Abstract
Background
Animal model experiments have suggested a role of the DNA repair protein ERCC1 (Excision Repair Cross-Complementation Group 1) in prostate cancer progression.
Methods
To better understand the impact of ERCC1 protein expression in human prostate cancer, a preexisting tissue microarray (TMA) containing more than 12,000 prostate cancer specimens was analyzed by immunohistochemistry and data were compared with tumor phenotype, PSA recurrence and several of the most common genomic alterations (TMPRSS2:ERG fusions: deletions of PTEN, 6q, 5q, 3p).
Results
ERCC1 staining was seen in 64.7% of 10,436 interpretable tissues and was considered weak in 37.1%, moderate in 22.6% and strong in 5% of tumors. High-level ERCC1 staining was linked to advanced pT stage, high Gleason grade, positive lymph nodes, high pre-operative serum PSA, and positive surgical margin status (p < 0.0001 each). High ERCC1 expression was strongly associated with an elevated risk of PSA recurrence (p < 0.0001). This was independent of established prognostic features. A subgroup analysis of cancers defined by comparable quantitative Gleason grades revealed that the prognostic impact was mostly driven by low-grade tumors with a Gleason 3 + 3 or 3 + 4 (Gleason 4: ≤5%). High ERCC1 expression was strongly associated with the presence of genomic alterations and expression levels increased with the number of deletions present in the tumor. These latter data suggest a functional relationship of ERCC1 expression with genomic instability.
Conclusion
The results of our study demonstrate that expression of ERCC1 - a potential surrogate for genomic instability - is an independent prognostic marker in prostate cancer with particular importance in low-grade tumors.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-017-3489-9) contains supplementary material, which is available to authorized users.
Keywords: ERCC1, DNA repair, Prostate cancer, Prognosis
Background
Prostate cancer is the most common cancer in males in the western societies. While most patients will never suffer symptoms from their disease, prostate cancer is still the third most common cause of cancer related death of men in most Western countries [1]. The highly variable clinical course of the disease cannot be predicted reliably enough by currently available criteria such as Gleason grade, clinical stage and PSA value. Additional and better prognostic markers are needed to differentiate between aggressive high risk and non-aggressive low risk cancer subtypes in order to prevent unnecessary invasive treatments.
The DNA repair endonuclease ERCC1 (Excision Repair Cross-Complementation Group 1) catalyzes 5′ incision during nucleotide excision repair process (NER) [2, 3]. ERCC1 has been described to be physiologically expressed in several tissues including skin, breast, intestine, testis, and ovary [4]. Overexpression of ERCC1 has been found in many cancer types such as urothelial carcinoma [5], head and neck squamous cell carcinoma [6] and non-small cell lung cancer [7]. For these entities it has been proposed that ERCC1 overexpression may serve as a prognostic and/or predictive tumor marker [5–9].
ERCC1 is of potential interest in prostate cancer. Experimental data from a mouse model system suggested an altered ERCC1 function as potential driver for an invasive prostate cancer phenotype [10]. Moreover, a specific nucleotide polymorphism of the ERCC1 gene was linked to prostate cancer aggressiveness in a Spanish cohort study of 494 men [11]. The present study evaluates the clinical impact of ERCC1 expression in human prostate cancer. For this purpose, a preexisting prostate cancer tissue microarray was examined for ERCC1 expression by immunohistochemistry.
Methods
Patients
Twelve thousand four hundred twenty seven prostatectomy specimens were obtained from consecutive patients treated between 1992 and 2012 in the Department of Urology and the Martini Clinics at the University Medical Center Hamburg-Eppendorf. Tumor stage, Gleason grade, nodal stage and the resection margin status were recorded. Classical Gleason categories and “quantitative” Gleason grading was performed as described [12]. Follow-up data were available for a total of 12,344 patients (median 36 months, range 1 to 241 months; Table 1). Prostate specific antigen (PSA) recurrence was defined as a postoperative PSA of ≥0.2 ng / ml and increasing. All prostate specimens were embedded for histological analysis by a standard procedure [13]. The TMA was produced as described [14, 15]. In brief, one 0.6 mm core sample was taken from a representative tissue block and distributed among 27 TMA blocks, each with 144 to 522 samples. Each TMA block contained various control and normal prostate tissue. The molecular database attached to this TMA contained results on ERG expression, ERG break apart FISH analysis [16], deletion status of 5q21 (CHD1) [17], 6q15 (MAP3K7) [18], PTEN (10q23) [19–21] and 3p13 (FOXP1) [22]).
Table 1.
