3A to be critical for its high deaminase activity and high mC selectivity. This family-wide study using purified recombinant proteins provides the first side-by-side comparative deaminase activities on C and mC. The results would be valuable for future comprehensive understanding of the broad biological functions of the APOBEC family.
APOBEC Protein expression and purification
We cloned eleven known APOBEC family proteins as codon-optimized genes for E. coli expression. While A3A was highly soluble when expressed with a short His-tag. However, the other APOBECs were less soluble or mostly insoluble with the His-tag. Those insoluble APOBEC proteins became much more soluble when expressed as fusion proteins with either maltose binding protein (MBP) or glutathione S-transferase (GST), and their MBP- or GST fusion proteins were purified through corresponding affinity column chromatography (Fig. 1A, 1B). A3H has seven different haplotypes (hap I–VII) [18, 55, 56], and the representative hap II (referred to as A3H throughout the text) was used in this study. For all APOBEC members, their inactive mutant proteins with the catalytic center glutamate replaced with alanine (E to A) were prepared as negative controls for deamination assays (Fig. 1C). SDS-PAGE analysis showed that the purified proteins of the wild-type APOBECs and their E to A mutants contained little detectable contaminating proteins (Fig. 1B, 1C). These purified recombinant APOBEC proteins were used for the following activity assays.
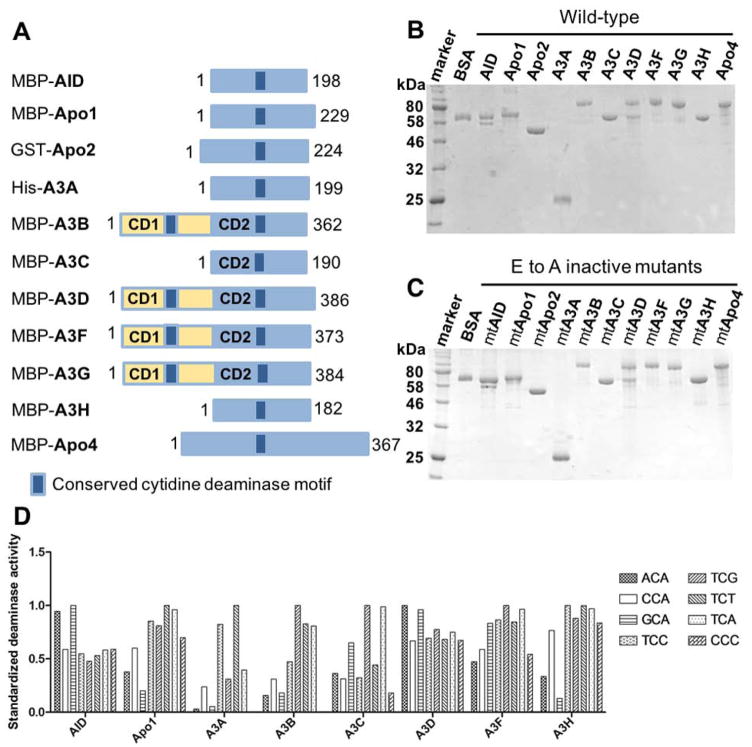
Figure 1. Protein purification and deaminase activity on various tri-nucleotide motifs of eleven APOBEC members.
(A) Schematic representation of the constructs for the eleven members of APOBEC family containing one or two Zn-coordinating conserved catalytic motifs (dark blue box) used in this study. The N-terminal fusion tags (His, GST, or MBP) for each purified APOBEC are indicated. (B, C) SDS-PAGE analyses of the purified proteins for the eleven wild-type (wt) (B) and catalytically inactive E to A active site mutants (C) of APOBECs with N-terminal affinity tags. APOBEC proteins were purified from the soluble fraction of E. coli cell lysate in lysis buffer (20 mM Tris-HCl, pH 8.0, 250 mM NaCl, and 2 mM DTT) by affinity column chromatography as previously described [50]. (D) Deaminase activity of the purified wt APOBECs on 8 different tri-nucleotide substrate motifs. The motifs include NCA and TCN, where N is any of the four nucleotides, as well as a CCC motif. The proteins not shown on the chart are Apo2 and Apo4 that had no detectible activity and A3G that has been shown to deaminate CCC as a preferred motif in our previous publications [4, 74]. Deaminase assay was performed as previously described [49, 50, 62]. Briefly, APOBECs were incubated with 600 nM 5′ 6FAM-labeled ssDNA (30 nt) in deamination buffer (25 mM HEPES, pH 6.5, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and 0.1 μg/ml RNase A) at 37°C for 2 hours. The reactions were terminated by heat inactivation at 90°C for 5 min. The deamination product uracil bases were subsequently cleaved by incubating with UDG (2 units, NEB) for 1 hr at 37°C The abasic sites were then hydrolyzed with 0.1 M NaOH at 90°C for 10 min. Deamination products were analyzed on 20% denaturing PAGE gels, and quantified with Molecular Imager FX Pro Plus System/Quantity One® analysis software (Bio-Rad). The activity shown for each APOBEC on the chart was normalized by dividing the activity on each motif by the maximum activity among the eight substrates (the most preferred motif).
