Abstract
Normal neuronal communication and synaptic plasticity at glutamatergic synapses requires dynamic regulation of postsynaptic molecules. Protein expression and protein post-translational modifications regulate protein interactions that underlie this organization. In this Review, we highlight data obtained over the last 20 years that have used qualitative and quantitative proteomics-based approaches to identify postsynaptic protein complexes. Herein, we describe how these proteomics studies have helped lay the foundation for understanding synaptic physiology and perturbations in synaptic signaling observed in different pathologies. We also describe emerging technologies that can be useful in these analyses. We focus on protein complexes associated with the highly abundant and functionally critical proteins: calcium/calmodulin-dependent protein kinase II, the N-methyl-D-aspartate, and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid glutamate receptors, and postsynaptic density protein of 95 kDa.
Keywords: Mass spectrometry, calcium/calmodulin-dependent protein kinase II, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor, N-methyl-D-aspartate receptor, postsynaptic density protein 95, postsynaptic density, signaling
Graphical abstract
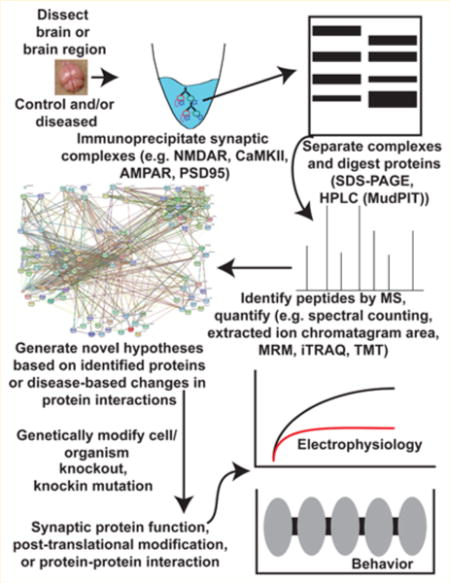
INTRODUCTION
While mass spectrometry based approaches have been used to quantify protein abundance changes that underlie long-term plasticity and learning and memory,1 these approaches are also well-suited to understand postsynaptic protein post-translational modifications, interactions, and protein function that allow for a rapid response to alterations in neuronal excitability. Following presynaptic release, glutamate diffuses across the synaptic cleft to activate multiple classes of glutamate receptors, including N-methyl-D-aspartate receptor (NMDARs) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs). Signaling molecules such as calcium/calmodulin-dependent protein kinase II (CaMKII) associate with glutamate receptors or voltage-gated calcium channels to respond to calcium that enters through these proteins. While CaMKII is one of the most abundant enzymes found in the postsynaptic density (PSD; Figure 1),2,3 many other abundant proteins act as scaffolds and are required for localizing glutamate receptors and other machinery directly opposed to release sites (Figure 1).4 While early studies employed electron microscopy and immunoblotting to isolated PSDs obtained via subcellular fractionation, shotgun-based proteomics approaches are exquisitely sensitive to identify and quantify components of the PSD. These approaches have utilized PSD enrichment strategies such as subcellular fractionation to isolate PSDs followed by mass spectrometry to identify and quantify PSD proteomes.1,5–18 It is important to note that different PSD preparations and mass spectrometry instrumentation may lead to distinct PSD proteomes.19–21 However, there is overlap between many of these approaches. For instance, integrating multiple approaches and studies, Grant and colleagues defined what they called the consensus PSD, which contained 466 total proteins. They compared 8 different studies and found overlap between the PSD proteomes that ranged between 31% and 100%.19 These approaches have identified highly abundant PSD proteins such as NMDARs, AMPARs, CaMKII, and PSD-95. Here we review how proteomics approaches have been used to interrogate postsynaptic protein complexes associated with the above proteins and discuss how this knowledge has increased our understanding of synaptic protein organization and function.
Figure 1.