Pathological and clinical data of the arrayed prostate cancers
| No. of patients (%) | ||
|---|---|---|
| Study cohort on TMA (n = 12,427) | Biochemical relapse among categories | |
| Follow-up (mo) | ||
| n | 11,665 (93.9%) | 2769 (23.7%) |
| Mean | 62.9 | − |
| Median | 50.0 | − |
| Age (y) | ||
| ≤50 | 334 (2.7%) | 81 (24.3%) |
| 51–59 | 3061 (24.8%) | 705 (23%) |
| 60–69 | 7188 (58.2%) | 1610 (22.4%) |
| ≥70 | 1761 (14.3%) | 370 (21%) |
| Pretreatment PSA (ng/ml) | ||
| <4 | 1585 (12.9%) | 242 (15.3%) |
| 4–10 | 7480 (60.9%) | 1355 (18.1%) |
| 10–20 | 2412 (19.6%) | 737 (30.6%) |
| >20 | 812 (6.6%) | 397 (48.9%) |
| pT stage (AJCC 2002) | ||
| pT2 | 8187 (66.2%) | 1095 (13.4%) |
| pT3a | 2660 (21.5%) | 817 (30.7%) |
| pT3b | 1465 (11.8%) | 796 (54.3%) |
| pT4 | 63 (0.5%) | 51 (81%) |
| Gleason grade | ||
| ≤3 + 3 | 2848 (22.9%) | 234 (8.2%) |
| 3 + 4 | 6679 (53.8%) | 1240 (18.6%) |
| 3 + 4 Tert.5 | 433 (3.5%) | 115 (26.6%) |
| 4 + 3 | 1210 (9.7%) | 576 (47.6%) |
| 4 + 3 Tert.5 | 646 (5.2%) | 317 (49.1%) |
| ≥4 + 4 | 596 (4.8%) | 348 (58.4%) |
| pN stage | ||
| pN0 | 6970 (91%) | 1636 (23.5%) |
| pN+ | 693 (9%) | 393 (56.7%) |
| Surgical margin | ||
| Negative | 9990 (81.9%) | 1848 (18.5%) |
| Positive | 2211 (18.1%) | 853 (38.6%) |
Percent in the column “Study cohort on TMA” refers to the fraction of samples across each category. Percent in column “Biochemical relapse among categories” refers to the fraction of samples with biochemical relapse within each parameter in the different categories. NOTE: Numbers do not always add up to 12,427 in different categories because of cases with missing data. Abbreviation: AJCC, American Joint Committee on Cancer
Immunohistochemistry
Newly cut sections of the complete TMA were stained on the same day in a single experiment. Slides were deparaffinized and antigen was retrieved by heat (5 min at 121 °C, pH 7.8 Tris-EDTA-citrate buffer). ERCC1 specific mouse monoclonal antibody clone UMAB8, BioCAT GmbH, Heidelberg; cat#UM500008; dilution 1:150) was applied at 37 °C for 60 min. Bound antibody was visualized with the EnVision Kit (Dako, Glostrup, Denmark). ERCC1 typically stained 100% tumor cell nuclei in a single tissue spot. Staining intensity was assessed semi-quantitatively as negative, weak, moderate and strong.
Statistics
Contingency tables were calculated to analyze associations between ERCC1 expression and clinico-pathological parameters. Chi-square (Likelihood) test was employed to identify significant relationships between these parameters. The F-test was used in analysis of variance to detect differences of the mean of groups. Kaplan-Meier curves were generated for the event of PSA recurrence free survival and the log-Rank test was applied to test for significant differences between stratified survival curves. The prognostic significance of pathological, molecular and clinical parameters was assessed by Cox proportional hazards regression analysis. All calculations were done with JMP® software (SAS Institute Inc., NC, USA).
Results
Technical issues
A total of 11,665 (93.9%) patients had follow up data and 10,436 (84%) of samples were interpretable in the TMA analysis (Table 1). Reasons for non-informative cases were lack of tissue samples (1991 spots; 16%), absence of unequivocal cancer tissue in the TMA spot or missing data.
ERCC1 immunohistochemistry
ERCC1 staining was negative or weak in basal and luminal cells of normal prostate glands. Positive nuclear ERCC1 staining was seen in 64.7% of 10,436 interpretable tissue samples, and was graded as weak in 37.1%, moderate in 22.6%, and strong in 5% of tumors. Representative images of ERCC1 immunohistochemistry in prostate cancer samples are shown in Fig. 1. Strong ERCC1 staining was linked to advanced pT stage, high Gleason grade, positive lymph nodes, high pre-operative serum PSA measurement, and positive surgical margin status (p ≤ 0.0078; Table 2).
Fig. 1.
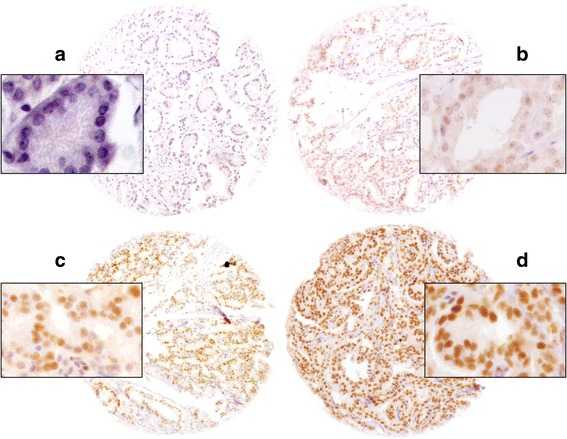
Representative pictures of a) negative, b) weak, c) moderate and d) strong ERCC1 staining in prostate cancer
Table 2.