Deaminase activity of purified APOBECs on various DNA motifs
It is known that different APOBEC proteins have specific substrate preferences for certain tri-nucleotide sequences. To evaluate the deaminase activity of the purified APOBEC proteins from E. coli, eight different tri-nucleotide sequence motifs (Fig. 1D inset), each embedded within a 30 nucleotide (nt) ssDNA, were used for the deamination assay. Nine of eleven APOBEC members had detectable deaminase activity on various tri-nucleotide motifs, and the results from the eight catalytically active APOBEC members are shown in Fig. 1D. Note that A3G was excluded from the chart as deaminase activity of A3G was confirmed by using a substrate containing the known -CCC- motif (data not shown). Each APOBEC member had distinctive substrate preference for tri-nucleotide sequence, and they are largely consistent with the previously reported sequence motif preferences, some of which were identified by analyzing DNA mutations in cell-based assays [4, 47, 49, 50, 57–62]. As expected, Apo2 and Apo4 had no detectable deaminase activity with the given ssDNA substrates, so as the catalytic E to A mutants of all eleven APOBEC members (data not shown). Based on these results, we chose one preferred tri-nucleotide motif as the substrate for each APOBEC member, which is as follows: -GCA- for AID, -ACA- for A3D, -CCC- for A3G, and -TCA- for all the other members (i.e. Apo1, A3A, A3B, A3C, A3F, A3H) for the subsequent evaluation of deamination. Apo2 and Apo4, together with the catalytic E to A inactive mutants of all APOBEC proteins, were excluded from the following assays.
Effect of pH on deaminase activity of catalytically active APOBECs
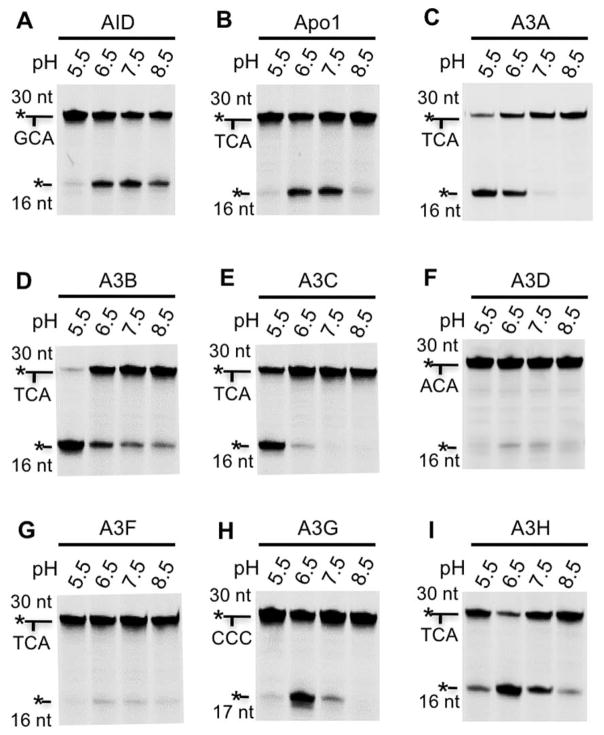
Previous reports suggest that different APOBEC proteins may have different optimum pH for their optimal deamination [63]. Therefore, the deamination activities of the nine catalytically active APOBECs were tested at several pH conditions (pH 5.5, 6.5, 7.5, and 8.5) by using their respective preferred tri-nucleotide motifs described above. The results showed that, while these APOBEC proteins all displayed certain detectable activity between pH 5.5–8.5, they had distinctive optimal pH (Fig. 2). For example, AID showed activity between pH 6.5 to 8.5, with its maximum activity at pH 7.5 (Fig. 2A). Apo1 was most active at both pH 6.5 and pH 7.5 (Fig. 2B). A3A, A3B and A3C all showed highest activity at pH 5.5 among pH conditions tested (Fig. 2C, D, E). Additional activity assay at pH 4.5 and 4.0 for A3B revealed little activity at pH 4.5, and no detectable activity at pH 4.0 (data not shown), thus no further assay was carried out at these pH conditions for A3A and A3C. A3D, A3F and A3G, and A3H showed increased activity at pH 6.5 and 7.5 (Fig. 2F, G, H, I). Based on these results, the pH conditions that yielded the highest activity of C deamination for each APOBEC were used in the following assays. In particular, pH 7.5 was used for AID; pH 6.5 for Apo1, A3D, A3F, A3G, and A3H; pH 5.5 for A3A, A3B, and A3C.
Figure 2. The pH effect on deaminase activity.
The deaminase activity assay of the purified APOBECs were performed at pH 5.5, 6.5, 7.5, and 8.5. 30 nt ssDNA containing the preferred tri-nucleotide motifs for the respective APOBEC members were used. The tri-nucleotide motifs used for each protein, and the positions of the 30 nt substrates and the 16 or 17 nt products are indicated next to each gel image. The deaminase assay was performed as described in Fig. 1D.
mC deamination and the relative activities of mC/C deamination by different APOBECs
By using the optimal substrate motifs and pH conditions determined above, we first performed mC deamination assays for nine catalytically active APOBEC members. The raw gel imaging showed that A3A and A3H had particularly high activity, compared to the other APOBEC members (Supplementary Fig. S1), which is consistent with previous studies [46–50]. On the other hand, relatively weak deamination activity on mC was detected for six other APOBEC members (i.e. AID, Apo1, A3B, A3C, A3F, A3G) when high protein concentrations were used (Supplementary Fig. S1). A3D was the only member that did not show consistently measurable activity for mC under our experimental conditions.
