Abstract
Micro-ribonucleic acids (miRNAs) are small functional non-coding RNAs that downregulate gene expression at the post-transcriptional level. Abnormal expression of specific miRNAs has been recorded in chronic lymphocytic leukaemia, other non-Hodgkin B-cell lymphomas, lung cancer and chronic myeloid leukaemia (CML). The aim of this study was to compare miRNA expression profiles among patients with newly diagnosed CML, those on established therapy with imatinib mesylate, and healthy individuals. The expression of 88 miRNAs was evaluated in a total of nine samples divided into three groups: Group 1 comprised three samples collected from newly diagnosed CML patients; group 2 consisted of three samples collected from patients on therapy; the remaining three samples were collected from healthy volunteers (control group). Total RNA was extracted from whole blood and reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) was performed on the LightCycler® 480 platform using Human Serum & Plasma miRNA PCR Arrays. In group 1, only SNORD44 was downregulated, while hsa-miR-372 and hsa-miR-375 were found to be significantly upregulated compared with the control group. By contrast, 49 miRNAs were significantly upregulated in group 2 compared with the control group. miRNAs hsa-miR-106b, hsa-miR-21, hsa-miR-221, hsa-miR-10a, hsa-miR-193a-5p and hsa-miR-30e were expressed in group 2. Therefore, miRNA expression profiles differed between the two patient groups.
Keywords: chronic, expression, leukaemia, microRNA, myeloid
Introduction
Chronic myeloid leukaemia (CML) is an acquired disease resulting from the malignant transformation of a single stem cell, characterized by anaemia extreme blood granulocytosis and granulocytic immaturity (1). CML is triggered by a reciprocal translocation of the long arms of chromosomes 9 and 22, referred to as the Philadelphia chromosome (2). The breakpoint cluster region (BCR) gene on chromosome 22 and the Abelson murine leukaemia viral oncogene homolog 1 (ABL1) gene on chromosome 9 are involved in this t(9;22)(q34;q11) translocation leading to the formation of a new chimeric fusion oncogene, BCR-ABL1. Expression of this fusion oncogene produces an abnormal, elongated 8-kb ribonucleic acid (RNA) fragment, which is translated into a unique tyrosine phosphoprotein kinase, considered to be essential for the development of the leukaemic phenotype (1,3).
Imatinib mesylate (Gleevec®; Novartis, Basel, Switzerland) has been established as the standard first-line therapy for CML (4,5). Response to therapy is defined as optimal, suboptimal, or failure of treatment. Optimal response is defined as follows: Complete haematological response [normal full blood count, minimal cytogenetic response (CyR) at 3 months]; partial cytogenetic response at 6 months; complete CyR (CCyR) at 12 months and major molecular response (≥3-log reduction in BCR-ABL1 transcripts) at 18 months (6).
MicroRNAs (miRNAs) have recently been identified as a class of small functional non-coding RNAs of 18–25 nucleotides (7,8) which bind to the 3′-untranslated region of mRNAs, downregulating gene expression at the post-transcriptional level. Recent research suggests that miRNAs may play an important role in the pathology of haematological malignancies (9).
miRNAs are found in the genomes of C. elegans, Drosophila, mice, as well as humans, often with highly conserved sequences. However, in mammals, no genomic sequences that are perfectly complementary to known miRNAs have yet been identified (10). The miRNA stem-loop precursor, pre-miRNA, may be transcribed as part of a longer primary RNA termed pri-miRNA (11). miRNAs play critical roles in cellular processes, including metabolism, apoptosis, differentiation and development (8).
miRNAs are often found in polycistronic clusters containing an average of 2–3 miRNAs. These polycistronic clusters are usually located in fragile regions prone to deletions and other molecular damaging alterations (12). miRNAs are expressed in specific haematological cell types with a regulatory function in early haematopoietic differentiation, erythropoiesis, granulopoiesis, megakaryopoiesis and lymphoid development (12). An example was the demonstration of a cluster of miRNAs found to be transcribed from an intron of the C13-25 open reading frame locus at 13q31-q32. This polycistron (hsa-miR-17-92) is amplified and overexpressed in different types of B-cell lymphoma (12). Lymphogenesis was found to be promoted by hsa-miR-17-92 in a murine stem cell transplantation model (13). In addition, it was determined that this polycistron is also transcriptionally regulated by c-MYC, which may be of great significance, as oncogenic variants of ABL1 are upregulated by c-MYC expression; furthermore, c-MYC is required and acts synergistically with BCR-ABL1, resulting in transformation to CML (12).