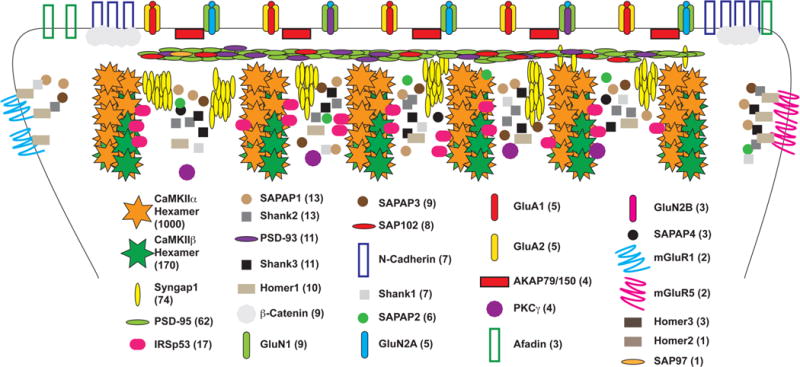
Relative expression of PSD proteins. Schematic representation of multiple PSD-enriched proteins shown at relative abundance based on previous quantitative data.4 Values in parentheses represent approximate arbitrary expression of PSD molecules with CaMKII, the most abundant PSD protein, set to 1000 individual subunits. The number of molecules shown is representative of this arbitrary expression.
Understanding how synaptic proteins are organized is important in decoding physiological changes that underlie normal learning and memory. These learning and memory changes are encoded via a process termed synaptic plasticity. Long-term potentiation (LTP) and long-term depression (LTD) are two molecular correlates of synaptic plasticity. Reorganization of synaptic protein components occurs during LTP and LTD. This reorganization can be achieved by changes in the levels or activity of signaling molecules such as kinases and phosphatases as well as changes in synaptic ion channel number or conductance. Throughout this Review, we focus on how synaptic protein modifications and interactions convey proper postsynaptic organization and facilitate normal synaptic plasticity both acutely and long-term. Moreover, we describe how perturbations in synaptic protein modifications and interactions are associated with pathological conditions, such as stroke, autism spectrum disorders like Angelman syndrome, and Parkinson disease. Finally, we describe emerging technologies and approaches that can be used to enhance detection of low-abundance synaptic protein complexes and post-translational modifications.
N-METHYL-D-ASPARTATE RECEPTORS (NMDARs)
The NMDARs are ionotropic glutamate receptors that allow for calcium and sodium influx into the postsynapse. The NMDAR is made up of an obligate GluN1 subunit and different GluN2 (or less commonly GluN3) subunits. In certain brain regions, there is a developmental shift from GluN2B to GluN2A such that GluN2B expression decreases and GluN2A expression increases.22 In rats, GluN2B levels decreased in both Cornu Ammonis (CA)1 and CA3 regions of the hippocampus beginning 4 days after birth and reaching adult levels (∼40% decrease) by postnatal day 21.23 Similar results were observed in humans, where the GluN2A:GluN2B mRNA ratio increases in the CA1 and CA3 region, but not the dentate gyrus.24 Despite age-dependent reductions in GluN2B levels, recent studies suggest that triheteromeric NMDARs containing GluN1/GluN2A/GluN2B may be the most abundant form in dissociated hippocampal neurons and adult hippocampal synapses.25,26 Accordingly, global knockout (KO) of the GluN1 subunit in mice resulted in loss of NMDAR activity, reduction in expression of the GluN2B subunit of the NMDAR, and death of animals by 15 h after birth.27 KO of the GluN2B subunit in CA3 pyramidal cells of the hippocampus abrogates NMDAR-dependent currents, decreases spine density, and attenuates hippocampal LTP.28 In contrast to GluN2B KO animals, loss of GluN2A attenuated CA1 hippocampal LTP but had fewer additional effects.29 The above data demonstrate that NMDAR subunit composition is developmentally and spatially organized. Moreover, the NMDAR is critical in different forms of LTP and LTD. However, understanding the full complement of NMDAR interactions and how those differ under physiological and pathological changes is well-suited for proteomics-based studies.