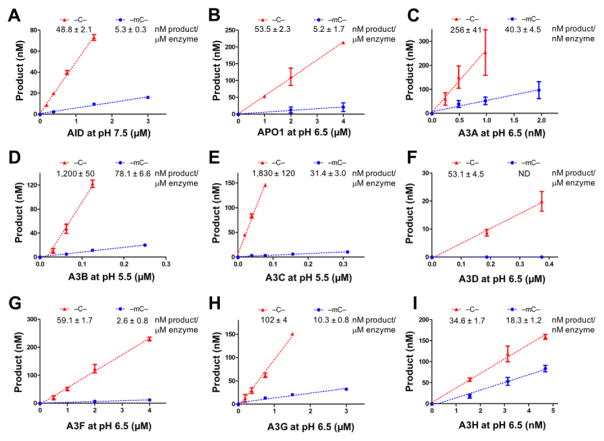
For a quantitative comparison of the deaminase activities on mC and C of different APOBEC members, dose response deaminase assays were performed, where deamination product formation was plotted against enzyme concentration (nM product/μM enzyme) as previously described [50, 51]. The specific activity was calculated from the initial range where product formation is linearly dependent on enzyme concentration for each APOBEC member (Fig. 3, Table 1). Again, except for A3D that did not show measurable mC deaminase activity, the other eight APOBECs showed the dose responsive mC deaminase activity (Fig. 3A–I). Our mC deamination activity of A3G contradicts a previous study that reported no activity on mC [47]. This difference may be the result of different experimental conditions, such as the type of the fusion proteins used in the study and/or the range of protein concentrations tested.
Figure 3. Dose dependent activity for C and mC deamination of APOBECs.
The deaminated products were plotted as a function of increasing concentrations of APOBECs (panels A–I) assayed with substrates containing a target C (red line) or target mC (blue line). The slope of each plot in the linear range is calculated as deaminase activity (nM product/μM enzyme) and listed at the top of each chart. Dose response assays on C/mC were performed as previously described [49, 50, 62]. Briefly, various concentrations of APOBECs at their respective linear range were incubated with 600 nM 5′ 6FAM-labeled ssDNA (30 nt) containing a target C/mC in deamination buffer (25 mM buffer at the indicated pH, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and 0.1 μg/ml RNase A) at 37°C for 2 hours. The reactions were terminated by heat inactivation at 90°C for 5 min. The bases of the deamination products uracil or thymine were subsequently cleaved by incubating with UDG (2 units, NEB) at 37°C for 1 hour or with TDG (2 units) and 3-fold excess amount of the complementary ssDNA at 42°C for 12 hours, respectively. The abasic sites were then hydrolyzed with 0.1 M NaOH at 90°C for 10 min. S.D. was estimated from data collected in three independent experiments.
Table 1.
Deaminase activity on C and mC of all APOBEC proteins assayed at the indicated pH with the preferred substrate motifs.
| Enzyme | C deaminase activity (nM prod/μM enz) | mC deaminase activity (nM prod/μM enz) | mC selectivity factor (mC/C) x 100 | pH | Substrate motifs |
|---|---|---|---|---|---|
| AID | 48.8 ± 2.1 | 5.3 ± 0.3 | 10.9 | 7.5 | GCA/GmCA |
| Apo1 | 53.5 ± 2.3 | 5.2 ± 1.7 | 9.7 | 6.5 | TCA/TmCA |
| Apo2 | – | – | – | – | – |
| A3A | 256,000 ± 41,000 | 40,300 ± 4,500 | 15.7 | 6.5 | TCA/TmCA |
| A3B | 1,200 ± 50 | 78.1 ± 6.6 | 6.5 | 5.5 | TCA/TmCA |
| A3C | 1,830 ± 120 | 31.4 ± 3.0 | 1.7 | 5.5 | TCA/TmCA |
| A3D | 53.1 ± 4.5 | ND | – | 6.5 | ACA/AmCA |
| A3F | 59.1 ± 1.7 | 2.6 ± 0.8 | 4.4 | 6.5 | TCA/TmCA |
| A3G | 102 ± 4 | 10.3 ± 0.8 | 9.9 | 6.5 | CCC/CCmC |
| A3H | 34,600 ± 1,700 | 18,300 ± 1,200 | 53.0 | 6.5 | TCA/TmCA |
| Apo4 | – | – | – | – | – |
Note: Deaminase activity for C and mC was calculated from the initial linear range of dose dependent product yield for each APOBEC protein (Fig. 3). S.D. was estimated from three independent deaminase assay experiments. The mC selectivity factor was calculated as mC/C activity x 100. ND indicates not determined.
We next compared the selectivity for mC among APOBEC members by using mC selectivity factor [mC/C specific activity x 100] as previously defined (Table 1) [49, 50]. The mC selectivity factor for the second most active A3H is 53.0 (Table 1), which is over 3-fold higher than 15.7 for A3A that showed the highest activity on C and mC (Table 1). The mC selectivity factor of AID, which has been previously linked to epigenetic regulation through mC demethylation [52, 53, 64, 65], is 10.9 (Table 1). The mC selectivity factors for the rest of the APOBEC members with detectable mC deamination activity are all less than 10 (Table 1). Interestingly, A3C has the lowest mC selectivity factor of 1.7 despite the fact that it has the third highest C deamination activity of 1,830 nM product/μM enzyme (Table 1). Taken together, each active APOBEC protein has distinctive specificity for mC. Given the fact that mC deamination activity and mC selectivity for A3A and A3H are much higher than those of AID, it raises an intriguing question whether A3A and A3H would be able to deaminate mC on the genomic DNA when these proteins gain access to the nucleus.