Association between ERCC1 staining results and prostate cancer clinical characteristics
| ERCC1 (%) | ||||||
|---|---|---|---|---|---|---|
| Parameter | n evaluable | Negative | Weak | Moderate | Strong | p value |
| All cancers | 10,436 | 35.4 | 37.1 | 22.6 | 5.0 | |
| Tumor stage | ||||||
| pT2 | 6790 | 38.4 | 37.3 | 20.1 | 4.2 | <0.0001 |
| pT3a | 2299 | 31.6 | 35.5 | 26.4 | 6.6 | |
| pT3b-pT4 | 1308 | 26.4 | 38.8 | 28.5 | 6.3 | |
| Gleason grade | ||||||
| ≤3 + 3 | 2363 | 46.3 | 34.2 | 16.5 | 3.0 | <0.0001 |
| 3 + 4 | 5630 | 34.7 | 37.4 | 22.9 | 5.0 | |
| 3 + 4 Tert.5 | 368 | 33.4 | 40.8 | 22.0 | 3.8 | |
| 4 + 3 | 1040 | 25.6 | 38.8 | 28.4 | 7.3 | |
| 4 + 3 Tert.5 | 563 | 23.4 | 40.0 | 29.1 | 7.5 | |
| ≥4 + 4 | 466 | 25.8 | 38.2 | 29.0 | 7.1 | |
| Lymph node metastasis | ||||||
| N0 | 5856 | 32.7 | 37.6 | 23.9 | 5.8 | 0.0037 |
| N+ | 585 | 25.6 | 39.8 | 27.9 | 6.7 | |
| Preoperative PSA level (ng/ml) | ||||||
| <4 | 1293 | 32.6 | 40.5 | 21.9 | 4.9 | 0.0078 |
| 4–10 | 6256 | 35.2 | 37.8 | 22.1 | 4.8 | |
| 10–20 | 2058 | 37.5 | 33.9 | 23.5 | 5.1 | |
| >20 | 714 | 35.6 | 33.9 | 24.8 | 5.7 | |
| Surgical margin | ||||||
| Negative | 8294 | 36.1 | 37.5 | 21.8 | 4.7 | <0.0001 |
| Positive | 1953 | 32.7 | 35.5 | 25.9 | 5.9 | |
Association with TMPRSS2:ERG fusion status
ERCC1 expression was massively linked to the presence of ERG expression and rearrangement. At least weak ERCC1 staining was found in 85.4% of cancers with immunohistochemically detected ERG expression and in 81.4% of tumors with ERG-rearrangement, but only in 52.6% (IHC) or 61.8% (FISH) of ERG-negative cancers (p < 0.0001 each, Fig. 2). ERCC1 staining was similarly linked to unfavorable tumor phenotype in subsets of both ERG-negative and ERG-positive cancers (Additional file 1: Tables S1 and S2).
Fig. 2.
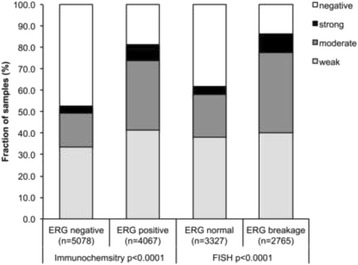
Positive ERCC1 staining correlates with ERG staining in immunochemistry or ERG breakage in fluorescence in situ hybridization (FISH)
Associations with key genomic changes of prostate cancer
Chromosomal deletions represent the most frequent genomic changes in prostate cancer next to TMPRSS2:ERG fusion. To study whether ERCC1 expression might be particularly linked to any of the most common deletions, ERCC1 data were compared to preexisting findings on 10q23 (PTEN), 3p13 (FOXP1), 6q15 (MAP3K7) and 5q21 (CHD1) deletions (Fig. 3). These analyses showed that ERCC1 expression was strongly linked to all examined deletions. This was particularly evident for ERG negative carcinomas (Fig. 3b) and only marginally visible in ERG positive carcinomas (Fig. 3c). Moreover, the level of ERCC1 expression was also related to the number of deletions found in all cancer (deletion load, Fig. 4; p < 0.0001). This held true also in the subset of ERG-negative and ERG-positive cancers (p < 0.0001; data not shown).
Fig. 3.
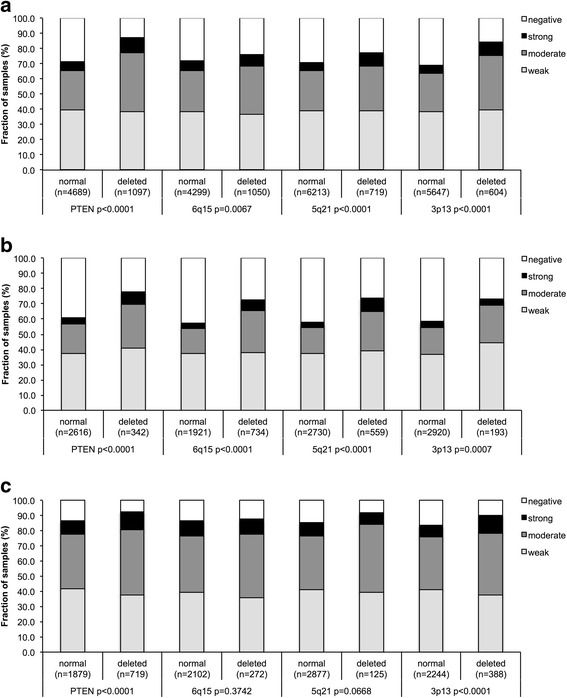
Association between positive ERCC1 staining and 10q23 (PTEN), 5q21 (CHD1), 6q15 (MAP3K7), and 3p13 (FOXP1) deletion in a) all cancers, b) the ERG-negative and c) ERG-positive subset
Fig. 4.
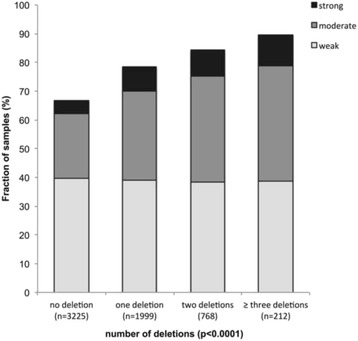
Association between positive ERCC1 staining and number of deletions in the ERG-positive subset
Association with cell proliferation
High levels of ERCC1 staining were significantly linked to increased tumor cell proliferation measured as Ki67 labeling index (Ki67LI) (Table 3, p < 0.0001). This association held also true in almost all subgroups of cancers with identical Gleason grade (≤3 + 3; 3 + 4; 4 + 3; p < 0.0001 each).
Table 3.