NMDAR-dependent synaptic plasticity requires proper expression, localization, and function of the NMDAR. In one of the first neuroproteomics studies ever performed, multiple affinity isolation techniques, including immunoprecipitation and immunoaffinity purification of GluN1 complexes, and peptide isolation of GluN2B complexes, were used to identify 66 different proteins that coimmunoprecipitate with the NMDAR.30 This approach allowed for identification of proteins that associate with both GluN1 and GluN2B or that may be enriched in one complex or another. For instance, SAP102 was weakly detected in the GluN1 immunoprecipitates and immunoaffinity purification, but was strongly detected in the GluN2B peptide isolation.30 Using the string database (www.string-db.org)31 to analyze a subset of these proteins, we highlight the interconnectivity of these molecules (Figure 2). We mapped those interactions that were experimentally validated and had medium to high confidence (0.4–0.9) to generate a connectivity map between the proteins that were detected in the GluN2B immunoprecipitates (Figure 2). NMDARs interact either directly or indirectly with kinases such as CaMKII, PKA, and PKC and phosphatases such as protein phosphatase 1, 2A, and 5, calcineurin, and tyrosine-protein phosphatase nonreceptor type 11 (Figure 2), signaling molecules that are known to regulate NMDAR function.32 PKA interacts with scaffolding/regulatory subunits termed A-kinase anchoring proteins (AKAPs), whereas PKC interacts with regulatory subunits for C-kinase (RACKs) for PKC. These subunits help target them to the NMDAR.33 CaMKII per se, AKAPs, or RACKs can target the kinase and/or interacting proteins to the NMDAR. In addition to kinases, the NMDAR was also found to associate with multiple scaffolding proteins including PSD-93, PSD-95, SAP-102, Homer 1, and Shank 1 and 2 (Figure 2), synaptic proteins that can modulate NMDAR targeting, localization at the PSD, and channel function.34–41 The scaffolding and GTPase activating protein, SynGAP1, also associates with the NMDAR. The association between the GluN2B subunit of the NMDAR and SynGAP1 modulates synaptic transmission through structural roles and/or RAS-GTPase activity that, together with NMDAR function, modulate ERK/MAPK signaling.42–45 Cytoskeletal proteins including actin, α-actinin, tubulin, and myosins also associate with the NMDAR. Interestingly, NMDAR activity regulates phosphorylation of cytoskeletal proteins such as MAP2 to modulate the synaptic cytoskeleton.46 Moreover, cytoskeletal proteins such as actin and myosins regulate NMDAR properties.47,48 Proteomics studies to identify novel interacting proteins do not require a priori knowledge of those interactions and the data obtained in this initial proteomics study helped direct many subsequent studies probing the function of specific interactions. However, it is important to address some caveats of these interactome maps. Using the above criteria, only two proteins were not connected to any other protein. These include Ppp1ca (the catalytic subunit of PP1) and Hspa1a. However, PP1 is known to modulate NMDARs49 and other synaptic proteins, suggesting a functional association with the NMDAR and additional proteins. Therefore, the fact that an interaction is not observed does not mean it does not exist. Moreover, while we restricted these maps to those associations for which experimental data is available, one can also utilize the string-db to glean information from other sources, such as data mining and co-occurrence in the literature. However, while these approaches are useful to generate hypotheses, specific experimental approaches need to be performed to validate these large interactome maps as well as to test specific hypotheses.
Figure 2.
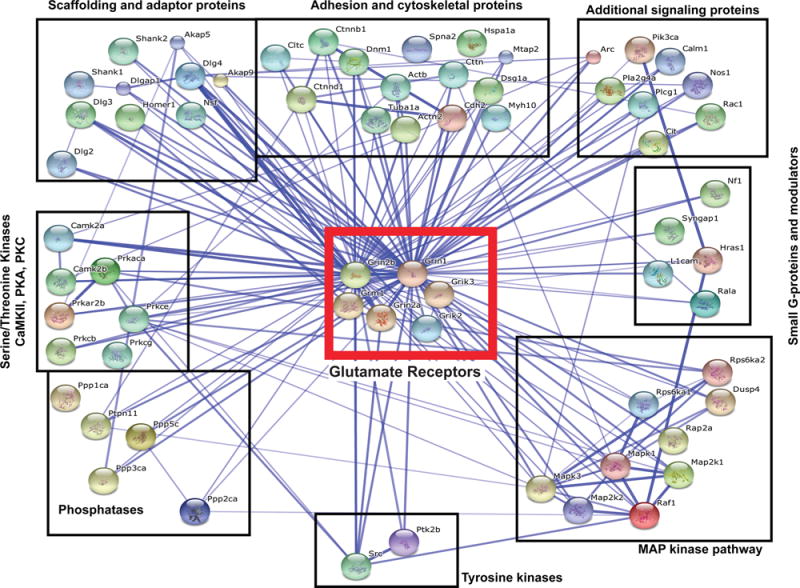
Interconnectivity of the NMDAR interactome. The NMDAR interactome30 was input into the string database (http://string-db.org/) to visualize interactions between the identified components. 66 proteins were evaluated. We set parameters to only detect interactions that were validated experimentally. The thickness of the line corresponds to the confidence of interaction (thin lines, >0.4; medium lines, >0.7; thick lines, >0.9).31