Effect of pH on selectivity between mC and C
We tested if different pH affects mC selectivity factor for AID, A3A, A3B, and A3H which displayed relatively high deaminase activity or high mC selectivity. The specific activities obtained at pH 5.5, 6.5 and 7.5 revealed that different pH conditions yielded different mC selectivity factors (Table 2). The mC selectivity factor of AID had the highest value of 10.9 at pH 7.5, which decreases to 5.3 at pH 6.5, and to undetectable level at pH 5.5. The lowered mC selectivity factor of AID at pH 6.5 is due to both the increased C deamination as well as the decreased mC deamination activity. Similarly, A3H showed pH dependence for mC selectivity factor, varying between 33.7–53.0 within pH 5.5–7.5, with the highest mC selectivity factor observed at pH 6.5 (Table 2). Interestingly, A3A showed the highest mC selectivity factor of 29.8 at pH 5.5, which is almost 2-fold higher that the 15.7 at pH 6.5. In this case, the increased mC selectivity factor at pH 5.5 is mainly because of the increased mC deaminase activity, as there is little change in C deaminase activity under these two pH conditions (Table 2). The physiological significance for A3A and A3H to have strong deamination activity at lower pH is unclear. Nonetheless, A3A from several different mammalian species were shown to deaminate both mC and C residues [66], and A3A is proposed to play a role in DNA catabolism [66, 67]. One possibility for A3A to function at lower pH could help to clear out foreign junk DNA that may end up in lysosomes with acidic environment.
Table 2.
Ddeaminase activity of AID, A3A, A3B, and A3H on C and mC and the resulting mC selectivity factor obtained at three different pH.
| Enzyme | C deaminase activity (nM prod/μM enz) | mC deaminase activity (nM prod/μM enz) | mC selectivity factor (mC/C) x 100 | pH | Substrate motifs |
|---|---|---|---|---|---|
| AID | 6.6 ± 0.2 | ND | – | 5.5 | GCA/GmCA |
| 70.9 ± 2.1 | 3.7 ± 0.4 | 5.2 | 6.5 | ||
| 48.8 ± 2.1 | 5.3 ± 0.3 | 10.9 | 7.5 | ||
| A3A | 260,000 ± 3,000 | 77,400 ± 1,500 | 29.8 | 5.5 | TCA/TmCA |
| 256,000 ± 40,600 | 40,300 ± 4,500 | 15.7 | 6.5 | ||
| 82,800 ± 4,200 | 9,850 ± 620 | 11.9 | 7.5 | ||
| A3B | 1,200 ± 50 | 78.1 ± 6.6 | 6.5 | 5.5 | TCA/TmCA |
| 240 ± 6 | 10.6 ± 0.4 | 4.4 | 6.5 | ||
| 103 ± 5 | ND | - | 7.5 | ||
| A3H | 4,920 ± 440 | 1,660 ± 100 | 33.7 | 5.5 | TCA/TmCA |
| 34,600 ± 1,700 | 18,300 ± 1,200 | 53.0 | 6.5 | ||
| 10,900 ± 1,500 | 4,600 ± 1,660 | 42.1 | 7.5 |
Note: Deaminase activity for C and mC was calculated from the initial linear range of dose dependent product formation for each APOBEC protein at the specified pH. S.D. was estimated from three independent deaminase assay experiments. The mC selectivity factor was calculated as mC/C activity x 100. ND indicates not determined.
Identification of A3A regions important for the deaminase activity and mC selectivity
A3A and A3H showed the highest deaminase activities on both C and mC as well as mC selectivity factors among the APOBEC family members. Even though C deaminase activity of A3A is about one order of magnitude higher than that of A3H, mC deaminase activity of A3A and A3H are similar level (Table 2), making A3H to have higher mC selectivity factor. Given the relatively high sequence homology among APOBEC family members, an intriguing question is which region(s)/sequences of these APOBECs are important for determining the efficiency of deaminase activity and mC selectivity. In order to address this question, we focused on A3A by mutating different regions of its sequence to examine their effects on the general deaminase activity and the mC selectivity.