Gordon et al found hsa-miR-10a, hsa-miR-17/92, hsa-miR-150, hsa-miR-203 and hsa-miR-328 to be the most commonly deregulated miRNAs in CML (14). A miRNA may function as a tumour suppressor or an oncogene (15). An example of this is hsa-miR-17/92, which has been shown to target BCL2, STAT5 and JAK2 (tumour suppressor), as well as the CDK inhibitor CDKN1A (p21), functioning as an oncogene. This cluster of miRNAs also exhibits increased expression in the early chronic phase of CML, but not in blast phase (BP), making it a possible therapeutic target. Downregulation of hsa-miR-328 has also been described as a crucial stage of disease progression to BP (15).
The present pilot study was performed to determine the miRNA expression profiles in a South African population of CML patients compared with the miRNA signature in CML described in the literature. This study was approved by the Research Ethics Committee of the University of Pretoria (38/2007).
Materials and methods
Patients
The expression profiles of 88 miRNAs were evaluated in nine blood samples, which consisted of samples from three healthy individuals (control group), three newly diagnosed CML patients (group 1) and three CML patients on imatinib mesylate therapy (group 2). The samples were collected from the Department of Medical Oncology at the Steve Biko Academic Hospital in 2010.
miRNA extraction
Whole blood was collected in PAXgene® Blood RNA Tube IVD (Qiagen, Hilden, Germany) or EDTA BD Vacutainer® (Becton Dickinson and Company, Franklin Lakes, NJ, USA) collection tubes. The PAXGene® Blood miRNA kit (Qiagen) was used to extract total RNA from three samples and the miRNeasy® Mini kit (Qiagen) was used to extract total RNA from the remaining six samples. Extracted products were analysed to ensure adequate quality and purity using the Agilent Bioanalyser (Agilent Technologies, Santa Clara, CA, USA).
Reverse transcriptase quantitative polymerase chain reaction (RT-qPCR)
miRNAs were transcribed to cDNA using the RT2 miRNA First Strand kit (Qiagen). RT-qPCR was performed on the LightCycler® 480 platform (Roche, Basel, Switzerland) using Human Serum miRNA PCR Arrays (Qiagen), which contained the miRNAs associated with CML in the literature. This was the only array at the time recommended for blood and/or blood products. The RT2 qPCR SYBR Green master mix (Qiagen) was used to amplify the cDNA according to the reaction mixture composition and the cycling conditions listed in Tables I and II. Post-amplification a melting curve program was run at 95°C for 1 min, 65°C for 2 min followed by 65–95°C at 2°C/min (data collection step).
Table I.
Reaction mix composition.
| Reagent description | Volume (µl) |
|---|---|
| 2X RT2 qPCR SYBR Green mastermix | 1,275 |
| Diluted first strand reaction (1:10) | 100 |
| RNase-free H2O | 1,175 |
| Total volume | 2,550 |
RT, reverse transcriptase; qPCR, quantitative polymerase chain reaction.
Table II.
RT-qPCR cycling conditions.
| Number of cycles | Duration (sec) | Temperature (°C) | Optic detection |
|---|---|---|---|
| 1 | 600 | 95 | Off |
| 45 | 15 | 95 | Off |
| 30–40 | 60 | Off | |
| 30 | 72 | On |
RT, reverse transcriptase; qPCR, quantitative polymerase chain reaction.
Data analysis
The relative quantitation method ΔΔ2Cq (16) was used to analyse and normalise the expression profile data.