Association between ERCC1 expression and Ki67-labeling index in all, low- grade and high-grade prostate cancers
| Gleason grade | ERCC1 | n | Ki67 LI (mean ± SD) | p value |
|---|---|---|---|---|
| All grades | Negative | 2189 | 1.94 ± 0.06 | <0.0001 |
| Weak | 2210 | 2.95 ± 0.06 | ||
| Moderate | 1422 | 3.38 ± 0.07 | ||
| Strong | 319 | 4.02 ± 0.14 | ||
| ≤3 + 3 | Negative | 672 | 1.68 ± 0.08 | <0.0001 |
| Weak | 471 | 2.46 ± 0.09 | ||
| Moderate | 233 | 3.04 ± 0.13 | ||
| Strong | 44 | 2.77 ± 0.31 | ||
| 3 + 4 | Negative | 1200 | 1.89 ± 0.07 | <0.0001 |
| Weak | 1308 | 2.86 ± 0.06 | ||
| Moderate | 866 | 3.27 ± 0.08 | ||
| Strong | 190 | 3.70 ± 0.17 | ||
| 4 + 3 | Negative | 241 | 2.51 ± 0.22 | <0.0001 |
| Weak | 342 | 3.57 ± 0.18 | ||
| Moderate | 240 | 3.87 ± 0.22 | ||
| Strong | 65 | 4.98 ± 0.42 | ||
| ≥4 + 4 | Negative | 62 | 3.79 ± 0.56 | 0.05 |
| Weak | 80 | 4.8 ± 0.49 | ||
| Moderate | 74 | 4.28 ± 0.51 | ||
| Strong | 19 | 6.89 ± 1.01 |
Associations with prostate-specific antigen recurrence
The prognostic impact of pT stage (Fig. 5a), traditional Gleason grade (Fig. 5b), and quantitative Gleason grade (Fig. 5c) were strongly linked to PSA recurrence. There was a significant association between high ERCC1 staining levels and early PSA recurrence (p< 0.0001; Fig. 5d). This held also true for the subgroups of ERG-negative (p< 0.0001; Fig. 5e) and ERG-positive (p< 0.0001; Fig. 5f) cancers. Analyzing subsets of tumors with comparable traditional and quantitative Gleason grades revealed that ERCC1 expression measurement did not provide very much additional prognostic impact in morphologically characterized tumor sets. Significant associations with PSA recurrence were seen in Gleason 3 + 3 = 6 (p = 0.061), Gleason 3 + 4 (p = 0.0021) and 4 + 3 carcinomas (p = 0.0494) but not in tumors with a Gleason ≥4 + 4 (Fig. 6a). A further refined subgroup analysis by quantitative Gleason grading showed that high ERCC1 expression identified cancers with worse outcome only in those 3 + 4 carcinomas with a minimal fraction of Gleason 4 (≤5%) (Fig. 6b; p = 0.0004). None of the other groups with a comparable quantitative Gleason grade showed outcome differences according to the ERCC1 status (Fig. 6c-f).
Fig. 5.
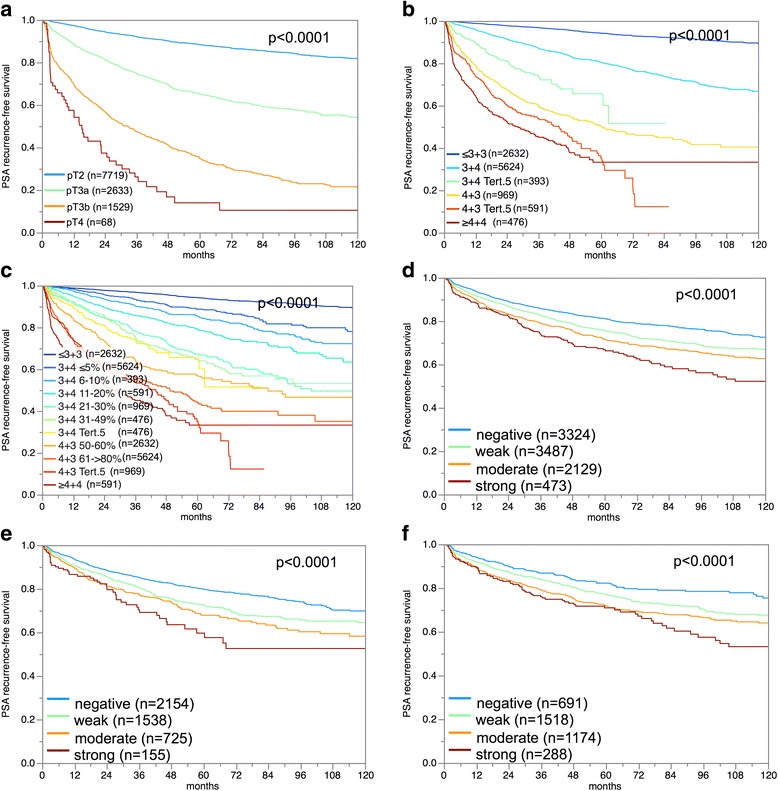
Prostate specific antigen (PSA) recurrence free survival correlates with a) pathological stage, b) classical Gleason grade, c) quantitative Gleason grade, and ERCC1 expression in d) all cancers, e) the ERG-fusion negative and f) positive subset
Fig. 6.