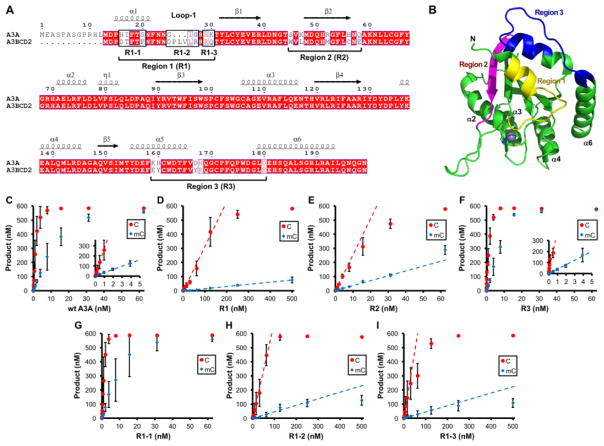
A previous study on the active A3BCD2 domain has shown that the residues around its α1/loop-1 play important roles in determining the overall deaminase activity and the mC selectivity [49]. Mutating certain loop-1 residues of A3BCD2 generate mutants that gained about 100 times in the deaminase activity on C and mC, and increased the mC selectivity factor by about 8 times [49]. Here in order to identify which region(s) of A3A is important for its high deaminase activity and relatively high mC selectivity, three different regions of A3A were replaced with the corresponding sequences from its close homolog A3BCD2 (Fig. 4A, 4B), and C and mC deaminase activity of the three region mutants, R1, R2, and R3, were examined (Fig 4C–F, Supplementary Fig. S2). Among three region mutants, only R1 mutant of A3A showed both reduced deaminase activity and mC selectivity (Fig. 4C–F, Table 3). The reduction of C and mC deaminase activity were about 85-fold and 200-fold, respectively, lowering the mC selectivity factor to 4.8 (Table 3). On the other hand, R2 showed relatively minor effects, with about 10-fold reduction for both C- and mC-deamination activity, and thus maintaining a similar mC selectivity factor of 13.5. R3 had no obvious effects on the deaminase activity and the mC selectivity (Table 3). These results clearly indicate that, among the three regions showing sequence variability between A3A and A3BCD2, region 1 of A3A (α1/loop-1) is critical for its high deaminase activity and high mC selectivity.
Figure 4. Identification of A3A regions important for deaminase activity and mC selectivity.
(A) Sequence alignment of A3A and A3B CD2. The three regions (Region 1–3) of A3A that show sequence differences with A3B CD2 are mutated to the amino acids of A3B CD2 to generate mutants R1, R2 and R3. (B) The three mutated regions (with distinct colors) are mapped onto A3A structure (5SWW) [75], in which the active center Zn and its coordinating histidine and cysteines are shown as a sphere and sticks, respectively. Only region 1 (loop-1 region) is next to the Zn-active center. Region 2 is on the opposite end of the active center, and region 3 is on loop 10 that is also far away from the active center. (C–I) Dose dependent activity assay for C and mC deamination for wt A3A and the six A3A mutants (R1, R2, R3, R1-1, R1-2, R1-3) shown in panel-A. Dose response deaminase assays were performed as described in Fig. 3 legends. S.D. was estimated from data collected in three independent experiments.
Table 3.
Deaminase activity on C and mC of wt and mutant A3As.
| Enzyme | C deaminase activity (nM prod/μM enz) | mC deaminase activity (nM prod/μM enz) | mC selectivity factor (mC/C) x 100 | |
|---|---|---|---|---|
| A3A | wt | 266,000 ± 6,000 | 33,900 ± 400 | 12.7 |
| R1 | 3,180 ± 150 | 153 ± 3 | 4.8 | |
| R2 | 25,500 ± 600 | 3,440 ± 70 | 13.5 | |
| R3 | 322,000 ± 79,000 | 42,300 ± 1,100 | 13.1 | |
| R1-1 | 352,000 ± 14,000 | 42,200 ± 800 | 12.0 | |
| R1-2 | 6,400 ± 480 | 391 ± 19 | 6.1 | |
| R1-3 | 9,430 ± 350 | 444 ± 7 | 4.7 | |
Note: Deaminase activity for C and mC was calculated from the initial linear range of dose dependent product formation for each A3A variant (Fig. 4C–I). S.D. was estimated from data collected in three independent deaminase assay experiments. The mC selectivity factor was calculated as mC/C activity x 100.
To identify the specific residues around region 1 (α1/loop-1) that are important for the high deaminase activity and mC selectivity of A3A, we mutated a subset of residues around region 1 to generate A3A mutants R1-1 (H16D/I17T/S20F), R1-2 (replacing 25GIG27 with DPLVLR), and R1-3 (H29R/K30Q) (Fig. 4A, Supplementary Fig. S2). The activity assay showed that mutant R1-1 had no significant change in deaminase activity on both C and mC (Fig 4G, Table 3). Interestingly, mutant R1-2 showed a ~42-fold reduction in C deamination, and a more pronounced 87-fold reduction in mC deamination, lowering the mC selectivity factor to 6.1 (Fig 4H, Table 3). Similarly, mutant R1-3 displayed a 28-fold reduction for C deamination, and a more pronounced 76-fold reduction for mC deamination, reducing mC selectivity factor to 4.7 (Fig 4I, Table 3). These results indicate that the residues mutated in both R1-2 and R1-3, but not in R1-1, are important for the high deaminase activity and mC selectivity of A3A. The effects of the mutated residues in R1-2 and R1-3 on deaminase activity appear to be additive, because the reduction of the deaminase activity of mutants R1-2 and R1-3 was less severe than that of mutant R1 that contains the mutations in both R1-2 and R1-3 (Fig. 4A, Table 3). The loss of function effect observed here for A3A by replacing residues on loop-1 corroborates well with a prior report that suggested a similar important role of the loop-1 of A3BCD2 through a gain of function effect [49].