Raw data were captured on an Excel® template and uploaded on the free PCR Array Data Analysis Web Portal (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php; accessed, 22/01/2015) (Qiagen). A cut-off cycle threshold (Ct) value of 35°C was used for miRNA expression. This cut-off value was recommended in the protocol for data analysis by the supplier of the program used. This program automatically calculated the average Ct, ΔCt, as well as the 2-ΔΔCt. Fold changes were then calculated with a 95% confidence interval. The P-value was automatically calculated and translated to fold regulation. This was plotted on a volcano plot combining the P-value and the fold regulation expression data from the two groups.
All reagents and kit protocols were used according to the manufacturer's guidelines, except for the Human Serum miRNA PCR Arrays, where total RNA extracted from whole blood instead of serum or plasma was used.
Results
Patient characteristics
The miRNA expression profiles of 88 miRNAs were analysed in a total of 9 patients divided into three different groups. The white cell count (WCC) of the samples was crucial for determining the amount of whole blood to be used for the miRNA extraction protocol. The WCC of the newly diagnosed group ranged from 253.73 to 282.12×109/l (median, 269.43×109/l). The WCC of the CML patients on therapy ranged from 5.32 to 166.67×109/l (median 33.79×109/l). The age of the study population ranged from 16 to 69 years (median, 33 years). The patient sample consisted of five male and four female individuals, including the age-matched control group.
Differential miRNA expression
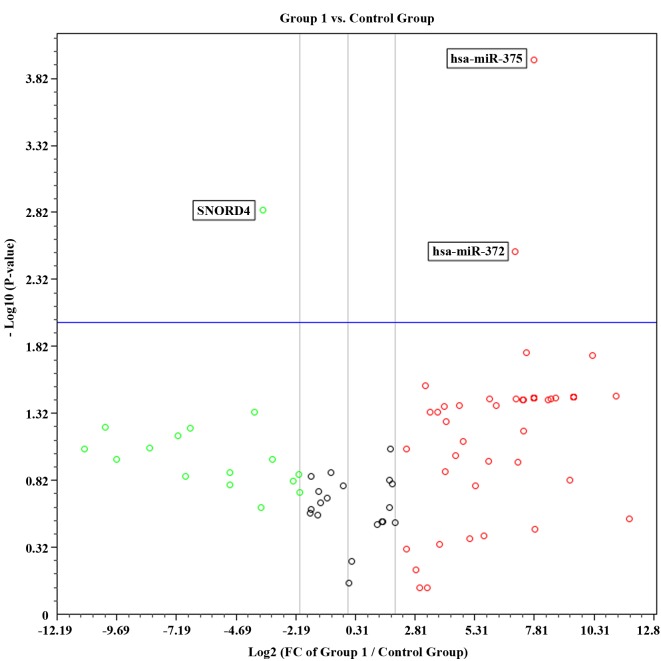
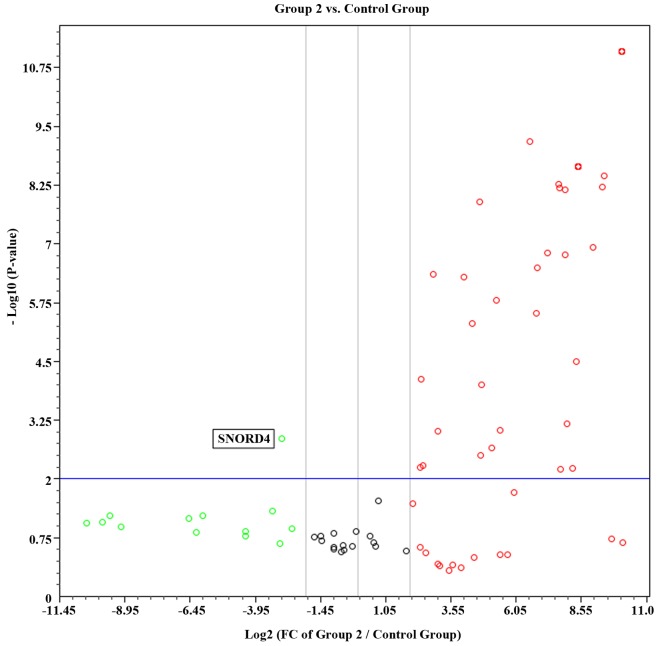
Compared with the control group, there was a significant decrease in the number of miRNAs expressed in newly diagnosed patients and CML patients receiving therapy. Of the 88 miRNAs investigated, 50 (56%) were expressed in the control group, 8/88 (9%) were expressed in the newly diagnosed group and 17/88 (19%) were expressed in the group of patients receiving imatinib mesylate therapy. Volcano plots combining the P-value (P<0.01) and the fold regulation expression data of the two groups are shown in Figs. 1 and 2. In group 1, only two miRNAs (hsa-miR-372 and hsa-miR-375) were significantly up regulated compared with the control group. By contrast, 49 miRNAs were significantly up regulated in group 2. These results suggest that the expression profiles may differ between the two groups of patients.