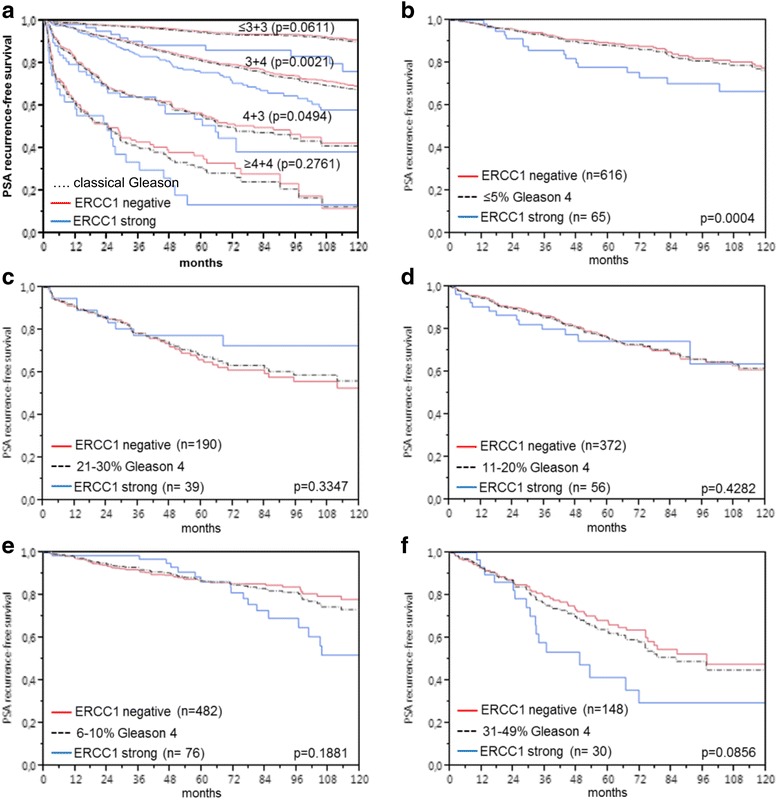
Limited prognostic impact of ERCC1 expression in cancers grouped by a) classical and b-f) quantitative Gleason score. The quantitative Gleason score is defined by the percentage of Gleason 4 patterns. Black dotted line denotes Gleason score category result, red line negative and blue line strongly positive ERCC1 cancers within the respective category
Multivariate analysis
Four different scenarios were tested. All these analyses were also done in the ERG-negative and ERG-positive subset (Table 4). Scenario 1 used the post-operatively available parameters (pathological tumor stage (pT), lymph node status (pN), surgical margin status, pre-operative PSA value and classical Gleason grade). In Scenario 2 the nodal status was dropped to reduce missing data as lymph node dissection is not yet standardized in radical prostatectomy. Scenario 3 included ERCC1 expression, pre-operative PSA, clinical tumor stage (cT stage) and Gleason grade obtained on the prostatectomy specimen. Since post-operative determination of a tumors Gleason grade is more precise than the preoperatively determined Gleason grade [23], scenario 4 was added to better model the pre-operative situation. Here, the pre-operative biopsy Gleason grade was combined with pre-operative PSA, cT stage and ERCC1 expression. Overall, these scenarios suggest a relevant role of ERCC1 expression as a prognostic factor, which tended to be - especially in the pre-operative setting - independent of established factors (scenario 3 and 4).
Table 4.
Multivariate analysis including ERCC1 expression in all cancers, ERG-negative and ERG-positive subset
| Tumor subset | Scenario | n analyzable | p value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Preoperative PSA-Level | pT Stage | cT Stage | Gleason grade prostatectomy | Gleason grade biopsy | pN Stage | R Stage | ERCC1-Expression | |||
| All cancers | 1 | 5644 | 0.0009 | <0.0001 | - | <0.0001 | - | <0.0001 | 0.0008 | 0.0746 |
| 2 | 9193 | <0.0001 | <0.0001 | - | <0.0001 | - | - | <0.0001 | 0.0052 | |
| 3 | 9062 | <0.0001 | - | <0.0001 | <0.0001 | - | - | - | 0.0045 | |
| 4 | 8926 | <0.0001 | - | <0.0001 | - | <0.0001 | - | - | <0.0001 | |
| ERG-negative subset | 1 | 2829 | <0.0001 | <0.0001 | - | <0.0001 | - | 0.0001 | 0.0846 | 0.1110 |
| 2 | 4471 | <0.0001 | <0.0001 | - | <0.0001 | - | - | 0.0002 | 0.0184 | |
| 3 | 4432 | <0.0001 | - | <0.0001 | <0.0001 | - | - | - | 0.0526 | |
| 4 | 4368 | <0.0001 | - | <0.0001 | - | <0.0001 | - | - | <0.0001 | |
| ERG-positive subset | 1 | 2242 | 0.0057 | <0.0001 | - | <0.0001 | - | 0.0763 | 0.0092 | 0.0626 |
| 2 | 3584 | <0.0001 | <0.0001 | - | <0.0001 | - | - | <0.0001 | 0.0613 | |
| 3 | 3506 | <0.0001 | - | <0.0001 | <0.0001 | - | - | - | 0.0746 | |
| 4 | 3451 | <0.0001 | - | <0.0001 | - | <0.0001 | - | - | 0.0009 | |
Discussion
In this study increased expression of the DNA repair factor ERCC1 was identified as a strong prognostic marker in prostate cancer, in particular for low-grade tumors. Under the selected experimental conditions, detectable ERCC1 staining was found in 65% of prostate tumors. ERCC1 expression was virtually not detected in normal prostate epithelium. This finding suggests an up-regulation of ERCC1 during tumor development in a proportion of prostate cancers. So far, comprehensive studies on ERCC1 expression in clinical prostate cancer samples are lacking. However, high-level ERCC1 expression has been reported from the prostate cancer cell lines DU-145 and LNCaP [24]. Also, the 12 prostate cancer samples, included in the Human Protein Atlas, showed ERCC1 staining in 83–100% of cases depending on the antibody used [25].