This initial NMDAR proteomics study was performed under basal conditions. NMDAR activity and membrane localization is regulated by multiple factors, including phosphorylation. For example, CaMKII, PKC, and death-associated protein kinase 1 (DAPK1) phosphorylate GluN2B at Ser1303. Phosphorylation at Ser1303 and/or Ser1323 by PKC or DAPK1 can enhance channel conductance.50 However, other studies have suggested that Ser1303 phosphorylation by CaMKII reduce channel conductance by enhancing desensitization in HEK293 cells.51 Recent data suggest that the differential role of Ser1303 phosphorylation by different kinases is actually due to Cl− levels in the cells, as Ser1303 phosphorylation enhances desensitization in the context of low intracellular Cl−, but decreases desensitization in the context of high intracellular Cl−.52 In addition to Ser1303 phosphorylation, casein kinase 2 (CK2) phosphorylates GluN2B at Ser1480, which decreases GluN2B binding to synaptic scaffolding proteins such as PSD-95 and SAP-102 and attenuates surface expression of the receptor.53 Increased CK2 activity during development decreased synaptic GluN2B expression.54 Together, these data have identified multiple NMDAR-specific interacting proteins and have determined phosphorylation sites that modulate these interactions. However, many of these studies use phosphorylation-specific mutants such as alanine (to prevent phosphorylation) or aspartate/glutamate (to mimic phosphorylation). One of the benefits of proteomics approaches is that one can survey multiple phosphorylation sites on a protein of interest in an unbiased manner and without potentially perturbing additional sites. Interestingly, this type of approach uncovered multiple novel phosphorylation sites on GluN2A and GluN2B.55 Moreover, coupling proteomics approaches to biochemical preparations that isolate different subcellular fractions can allow for tracking of specific phosphorylation sites in different parts of the neuron. This type of approach can also be used to determine how different pathological or developmental conditions impact phosphorylation at multiple sites at one time.
Proteomics experiments have revealed pathological perturbations in the NMDAR interactome. For instance, the GluN2B subunit of the NMDAR has enhanced association with synaptic proteins during aging.56 In addition, withdrawal from chronic intermittent ethanol (CIE) treatment increased the association of GluN2B with activity-regulated cytoskeleton-associated protein (Arc) and Homer1,57 two NMDAR interacting proteins that were detected in the initial NMDAR proteomics screen.30 Translocation of Arc mRNA to synapses has been shown to require NMDAR activity.58 Also, Arc mRNA synthesis is enhanced by epileptic seizures.58–60 Interestingly, the observed CIE-dependent increased association of Arc and Homer with the NMDAR occurred in a synaptic subcellular fraction. Previous studies have found that synaptic and extrasynaptic NMDAR activation differentially modulate ischemia-induced injuries and neurodegenerative diseases.61–63 Using proteomics to delineate unique subcellular differences in the NMDAR interactome in different disease states will greatly inform the function of different complexes and determine how alterations in these complexes may underlie specific pathologies.
CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE II (CaMKII)
NMDARs allow for ions such as Ca2+ to enter the postsynaptic spine. Ca2+ entry via NMDARs and other sources activates molecules such as CaMKII. When the calcium signal is robust and persistent, the kinase autophosphorylates at Thr286 on CaMKIIα which allows the kinase to retain Ca2+-independent activity and regulates CaMKII interactions and subcellular localization.64–67 CaMKIIα expression and autophosphorylation are required for NMDAR-dependent synaptic plasticity. Both complete loss of CaMKIIα and mutant CaMKIIα mice that cannot autophosphorylate at Thr286 (T286A) have abrogated LTP and LTD and behavioral deficits.68–72 It is important to note that there are four different isoforms of CaMKII: α, β, γ, and δ. While CaMKIIα is the most abundant CaMKII isoform in the forebrain, CaMKIIβ is the second most abundant CaMKII isoform in the forebrain with one CaMKIIβ molecule per three CaMKIIα molecules. In contrast to forebrain, CaMKIIβ is three times more abundant than CaMKIIα in the cerebellum73 and CaMKIIβ KO mice have altered cerebellar plasticity that leads to deficits in motor coordination.74 The above studies demonstrate that kinase expression and/or autophosphorylation are critical for normal synaptic function in different parts of the brain.