Figure 1.
Volcano plot representing statistical significant fold regulation data from the control group and group 1. FC, fold change.
Figure 2.
Volcano plot representing statistical significant fold regulation data from the control group and group 2. FC, fold change.
The miRNA expression of the three samples in the group of CML patients on therapy (group 2) was separately analysed to determine whether therapy affects miRNA expression (data not shown). Although there was a correlation of miRNA expression among samples C2, C7 (not in remission) and C9 (achieved CCyR) (6), it should be noted that a number of miRNAs were expressed in sample C9 (in CCyR) and not expressed in the other two samples, including hsa-miR-122, hsa-miR-150, hsa-miR-210 and hsa-miR-23a. Of these, only hsa-miR-23a and hsa-miR-150 were also expressed in the control group.
Discussion
Complete haematological response, CCyR and major molecular response are currently used as prognostic indicators. The RT-qPCR used to detect and measure BCR-ABL transcript levels is required for this classification (6). A growing body of evidence suggests that miRNA expression may be used as a prognostic marker. In a study performed on miRNA expression in chronic lymphocytic leukaemia (CLL), Calin et al (17) observed a significant association between a group of miRNAs and the expression of ZAP-70 and the time between diagnosis and initial treatment. The authors believe that miRNA expression may be a valuable addition to existing prognostic markers for CLL (17). Cummins and Velculescu alluded to the possibility of using miRNAs as early indicators of cancer initiation or progression (18). Wu et al also concluded that miRNAs may be used as prognostic markers in cancer (19).
The aim of the present study was to compare miRNA expression profiles in patients with newly diagnosed CML, those on therapy with imatinib mesylate, and healthy individuals. The miRNA expression data demonstrated that a significant number of miRNAs were expressed in the control group compared with the other two groups. Our results were consistent with those of Ramkissoon et al (20), in that no hsa-miR155 expression was found in healthy individuals, but was present in both groups of CML patients. One discrepant finding was the expression of hsa-miR-223 by the malignant cells isolated by Ramkissoon et al (20). We were unable to identify expression of hsa-miR223 in either of the two groups of CML patients, whereas their study only focused on expression of miR-142, miR-155, miR-181 and miR-223 (20).
Focusing on the samples from the group of patients on therapy (group 2), samples C2 and C7 were from patients who were not in remission at the time. Sample C9 was from a patient who achieved CCyR at the time the sample was collected. Of note, the patient who provided sample C7 was not responding to therapy at the time the sample was collected. The expression profiles of these three samples suggest that hsa-miR-122 and hsa-miR210 may be used as possible indicators of response to therapy; however, due to the small sample size, definitive conclusions could not be reached.
