Abstract
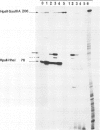
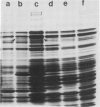
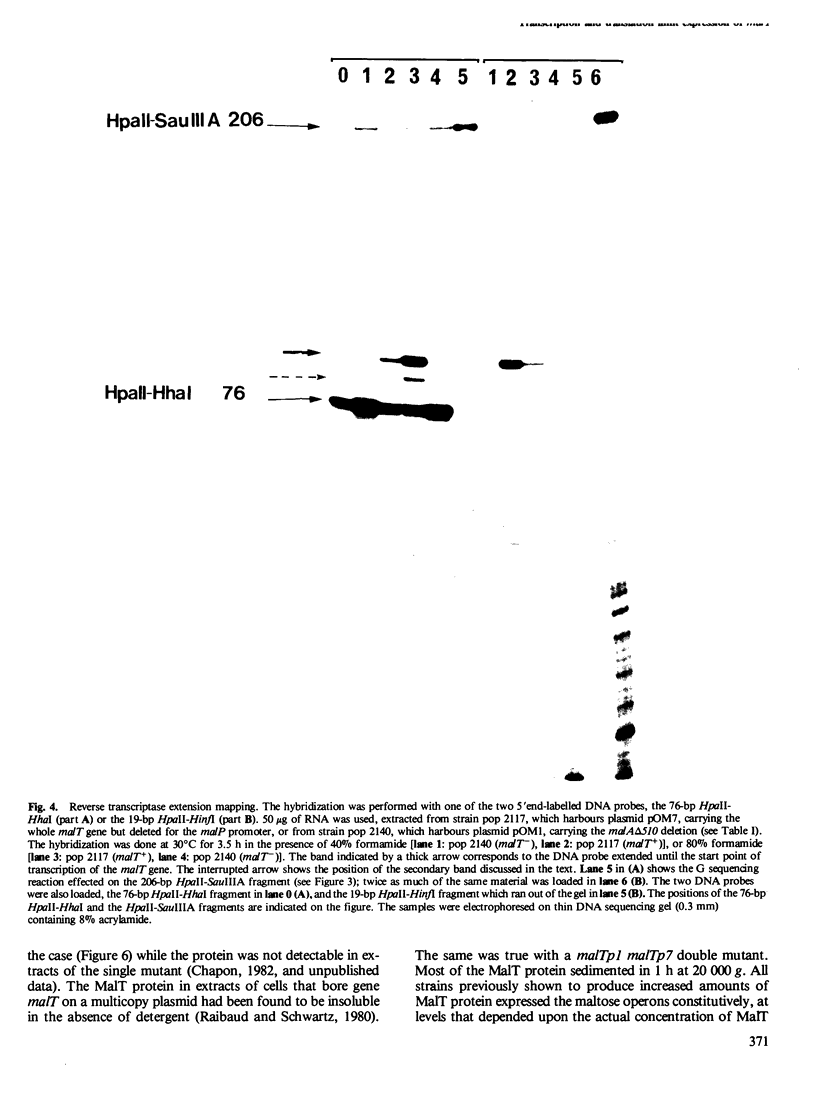
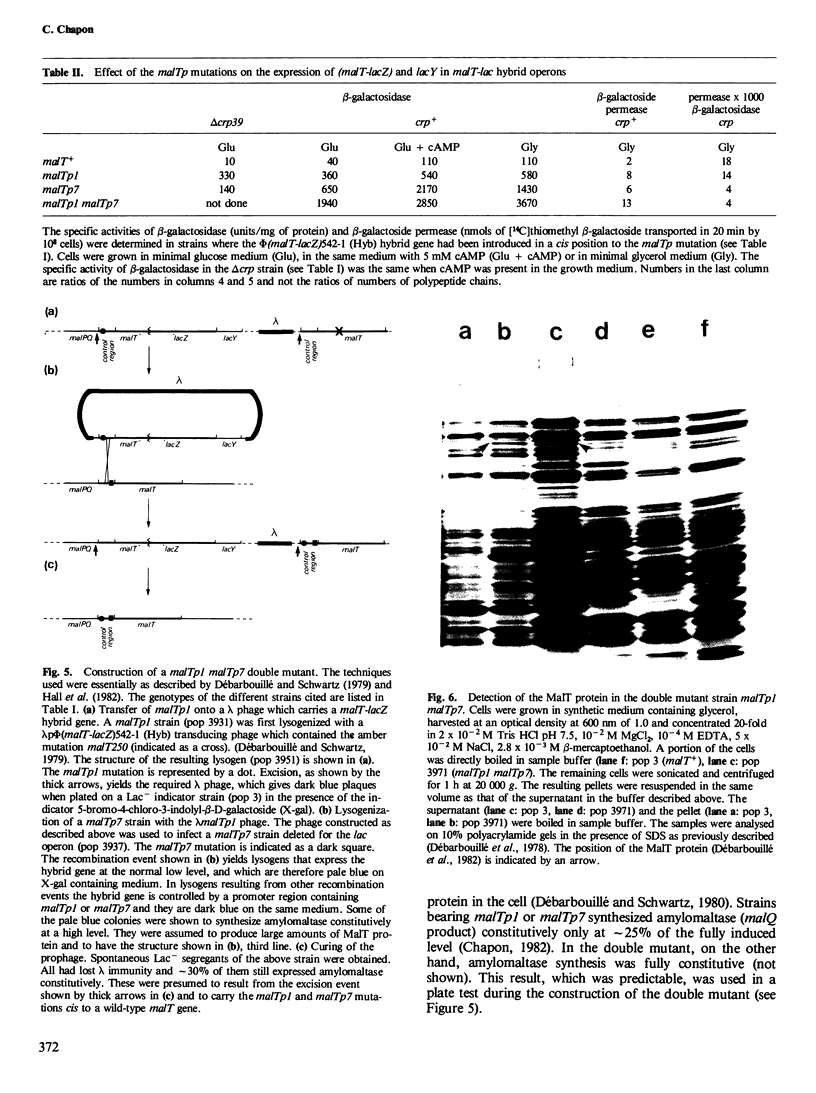
Six mutations, which lead to an increase in malT expression, were mapped by sequencing techniques. All of them had one or other of two base changes. Determination of the transcription start point by reverse transcriptase mapping localised the two base changes with respect to the elements that control malT expression. One of the base changes ( malTp1 ) is located in the Pribnow box of the promoter, and presumably results in an increase in the rate of transcription initiation. The other ( malTp7 ) is located in the Shine and Dalgarno sequence, which precedes the malT cistron. It probably created a more favourable ribosome binding site on malT mRNA. A correlate of these observations is that the promoter and the ribosome binding site are both inefficient in a wild-type malT gene. A malTp1 malTp7 double mutant was constructed, which produced equivalent to 30 times more MalT protein than the wild-type strain.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arditti R., Grodzicker T., Beckwith J. Cyclic adenosine monophosphate-independent mutants of the lactose operon of Escherichia coli. J Bacteriol. 1973 May;114(2):652–655. doi: 10.1128/jb.114.2.652-655.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTTIN G., COHEN G. N., MONOD J., RICKENBERG H. V. La galactoside-perméase d'Escherichia coli. Ann Inst Pasteur (Paris) 1956 Dec;91(6):829–857. [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Debarbouille M., Schwartz M. The use of gene fusions to study the expression of malT the positive regulator gene of the maltose regulon. J Mol Biol. 1979 Aug 15;132(3):521–534. doi: 10.1016/0022-2836(79)90273-0. [DOI] [PubMed] [Google Scholar]