CaMKII is the most abundant protein in the PSD, with levels > 1.5 pmol/μg protein across CaMKIIα and CaMKIIβ isoforms.4 This protein accounts for ∼6% of the total protein in the PSD.3,75 Biochemical estimates of the abundance of endogenous kinase suggest 80 dodecameric holoenzymes (∼960 CaMKII molecules) per typically sized (∼100 nm2) PSD.3,75 While this estimation may be conflated due to translocation and targeting postdecapitation during sample preparation,76 it remains interesting that a kinase is so highly concentrated in this subcellular compartment. While estimation of CaMKII abundance in the PSD may be high, CaMKII actively translocates in slices and cultured neurons (within 30 min of stimulation),77–80 indicating that activated CaMKII moves to preferentially associate with the PSD following glutamate signaling.
In contrast to Thr286 autophosphorylation enhancing CaMKII activity, Thr305/6 phosphorylation prevents Ca2+/calmodulin-stimulation of CaMKII. Whereas T286 phosphorylation enhances CaMKII at the PSD, Thr305/6 phosphorylation diminishes CaMKII targeting to the PSD.78,79 Functionally, Thr286 phosphorylation without Thr305/6 phosphor-ylation leads to Ca2+/calmodulin-independent CaMKII activity along with Ca2+/calmodulin binding. This form of the kinase favors enhanced synaptic strength.81,82 However, T286D mutants in combination with TT305/6DD mutants decrease synaptic strength.83 Proteomics-based approaches have recapitulated overexpression and immunoblotting data showing that CaMKIIα phosphorylation at Thr286 is greatest in the synaptic fraction whereas phosphorylation at Thr306 is greatest in a cytosolic fraction.66
As mentioned above, T286A mutant CaMKIIα had lower levels in a synaptic fraction; however, immunoprecipitation of the kinase followed by both immunoblotting and proteomics approaches revealed that the kinase that was detected in this fraction had a greater association with multiple synaptic scaffolding proteins and NMDARs.66,84 Previous studies have suggested that the NMDAR is critical in targeting CaMKII to the PSD and that autophosphorylation of the kinase enhances the association of CaMKII with the GluN2B subunit of the NMDAR.85–87 This discrepancy may be due to differences in using the T286A mutant kinase, which can regulate additional phosphorylation sites on CaMKII as well as calmodulin binding affinity.66,88 Together, the above data suggest that CaMKII binding to the NMDAR is critical in regulating autophosphorylation-dependent CaMKII targeting to the PSD; however, additional synaptic proteins may also play critical roles. For example, based on a recent proteomics study, the cytoskeletal protein, brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2, also known as IRSp53) had a decreased association with CaMKII in a synaptic fraction.66 BAIAP2 is a highly expressed PSD protein with an abundance of 23.5 fmol/μg protein.4 BAIAP2 KO mice have decreased PSD size, but increased insertion of NMDARs and enhanced hippocampal LTP.89 Moreover, proteomics studies determined that BAIAP2 expression is decreased in human Alzheimer disease patients.90 It is important to note that due to the multimeric structure of CaMKII, immunoprecipitation of CaMKII may isolate low levels of interacting proteins. If a single CaMKII molecule is bound to your protein of interest, immunoprecipitating that CaMKII molecule will copurify 11 other CaMKII molecules, but only one interacting protein. Therefore, low abundance CaMKII interacting proteins may be valid interactors and using technologies such as multidimensional protein identification technology (MudPIT) and other approaches to detect low abundance proteins will be useful in identifying novel interacting proteins (see Summary, Emerging Approaches, and Future Directions, below). Moreover, given the dodecameric structure and the high abundance of CaMKII in the PSD, as well as the panoply of CaMKII interacting proteins, CaMKII may also function as a structural or scaffolding protein in dendritic spines.91
Many of the studies that have delineated the function of CaMKII autophosphorylation at Thr286 and Thr305/6, have done so using phosphorylation mutants. These mutants do not fully recapitulate a nonphosphorylated residue (e.g., T286A) or a phosphorylated residue (e.g., T286D). Moreover, phosphorylation mutants at one site may influence the phosphorylation status of additional sites.66 While Thr286 and Thr305/6 are the most well-studied sites of autophosphorylation, CaMKIIα is autophosphorylated at multiple other sites, including Thr253, Ser279, Thr305, Thr306, Ser314, Ser318, Ser331, Ser333, Thr334, Thr336, and Thr337 (Figure 3).92–102 In addition to these sites on CaMKIIα, CaMKIIβ contains an actin-binding domain and can be phosphorylated at Ser315 and Thr320/1 nearby and within this domain. Phosphorylation at these CaMKIIβ sites is associated with the cytosolic fraction66 suggesting phosphorylation at these sites may modulate CaMKIIβ interactions and synaptic targeting. While many of these sites have been identified as autophosphorylation sites, the interplay between them in vivo is not completely understood. Many of the above phosphorylation sites have been identified in vivo using proteomics approaches and future proteomics-based studies will be needed to track phosphorylation changes at multiple sites under different pathological conditions and/or subcellular fractions.