ssDNA binding and motif specificity of R1 mutant of A3A
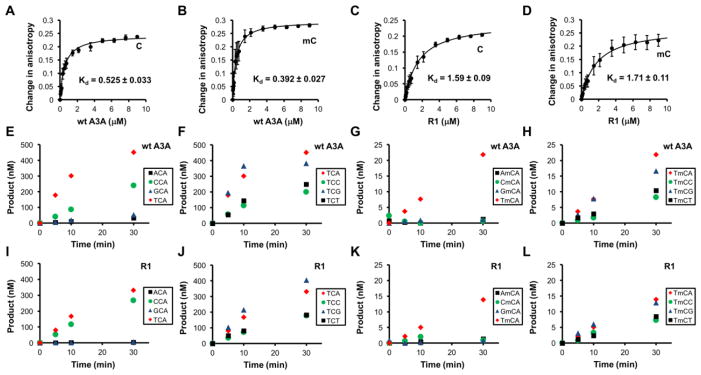
To address the question whether the decreased deaminase activity and mC selectivity of A3A loop-1 mutant R1 could be caused by altered ssDNA substrate binding, we measured the ssDNA binding affinity by rotational anisotropy with 30 nt FAM-labeled ssDNA containing 5′-TCA or 5′-TmCA. The changes in rotational anisotropy with increasing concentrations of proteins were fitted to a simple one-site specific binding model (Fig. 5A–D). The wt A3A showed a similar affinity for both C and mC ssDNA substrates with Kd of 0.525 μM and 0.392 μM, respectively (Fig. 5A, 5B). R1 mutant showed about 3–4 fold lower affinity for C and mC substrates with Kd of 1.59 μM and 1.71 μM, respectively (Fig. Fig 5C, 5D). These results suggest that such reduction of DNA binding affinity may provide partial explanation for the significant decrease of the overall deaminase activity (85-fold decrease in C deamination and the 200-fold decrease in mC deamination). Other factors, such as the orientation of target C and mC at the active center pocket, may also affect the deaminase activity and the mC selectivity of the R1 mutant.
Figure 5. Comparison of the substrate binding affinity and substrate motif specificity of wt and R1 mutant of A3A.
(A–D) Substrate ssDNA binding of wt and R1 mutant. Binding of proteins to FAM-labeled 30-nt ssDNA containing –TCA- or –TmCA- motif was measured by rotational anisotropy. Binding mixtures contained 50 nM ssDNA and various concentrations of proteins. The plots of changes in anisotropy were fitted by one-site specific binding model. (E–H) Deaminase activity of wt A3A for C (E, F) and mC (G, H) on different DNA sequence motifs. Deamination reaction mixture contains 10 nM A3A and 600 nM substrates that contain the listed tri-nucleotide motifs. (I–L) Deaminase activity of R1 mutant of A3A for C (I, J) and mC (K, L) on different DNA sequence motifs. Reaction mixture contains 100 nM R1 mutant and 600 nM substrates that contain the listed tri-nucleotide motifs. These results demonstrate that the substrate motif specificity of R1 mutant is similar to that of wt A3A, both of which show a strong preference for TCA/G or TmCA/G. S.D. was estimated from data collected in three independent experiments.
To ascertain that the reduced deaminase activity and selectivity for mC of region 1 mutants are not the result of altered sequence motif specificity, we examined the deaminase activity of the wild type and R1 mutant with a set of 30 nt FAM-labeled ssDNA substrates containing motifs of 5′-NCA and 5′-TCN, as well as 5′-NmCA and 5′-TmCN, where N is any of the four nucleotides (Fig. 5E–L). Overall, R1 mutant showed the same sequence motif preference as the wt A3A for both normal C and mC containing substrates (Fig. 5I–L). These results indicate that the decreased overall deaminase activity and the reduced mC selectivity factor observed for the R1 mutant of A3A are not due to altered sequence motif preference.
In summary, APOBEC cytidine deaminases play important roles in immunity and other biological processes including genomic mC modification and potential epigenetic regulation. However, their aberrant deamination can also generate malignant mutations leading to cancer [59, 60, 68–73]. Even though the eleven known APOBEC members are evolutionarily conserved and share the same core structure, their deamination activities on C and mC and resulting specificity for mC are different. Here we performed family-wide comparative activity assays on C and mC deamination using purified APOBEC proteins. Our results showed that nine APOBECs (except for Apo2, Apo4) had strong to readily detectible deaminase activity on C. Four of these nine members (A3A, A3H, A3B, A3C) showed strong to readily detectable mC deaminase activity, and another four (AID, Apo1, A3F, A3G) showed weak mC deaminase activity, with A3D having no detectible mC deaminase activity. Notably, A3A and A3H had robust deaminase activity on C and mC, and also high mC selectivity factor. On the other hand, A3C showed the third highest C deaminase activity, but had the lowest mC selectivity factor. By mutating different regions of A3A, we show that loop-1 residues next to the active center contribute critically to the observed high deaminase activity and high mC selectivity factor. This study provides the first direct comparison of the deaminase activity on C and mC of eleven known APOBEC family members. A caveat is that the activity data are obtained from in vitro studies using purified proteins from E. coli. While the activities may reflect the intrinsic properties of the APOBECs in vitro, the in vivo situation may vary due to potential post-translational modification, co-factor binding, and subcellular localization. However, this report on the family-wide comparison of the activity and mC selectivity would be valuable for further understanding of the various biological functions of different APOBEC family members.