- Débarbouillé M., Shuman H. A., Silhavy T. J., Schwartz M. Dominant constitutive mutations in malT, the positive regulator gene of the maltose regulon in Escherichia coli. J Mol Biol. 1978 Sep 15;124(2):359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Horwitz A. H., Morandi C., Wilcox G. Deoxyribonucleic acid sequence of araBAD promoter mutants of Escherichia coli. J Bacteriol. 1980 May;142(2):659–667. doi: 10.1128/jb.142.2.659-667.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell D., Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977 Jul;114(1):1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. lac Promoter mutation Pr115 generates a new transcription initiation point. J Mol Biol. 1980 May 25;139(3):551–556. doi: 10.1016/0022-2836(80)90146-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Roa M., Braun-Breton C., Schwartz M. Structure of the malB region in Escherichia coli K12. I. Genetic map of the malK-lamB operon. Mol Gen Genet. 1979 Jul 24;174(3):241–248. doi: 10.1007/BF00267796. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Restriction map of the Escherichia coli malA region and identification of the malT product. J Bacteriol. 1980 Aug;143(2):761–771. doi: 10.1128/jb.143.2.761-771.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Roa M., Débarbouillé M. Mutations that affect lamB gene expression at a posttranscriptional level. Proc Natl Acad Sci U S A. 1981 May;78(5):2937–2941. doi: 10.1073/pnas.78.5.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Brickman E., Bassford P. J., Jr, Casadaban M. J., Shuman H. A., Schwartz V., Guarente L., Schwartz M., Beckwith J. R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979 Jul 24;174(3):249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- Silverstone A. E., Magasanik B., Reznikoff W. S., Miller J. H., Beckwith J. R. Catabolite sensitive site of the lac operon. Nature. 1969 Mar 15;221(5185):1012–1014. doi: 10.1038/2211012b0. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. Interaction of the cAMP receptor protein with the lac promoter. Nucleic Acids Res. 1980 Feb 25;8(4):759–766. [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]