Figure 3.

CaMKII autophosphorylation sites.102 Those autophosphorylation sites that are enriched in nonsynaptic (green) or synaptic (red) locations are labeled. CD, catalytic domain; AD, association domain; LK, linker region; ABD, actin-binding domain.
CaMKII phosphorylation is modulated in different disease states. For example, Thr286 phosphorylation is increased in animal models of Parkinson disease103,104 and Thr305/6 phosphorylation is increased in an animal model of Angelman syndrome.105 Rescue of changes in CaMKII autophosphorylation/activity normalizes behavioral deficits observed in these animals.104,105 In addition to Parkinson disease and Angelman syndrome, CaMKII autophosphorylation and interaction with the NMDAR is increased in brain slices following oxygen/glucose deprivation, a model of ischemia.106 Furthermore, a recent phospho-proteomics study identified multiple phosphor-ylation sites on different synaptic proteins, including phosphorylation of Ser331 on CaMKIIα. Phosphorylation at this site was increased by cocaine-cue memory extinction, but decreased by cocaine-cue memory reconsolidation.107 While individual phosphorylation sites have been interrogated on CaMKII, understanding the interdependencies of phosphorylation on the kinase would be very interesting. As an emerging field, top-down proteomics is useful in identifying not only individual phosphorylation sites, but the phosphorylation signal or code on an intact protein molecule.108 For CaMKII, this approach would be useful in determining how a single kinase subunit within a holoenzyme is phosphorylated in a specific subcellular localization or in response to physiological or pathological changes in synaptic plasticity. Using proteomics to understand pathological changes in kinase autophosphorylation, protein interactions, and subcellular localization in different neurological diseases will increase understanding of the normal and pathological role of this multifunctional kinase and may uncover novel targets to treat multiple synaptopathies.
α-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID RECEPTOR (AMPAR)
AMPARs are ionotropic glutamate receptors that allow for Na+ (and in some cases Ca2+) to enter the postsynapse. There are four subunits of the AMPAR, GluA1−4, with the GluA2 subunit being the most abundant, followed by GluA1, then GluA3, and then GluA4 (Figure 4). NMDAR-dependent synaptic plasticity requires CaMKII activation. Activation of CaMKII and the NMDAR modulate AMPAR localization at the synapse, causing long-term plasticity changes. Increased AMPAR levels in the membrane are essential for LTP, whereas decreased AMPARs in the membrane underlies LTD. Multiple, immunoprecipitation-based studies have identified novel proteins that interact with the AMPAR to modulate AMPAR insertion into membranes and/or AMPAR channel conductance.109–111 Quantitative proteomics approaches have recently been used to determine absolute or relative abundance of AMPAR interacting proteins.110–112 Based on these quantitative data, we have generated a schematic model depicting the ratiometric expression of AMPARs and their interacting proteins (Figure 4). The transmembrane-AMPAR regulatory proteins (TARPs) are localized to the membrane (shown as inner components in Figure 4). Multiple TARP isoforms include γ-8, γ-3, γ-2, γ-7, and γ-4. Knockout of TARPγ-2 (aka stargazin) leads to deficits in basal AMPAR synaptic transmission in hippocampal neurons and also AMPAR insertion in cerebellar granule cells.113 Mice with a spontaneous mutation in what was later described as stargazin were found to have an ataxic gait and were prone to seizures.114,115 Stargazin and TARPγ-7 are the main TARPs expressed in the cerebellum,112 but other TARPs may compensate for stargazin activity at other synapses.116 TARPγ-8 is one of the most abundant TARPs detected in the whole brain (Figure 4).110 Recent studies have found that CaMKII phosphorylates TARPγ-8 at Ser277 and Ser281. Phosphorylation at these sites enhances AMPAR transmission and is required for LTP.117 Conversely, mutating similar phosphorylation sites on stargazin in the context of knocking out TARPγ-3 and TARPγ-4 had no effect on LTP,117 suggesting that other TARPs do not compensate for loss of TARPγ-8.