Jose-Eneriz et al investigated the expression profiles of 250 miRNAs in 8 CML patients, 3 of whom were considered to be resistant to therapy (4), and they identified 19 miRNAs differentially expressed between CML patients responding to therapy and those resistant to imatinib mesylate therapy. They found the following 18 miRNAs to be downregulated in CML patients not responding to therapy: hsa-miR-7, hsa-miR-23a, hsamiR-26a, hsa-miR-29a, hsa-miR-29c, hsa-miR-30b, hsamiR-30c, hsa-miR-100, hsa-miR-126#, hsa-miR-134, hsa-miR-141, hsa-miR-183, hsa-miR-196b, hsa-miR-199a, hsa-miR-224, hsa-miR-326, hsa-miR-422b and hsa-miR-520a. Only hsa-miR-191 was upregulated in CML patients resistant to therapy (4). In contrast, our results showed 49 miRNAs to be upregulated compared with the 18 miRNAs that were downregulated (4). However, the results of both studies were based on small sample sizes. In addition, although both studies had the same objectives, the methodology differed, making comparison difficult. However, the results of both studies suggest that there is a significant difference in the miRNA expression profile patterns between the two groups of CML patients.
As the sample size of this pilot study was small, more in-depth research on a large scale is required to validate these results. Further research in this field is required to elucidate the possible role of miRNA expression as a prognostic marker. Novel miRNAs may be identified that may be of value as prognostic markers in CML, which may lead to individualized therapy and improved patient outcome.
Acknowledgements
The authors are grateful to the National Health Laboratory Service Research Trust Fund for funding this study (Grant004_94200) and would especially like to thank the Department of Medical Microbiology for providing access to essential equipment.
Glossary
Abbreviations
- BCR
breakpoint cluster region
- CCyR
complete cytogenetic response
- CML
chronic myeloid leukaemia
- miRNA
microRNA
- RNA
ribonucleic acid
- RT-qPCR
reverse transcriptase quantitative polymerase chain reaction
- WCC
white cell count
References
- 1.Liesveld JL, Lichtman MA. Chronic Myelogenous Leukaemia and related disorders. In: Kaushansky K, Lichtman MA, Beutler E, Kipps TJ, Seligsohn U, Prchal JT, editors. Williams Hematology. Mcraw-Hill; New York: 2010. pp. 1331–1336. [Google Scholar]
- 2.Schüler F, Dölken G. Detection and monitoring of minimal residual disease by quantitative real-time PCR. Clin Chim Acta. 2006;363:147–156. doi: 10.1016/j.cccn.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 3.Stenthoft J, Pallisgaard N, Kjeldsen E, Holm MS, Nielsen JL, Hokland P. Kinetics of BCR-ABL fusion transcript levels in chronic myeloid leukemia patients treated with STI571 measured by quantitative real-time polymerase chain reaction. Eur J Haematol. 2001;67:302–308. doi: 10.1034/j.1600-0609.2001.00556.x. [DOI] [PubMed] [Google Scholar]
- 4.San J, osé-Enériz E, Román-Gómez J, Jiménez-Velasco A, Garate L, Martin V, Cordeu L, Vilas-Zornoza A, Rodríguez-Otero P, Calasanz MJ, Prósper F, Agirre X. MicroRNA expression profiling in imatinib-resistant chronic myeloid leukemia patients without clinically significant ABL1-mutations. Mol Cancer. 2009;8:69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger M, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 6.Hoffbrand AV, Moss PAH. Essential Haematology. Wiley-Blackwell; United Kingdom: 2011. Chronic Myeloid Leukaemia; p. 196. [Google Scholar]
- 7.Yeung ML, Bennasser Y, Jeang KT. miRNAs in the biology of cancers and viral infections. Curr Med Chem. 2007;14:191–197. doi: 10.2174/092986707779313417. [DOI] [PubMed] [Google Scholar]
- 8.Georgantas RW, III, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrie CH. MicroRNAs and haematology: Small molecules, big function. Br J Haematol. 2007;137:503–512. doi: 10.1111/j.1365-2141.2007.06611.x. [DOI] [PubMed] [Google Scholar]
- 10.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M, Scherr M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 13.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon JE, Wong JJ, Rasko JE. MicroRNAs in myeloid malignancies. Brit J of Haematol. 2013;162:162–176. doi: 10.1111/bjh.12364. [DOI] [PubMed] [Google Scholar]
- 15.Yeh CH, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016;15:37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 19.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 20.Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP, Jr, Sloand EM, Kajigaya S, Young NS. Hematopoietic-specific microRNA expression in human cells. Leuk Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]