Supplementary Material
The substrate motifs and pH conditions for mC deamination were the same as C deamination except that the target C is replaced with mC. Deaminase assay on mC was performed as previously described [49, 50, 62]. Briefly, various concentrations of APOBECs were incubated with 600 nM 5′ 6FAM-labeled ssDNA (30 nt) containing a target mC in deamination buffer (25 mM buffer at the indicated pH, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and 0.1 μg/ml RNase A) at 37°C for 2 hours. The protein concentrations used for each proteins are as follows: AID: 0.75, 1.5, 3, 6, 12 μM; Apo1: 0.5, 1, 2, 4, 8 μM; A3A: 3.75, 7.5, 15, 30, 60 nM; A3B: 0.125, 0.25, 0.5, 1, 2 μM; A3C: 0.14, 0.28, 0.56, 1.13, 2.25 μM; A3D: 0.09, 0.19, 0.38, 0.75, 1.5 μM; A3F: 0.25, 0.5, 1, 2, 4 μM; A3G: 0.38, 0.75, 1.5, 3, 6 μM; and A3H: 7.8, 15.6, 31.3, 62.5, 125 nM. The reactions were stopped by heating at 90°C for 5 min. The bases of the deamination products thymine were subsequently cleaved by incubating with 2 units of TDG and 3-fold excess amount of the complementary ssDNA at 42°C for 12 hours. The abasic sites were then hydrolyzed with 0.1 M NaOH at 90°C for 10 min. Deamination products were analyzed on 20% denaturing PAGE gels, and quantified with Molecular Imager FX Pro Plus System/Quantity One® analysis software (Bio-Rad).
All the constructs have His6-tag on the N-terminus.
Highlights.
Comparative study of the in vitro activities on cytidine (C) and methylcytidine (mC) deamination of the APOBEC protein family.
Eight of nine C-deaminase active APOBECs show deaminase activity on mC.
The top three APOBECs with the highest C-deamination activity is in the order of A3A, A3H, and A3C.
The top three APOBECs with the highest mC-deamination activity is in the order of A3A, A3H, and A3B.
The top three APOBECs with the highest mC selectivity factor is in the order of A3H, A3A, and AID. The lowest mC selectivity factor is observed in A3C.
Residues on loop 1 of A3A contribute critically to the high deaminase activity and high mC selectivity.
Acknowledgments
We thank the core laboratory in the Center of Excellence in NanoBiophysics at University of Southern California for support in the related characterization of the purified protein samples. This work was supported by a fellowship from Nakajima Foundation to F.I. and by the NIH grant R01GM087986 to X.S.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome biology. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macduff DA, Harris RS. Directed DNA deamination by AID/APOBEC3 in immunity. Curr Biol. 2006;16:R186–9. doi: 10.1016/j.cub.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 3.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–51. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 4.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, et al. Crystal structure of the antiviral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–4. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cell Mol Life Sci. 2009;66:3137–47. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–9. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 7.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–10. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 9.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–50. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siu KK, Sultana A, Azimi FC, Lee JE. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. Crystal Structure of the DNA Deaminase APOBEC3B Catalytic Domain. J Biol Chem. 2015;290:28120–30. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moris A, Murray S, Cardinaud S. AID and APOBECs span the gap between innate and adaptive immunity. Front Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol. 2014;426:1220–45. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23:279–87. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell host & microbe. 2008;4:249–59. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews Immunology. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 20.Prochnow C, Bransteitter R, Chen XS. APOBEC deaminases-mutases with defensive roles for immunity. Science in China Series C, Life sciences/Chinese Academy of Sciences. 2009;52:893–902. doi: 10.1007/s11427-009-0133-1. [DOI] [PubMed] [Google Scholar]
- 21.Prochnow C, Goodman MF, Chen XS. The prospct of APOBEC3G for the future of HIV therapy. HIV Ther. 2009;3(1):7–10. [Google Scholar]
- 22.Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hyper-mutational targeting by wild type and mutant AID. J Biol Chem. 2004;279:51612–21. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- 23.Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–68. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–8. doi: 10.1038/ni964. Epub 2003 Aug 10. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 26.Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–12. doi: 10.1038/ni1086. Epub 2004 Jun 13. [DOI] [PubMed] [Google Scholar]
- 27.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 28.Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–12. [PubMed] [Google Scholar]
- 29.Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, et al. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D, Liu J, Kang F, Guan W, Gao X, Wang Y, et al. Core-APOBEC3C chimerical protein inhibits hepatitis B virus replication. J Biochem. 2011;150:371–4. doi: 10.1093/jb/mvr086. [DOI] [PubMed] [Google Scholar]
- 31.Krisko JF, Martinez-Torres F, Foster JL, Garcia JV. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 2013;9:e1003242. doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–3. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Wakae K, Kitamura K, Aoyama S, Liu G, Koura M, et al. APOBEC3 deaminases induce hypermutation in human papillomavirus 16 DNA upon beta interferon stimulation. J Virol. 2014;88:1308–17. doi: 10.1128/JVI.03091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, et al. APOBEC3A Functions as a Restriction Factor of Human Papillomavirus. J Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–6. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumert TF, Rosler C, Malim MH, von Weizsacker F. Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology. 2007;46:682–9. doi: 10.1002/hep.21733. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez MC, Suspene R, Henry M, Guetard D, Wain-Hobson S, Vartanian JP. Human APOBEC1 cytidine deaminase edits HBV DNA. Retrovirology. 2009;6:96. doi: 10.1186/1742-4690-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei YC, Hao YH, Zhang ZM, Tian YJ, Wang BJ, Yang Y, et al. Inhibition of hepatitis B virus replication by APOBEC3G in vitro and in vivo. World J Gastroenterol. 2006;12:4492–7. doi: 10.3748/wjg.v12.i28.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X, Li SX, Yang H, Chen XS. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun. 2016;7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lada AG, Krick CF, Kozmin SG, Mayorov VI, Karpova TS, Rogozin IB, et al. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry (Mosc) 2011;76:131–46. doi: 10.1134/s0006297911010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 42.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–7. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–60. doi: 10.1074/jbc.M407695200. Epub 2004 Sep 24. [DOI] [PubMed] [Google Scholar]