The strong association of elevated ERCC1 expression with adverse morphological and clinical features of prostate cancer found in this study, argues for a role of ERCC1 overexpression/activation in prostate cancer progression. This assumption is supported by findings in other cancer types where associations between high ERCC1 expression levels and reduced overall survival had been found. This, for example, includes reports on NSCLC as well as in gastric and pancreatic cancers [7, 9, 26].
The large number of samples in this TMA and the associated database with numerous molecular features allowed us to draw conclusions on the mechanistic role of ERCC1 in prostate cancer. ERCC1-mediated endonucleolytic incision and homologous recombination (HR) have been implicated in the repair of DNA-interstrand crosslinks (ICLs) which induce a potent replication block followed by formation and repair of double strand breaks (DSBs) [27]. Defective DSB repair and faulty DNA replication are thought to be involved in the generation of chromosomal aberrations commonly seen in cancer cells [28]. The striking association found between elevated ERCC1 expression and chromosomal deletions as well as with a positive ERG status is suggestive of a link between ERCC1 activation and presence of chromosomal damage. ERCC1 may thus represent a surrogate for genomic instability in proliferative active prostate cancer cells. This hypothesis is further supported by the continuous increase of ERCC1 levels with the number of deletions detected, suggesting high level activity of replication associated DNA damage repair mechanisms in subsets of prostate cancer with generation of chromosomal aberration via DSB formation and faulty repair. TMPRSS2:ERG fusions were most strikingly linked to ERCC1 expression. The reason for this particular strong association remains unclear. Earlier studies had not implicated ERCC1 as a gene that is directly regulated by the transcription factor ERG [29–31]. The association between deletions and ERCC1 expression was less clear in ERG positive than in ERG-negative cancers, which is likely due to the (already) markedly elevated levels of ERCC1 in ERG-positive tumors. In case of additional deletions, this may not allow for a further elevation measurable under the experimental conditions applied in this study. The observed strong association between high levels of ERCC1 and rapid tumor cell proliferation, as determined by the Ki67 labeling index, is consistent with the involvement of ERCC1 in the repair of replication associated DNA damage [32] as rapidly proliferating cancer cells are subjected to high replication stress [33, 34].
ERCC1 was an independent predictor of poor outcome in most multivariate analyses suggesting a strong clinical utility of ERCC1 measurement. Remarkably, the analysis of the prognostic role of ERCC1 expression in subgroups of prostate cancer that were narrowly defined by identical quantitative Gleason grades suggested a limitation of the prognostic value of ERCC1 measurement to the earliest lesions, i.e. Gleason 3 + 3 or 3 + 4 with only minimal (≤5%) Gleason 4 fraction. This limitation of the prognostic impact to these subgroups is not a disappointment as these tumors are subject to the most difficult therapeutic decision making with options ranging from active surveillance to prostatectomy. That ERCC1 expression failed to provide prognostic information in most subgroups in cancers with comparable quantitative Gleason findings also demonstrates how high the bar lies for prognostic molecular tests in prostate cancer. The Gleason grading system is purely based on the simple distinction of architectural features, neglects any cytological criteria, but is extremely powerful. The prognostic power of the Gleason grade is much higher than the histologic grading in various other cancer types, such as for example kidney cancer [35] or invasive bladder cancer [36]. This holds true if the Gleason grading method is limited 5 prognostic subgroups [37]. Based on the analysis of a cohort of more than 10,000 prostate cancers available at our institution, we had recently shown, that using the percentage of Gleason 4 grades as a continuous variable could expand Gleason Grade information. Both in biopsies and in prostatectomy samples, prostate cancer prognosis deteriorates gradually with increasing percentage of Gleason 4 pattern (quantitative Gleason Grade) [12]. Given the high impact of pure morphologic information in prostate cancer, we believe that a further improvement of morphologic assessment going beyond architecture and also involving digital image analysis and deep machine learning will play a very important role in prostate cancer assessment in the future.
Conclusions
In summary, elevated expression of ERCC1 is strongly linked to unfavorable tumor phenotype and PSA recurrence in prostate cancer. In this study, an association between ERCC1 overexpression and chromosomal aberrations (including both ERG fusion and deletions) was observed. These findings suggest overexpression of ERCC1 in the context of replication associated DNA damage repair, genomic instability and generation of structural chromosomal alterations.
Acknowledgements
The authors appreciate the excellent technical support of Christina Koop, Sylvia Schnöger and Sasha Eghtessadi.
Funding
No specific funding was received for this study.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CHD1
Chromodomain-helicase-DNA-binding protein 1
- ERCC1
Excision repair cross-complementation
- ERG
Erythroblast transformation-specific (ETS) related gene
- FISH
Fluorescence in situ hybridization
- FOXP1
Forkhead box protein P1
- Ki67LI Ki67
Labeling index
- MAP3K7
Mitogen-activated protein kinase kinase kinase 7
- NER
Nucleotide excision repair
- PSA
Prostate specific antigen
- PTEN
Phosphatase and tensin homolog
- TMA
Tissue microarray
- TMPRSS2
Transmembrane protease, serine 2
Additional file
Association between ERCC1 immunostaining results and prostate cancer phenotype in ERG-negative tumors. Table S2. Association between ERCC1 immunostaining results and prostate cancer phenotype in ERG-positive tumors. (PDF 112 kb)
Authors’ contributions
FJ, CH, RS, and GS designed the study, and drafted the manuscript. TS, BB, SBR, and TSch participated in study design. IT, CS, BT, CW performed immunohistochemistry analysis and scoring. MK and RS participated in pathology data analysis. NM and RS performed statistical analysis. GS, SBR, and MK participated in data interpretation, and helped to draft the manuscript. WW, KM, SW, DP participated in data interpretation. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the ethics committee Ärztekammer Hamburg (WF-049/09 and PV3652). According to local laws, informed consent was not required for this study (HmbKHG, §12,1). Patient records were anonymized and de-identified prior to analysis. All work has been carried out in compliance with the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-017-3489-9) contains supplementary material, which is available to authorized users.