- 44.Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, et al. Methylation protects cytidines from AID-mediated deamination. Mol Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–8. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40:9206–17. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–8. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suspene R, Aynaud MM, Vartanian JP, Wain-Hobson S. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PloS one. 2013:8. doi: 10.1371/journal.pone.0063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Y, Ito F, Zhang G, Fernandez B, Yang H, Chen XS. DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B. Biochem J. 2015;471:25–35. doi: 10.1042/BJ20150382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu J, Chen Q, Xiao X, Ito F, Wolfe A, Chen XS. Biochemical Characterization of APOBEC3H Variants: Implications for Their HIV-1 Restriction Activity and mC Modification. J Mol Biol. 2016;428:4626–38. doi: 10.1016/j.jmb.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 52.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–5. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–7. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fritz EL, Rosenberg BR, Lay K, Mihailovic A, Tuschl T, Papavasiliou FN. A comprehensive analysis of the effects of the deaminase AID on the transcriptome and methylome of activated B cells. Nat Immunol. 2013;14:749–55. doi: 10.1038/ni.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–52. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 58.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 59.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saraconi G, Severi F, Sala C, Mattiuz G, Conticello SG. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome biology. 2014;15:417. doi: 10.1186/s13059-014-0417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–33. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Q, Xiao X, Wolfe A, Chen XS. The in vitro Biochemical Characterization of an HIV-1 Restriction Factor APOBEC3F: Importance of Loop 7 on Both CD1 and CD2 for DNA Binding and Deamination. J Mol Biol. 2016;428:2661–70. doi: 10.1016/j.jmb.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pham P, Landolph A, Mendez C, Li N, Goodman MF. A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer. J Biol Chem. 2013;288:29294–304. doi: 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhutani N, Decker MN, Brady JJ, Bussat RT, Burns DM, Corbel SY, et al. A critical role for AID in the initiation of reprogramming to induced pluripotent stem cells. FASEB J. 2013;27:1107–13. doi: 10.1096/fj.12-222125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar R, DiMenna L, Schrode N, Liu TC, Franck P, Munoz-Descalzo S, et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013;500:89–92. doi: 10.1038/nature12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caval V, Suspène R, Vartanian J-p, Wain-Hobson S, Caval V, Suspe R. Orthologous Mammalian APOBEC3A Cytidine Deaminases Hypermutate Nuclear DNA. Molecular biology and evolution. 2013:1–11. doi: 10.1093/molbev/mst195. [DOI] [PubMed] [Google Scholar]
- 67.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–9. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris RS. Cancer mutation signatures, DNA damage mechanisms, and potential clinical implications. Genome Med. 2013;5:87. doi: 10.1186/gm490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, et al. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 2013;73:7222–31. doi: 10.1158/0008-5472.CAN-13-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuong KJ, Loeb LA. APOBEC3B mutagenesis in cancer. Nat Genet. 2013;45:964–5. doi: 10.1038/ng.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chelico L, Prochnow C, Erie DA, Chen XS, Goodman MF. Structural model for deoxycytidine deamination mechanisms of the HIV-1 inactivation enzyme APOBEC3G. J Biol Chem. 2010;285:16195–205. doi: 10.1074/jbc.M110.107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi K, Carpenter MA, Banerjee S, Shaban NM, Kurahashi K, Salamango DJ, et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The substrate motifs and pH conditions for mC deamination were the same as C deamination except that the target C is replaced with mC. Deaminase assay on mC was performed as previously described [49, 50, 62]. Briefly, various concentrations of APOBECs were incubated with 600 nM 5′ 6FAM-labeled ssDNA (30 nt) containing a target mC in deamination buffer (25 mM buffer at the indicated pH, 100 mM NaCl, 0.1% Triton X-100, 1 mM DTT, and 0.1 μg/ml RNase A) at 37°C for 2 hours. The protein concentrations used for each proteins are as follows: AID: 0.75, 1.5, 3, 6, 12 μM; Apo1: 0.5, 1, 2, 4, 8 μM; A3A: 3.75, 7.5, 15, 30, 60 nM; A3B: 0.125, 0.25, 0.5, 1, 2 μM; A3C: 0.14, 0.28, 0.56, 1.13, 2.25 μM; A3D: 0.09, 0.19, 0.38, 0.75, 1.5 μM; A3F: 0.25, 0.5, 1, 2, 4 μM; A3G: 0.38, 0.75, 1.5, 3, 6 μM; and A3H: 7.8, 15.6, 31.3, 62.5, 125 nM. The reactions were stopped by heating at 90°C for 5 min. The bases of the deamination products thymine were subsequently cleaved by incubating with 2 units of TDG and 3-fold excess amount of the complementary ssDNA at 42°C for 12 hours. The abasic sites were then hydrolyzed with 0.1 M NaOH at 90°C for 10 min. Deamination products were analyzed on 20% denaturing PAGE gels, and quantified with Molecular Imager FX Pro Plus System/Quantity One® analysis software (Bio-Rad).
All the constructs have His6-tag on the N-terminus.