Contributor Information
Frank Jacobsen, Email: f.jacobsen@uke.de.
Billurvan Taskin, Email: billur9@hotmail.de.
Nathaniel Melling, Email: n.melling@uke.de.
Charlotte Sauer, Email: charlottesauer4@gmail.com.
Corinna Wittmer, Email: c.wittmer@uke.de.
Claudia Hube-Magg, Email: c.hube@uke.de.
Martina Kluth, Email: m.kluth@uke.de.
Ronald Simon, Phone: +49 40 7410 57214, Email: R.Simon@uke.de.
Dirk Pehrke, Email: pehrke@uke.de.
Burkhard Beyer, Email: b.beyer@uke.de.
Thomas Steuber, Email: steuber@uke.de.
Imke Thederan, Email: i.thederan@uke.de.
Guido Sauter, Email: g.sauter@uke.de.
Thorsten Schlomm, Email: t.schlomm@uke.de.
Waldemar Wilczak, Email: w.wilczak@uke.de.
Katharina Möller, Email: ka.moeller@uke.de.
Sören A. Weidemann, Email: s.weidemann@uke.de
Susanne Burdak-Rothkamm, Email: s.burdak-rothkamm@uke.de.
References
- 1.International Agency for Research on Cancer: Cancer Mortality Database [http://gco.iarc.fr].
- 2.Matsunaga T, Mu D, Park CH, Reardon JT, Sancar A. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J Biol Chem. 1995;270(35):20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 3.Sijbers AM, van der Spek PJ, Odijk H, van den Berg J, van Duin M, Westerveld A, Jaspers NG, Bootsma D, Hoeijmakers JH. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 1996;24(17):3370–3380. doi: 10.1093/nar/24.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng L, Guan Y, Li L, Legerski RJ, Einspahr J, Bangert J, Alberts DS, Wei Q. Expression in normal human tissues of five nucleotide excision repair genes measured simultaneously by multiplex reverse transcription-polymerase chain reaction. Cancer Epidemiol Biomark Prev. 1999;8(9):801–807. [PubMed] [Google Scholar]
- 5.Klatte T, Seitz C, Rink M, Roupret M, Xylinas E, Karakiewicz P, Susani M, Shariat SF. ERCC1 as a prognostic and predictive biomarker for Urothelial carcinoma of the bladder following radical Cystectomy. J Urol. 2015;194(5):1456–1462. doi: 10.1016/j.juro.2015.06.099. [DOI] [PubMed] [Google Scholar]
- 6.Ciaparrone M, Caspiani O, Bicciolo G, Signorelli D, Simonelli I, de Campora L, Mazzarella G, Mecozzi A, Pianelli C, Camaioni A, et al. Predictive role of ERCC1 expression in head and neck Squamous cell carcinoma patients treated with surgery and adjuvant Cisplatin-based Chemoradiation. Oncology. 2015;89(4):227–234. doi: 10.1159/000430447. [DOI] [PubMed] [Google Scholar]
- 7.Deng Q, Yang H, Lin Y, Qiu Y, Gu X, He P, Zhao M, Wang H, Xu Y, Lin Y, et al. Prognostic value of ERCC1 mRNA expression in non-small cell lung cancer, breast cancer, and gastric cancer in patients from southern China. Int J Clin Exp Pathol. 2014;7(12):8312–8321. [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Wu J, Chen Y, Tang W, Peng Q, Deng Y, Xie L, Wang J, Huang S, Li R, et al. ERCC1 expression levels predict the outcome of platinum-based chemotherapies in advanced bladder cancer: a meta-analysis. Anti-Cancer Drugs. 2014;25(1):106–114. doi: 10.1097/CAD.0000000000000021. [DOI] [PubMed] [Google Scholar]
- 9.Maithel SK, Coban I, Kneuertz PJ, Kooby DA, El-Rayes BF, Kauh JS, Sarmiento J, Staley CA, 3rd, Volkan Adsay N. Differential expression of ERCC1 in pancreas adenocarcinoma: high tumor expression is associated with earlier recurrence and shortened survival after resection. Ann Surg Oncol. 2011;18(9):2699–2705. doi: 10.1245/s10434-011-1610-x. [DOI] [PubMed] [Google Scholar]
- 10.Matoka DJ, Yao V, Harya DS, Gregg JL, Robinson AR, Niedernhofer LJ, Parwani AV, Maier C, Bacich DJ. Deficiency of DNA repair nuclease ERCC1-XPF promotes prostate cancer progression in a tissue recombination model. Prostate. 2012;72(11):1214–1222. doi: 10.1002/pros.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriquez-Hernandez LA, Valenciano A, Foro-Arnalot P, Alvarez-Cubero MJ, Cozar JM, Suarez-Novo JF, Castells-Esteve M, Fernandez-Gonzalo P, De-Paula-Carranza B, Ferrer M, et al. Single nucleotide polymorphisms in DNA repair genes as risk factors associated to prostate cancer progression. BMC Med Genet. 2014;15:143. doi: 10.1186/s12881-014-0143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter G, Steurer S, Clauditz TS, Krech T, Wittmer C, Lutz F, Lennartz M, Janssen T, Hakimi N, Simon R, et al. Clinical utility of quantitative Gleason grading in prostate biopsies and prostatectomy specimens. Eur Urol. 2016;69(4):592–598. doi: 10.1016/j.eururo.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Schlomm T, Iwers L, Kirstein P, Jessen B, Kollermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M, et al. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21(11):1371–1379. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 14.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 15.Heumann A, Kaya O, Burdelski C, Hube-Magg C, Kluth M, Lang DS, Simon R, Beyer B, Thederan I, Sauter G, et al. Up regulation and nuclear translocation of Y-box binding protein 1 (YB-1) is linked to poor prognosis in ERG-negative prostate cancer. Sci Rep. 2017;7(1):2056. doi: 10.1038/s41598-017-02279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minner S, Wittmer C, Graefen M, Salomon G, Steuber T, Haese A, Huland H, Bokemeyer C, Yekebas E, Dierlamm J, et al. High level PSMA expression is associated with early PSA recurrence in surgically treated prostate cancer. Prostate. 2011;71(3):281–288. doi: 10.1002/pros.21241. [DOI] [PubMed] [Google Scholar]
- 17.Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73(9):2795–2805. doi: 10.1158/0008-5472.CAN-12-1342. [DOI] [PubMed] [Google Scholar]
- 18.Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K, et al. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26(7):975–983. doi: 10.1038/modpathol.2012.236. [DOI] [PubMed] [Google Scholar]
- 19.Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, Kotschau D, Tennstedt P, Huang J, Gerhauser C, et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181(2):401–412. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Burdelski C, Menan D, Tsourlakis MC, Kluth M, Hube-Magg C, Melling N, Minner S, Koop C, Graefen M, Heinzer H, et al. The prognostic value of SUMO1/Sentrin specific peptidase 1 (SENP1) in prostate cancer is limited to ERG-fusion positive tumors lacking PTEN deletion. BMC Cancer. 2015;15:538. doi: 10.1186/s12885-015-1555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluth M, Runte F, Barow P, Omari J, Abdelaziz ZM, Paustian L, Steurer S, Christina Tsourlakis M, Fisch M, Graefen M, et al. Concurrent deletion of 16q23 and PTEN is an independent prognostic feature in prostate cancer. Int J Cancer. 2015;137(10):2354–2363. doi: 10.1002/ijc.29613. [DOI] [PubMed] [Google Scholar]
- 22.Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M, et al. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231(1):130–141. doi: 10.1002/path.4223. [DOI] [PubMed] [Google Scholar]
- 23.Ellis SD, Blackard B, Carpenter WR, Mishel M, Chen RC, Godley PA, Mohler JL, Bensen JT. Receipt of National Comprehensive Cancer Network guideline-concordant prostate cancer care among African American and Caucasian American men in North Carolina. Cancer. 2013;119(12):2282–2290. doi: 10.1002/cncr.28004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yacoub A, McKinstry R, Hinman D, Chung T, Dent P, Hagan MP. Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. Radiat Res. 2003;159(4):439–452. doi: 10.1667/0033-7587(2003)159[0439:EGFAIR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 26.Yamada Y, Boku N, Nishina T, Yamaguchi K, Denda T, Tsuji A, Hamamoto Y, Konishi K, Tsuji Y, Amagai K, et al. Impact of excision repair cross-complementing gene 1 (ERCC1) on the outcomes of patients with advanced gastric cancer: correlative study in Japan clinical oncology group trial JCOG9912. Ann Oncol. 2013;24(10):2560–2565. doi: 10.1093/annonc/mdt238. [DOI] [PubMed] [Google Scholar]
- 27.Vare D, Groth P, Carlsson R, Johansson F, Erixon K, Jenssen D. DNA interstrand crosslinks induce a potent replication block followed by formation and repair of double strand breaks in intact mammalian cells. DNA Repair (Amst) 2012;11(12):976–985. doi: 10.1016/j.dnarep.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Hasty P, Montagna C. Chromosomal rearrangements in cancer: detection and potential causal mechanisms. Mol Cell Oncol. 2014;1(1):e29904. [DOI] [PMC free article] [PubMed]
- 29.Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H, et al. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jhavar S, Brewer D, Edwards S, Kote-Jarai Z, Attard G, Clark J, Flohr P, Christmas T, Thompson A, Parker M, et al. Integration of ERG gene mapping and gene-expression profiling identifies distinct categories of human prostate cancer. BJU Int. 2009;103(9):1256–1269. doi: 10.1111/j.1464-410X.2008.08200.x. [DOI] [PubMed] [Google Scholar]
- 31.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naim V, Wilhelm T, Debatisse M, Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat Cell Biol. 2013;15(8):1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 33.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15(5):276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 34.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–448. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- 35.Ficarra V, Martignoni G, Maffei N, Brunelli M, Novara G, Zanolla L, Pea M, Artibani W. Original and reviewed nuclear grading according to the Fuhrman system: a multivariate analysis of 388 patients with conventional renal cell carcinoma. Cancer. 2005;103(1):68–75. doi: 10.1002/cncr.20749. [DOI] [PubMed] [Google Scholar]
- 36.Grignon DJ. The current classification of urothelial neoplasms. Mod Pathol. 2009;22(Suppl 2):S60–S69. doi: 10.1038/modpathol.2008.235. [DOI] [PubMed] [Google Scholar]
- 37.Pierorazio PM, Walsh PC, Partin AW, Epstein JI. Prognostic Gleason grade grouping: data based on the modified Gleason scoring system. BJU Int. 2013;111(5):753–760. doi: 10.1111/j.1464-410X.2012.11611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