Figure 4.
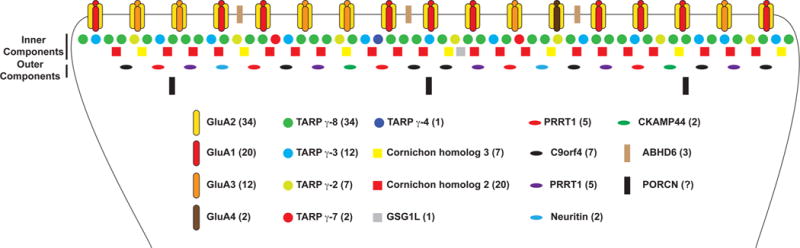
Relative expression of AMPAR interacting proteins. AMPAR interacting protein abundance110,111 was used to generate a schematic showing the relative abundance of AMPAR and proteomically quantified interacting proteins. Interactions are split into integral membrane proteins, inner components, and outer components. Ratiometric expression is normalized to the highest abundance protein, GluA2, which is set at an arbitrary value of 34.
Germ cell-specific gene 1-like protein (GSG1L) is a novel AMPAR interacting protein that was found using proteomics-based approaches and that uniquely modulates AMPAR function by altering desensitization kinetics.110,111 GSG1L has also been shown to suppress the calcium permeability of AMPARs.118 In addition to GSG1L, proteomics studies have uncovered other proteins such as Abhydrolase domain-containing 6 (ABHD6) and porcupine (PORCN), which modulate AMPAR trafficking to the membrane.119 Unbiased, proteomics-based approaches are well-suited for identifying interacting proteins such as those where there is no a priori knowledge that the protein may interact with the receptor.
In addition to identification of AMPAR components, proteomics approaches have also been utilized to determine developmental and region specific differences in AMPAR levels and interactions.112,120 The hippocampus, cortex, and cerebellum have the highest relative amounts of AMPAR levels.112 While the quantity of the AMPARs are fairly stable during development, the associated proteins tend to vary. TARPγ-4 is highest at birth but decreases as animals age to adults. In contrast, TARPγ-8 increases during development to adult levels in rat brain.112 Proline Rich Transmembrane Protein 1 (PRRT1) was expressed in, and interacts with, AMPARs most robustly in the hippocampus. GSG1L had robust interaction with AMPARs in the cortex and less association in the hippocampus. Ras-related protein Rap2B (RAP2B) and Abhydrolase domain-containing 12 (ABHD12) had robust interactions with AMPARs in the hippocampus. Characterization of AMPAR proteomes from different brain regions and/or subcellular fractions has greatly enhanced our knowledge of differential AMPAR complexes; however, it is currently unclear how different physiological (e.g., induction of LTP/LTD) or pathological perturbations may modulate these specific complexes. Recent work by Castillo, Tomita, and colleagues has shown that, in addition to its role in LTP in hippocampal slices (see above), phosphorylation of TARP γ-8 at Ser-277 and Ser-281, is critical for both cued and contextual fear memory.117 While this is one change, it is likely that changes in the phosphorylation of, or association of AMPARs with, additional AMPAR interacting proteins also occurs in normal plasticity and under pathological conditions. Understanding pathologies associated with interactions in specific brain regions and/or cell types may reveal novel disease targets that have enhanced specificity for treating diseases associated with plasticity deficits.
POSTSYNAPTIC DENSITY PROTEIN OF 95 kDa (PSD-95)
PSD-95 is a highly enriched scaffolding protein that has multiple organization domains. PSD-95 and other scaffolding proteins interact with multiple synaptic proteins including NMDARs, AMPARs, and CaMKII (Figure 5) to properly localize them at the PSD. PSD-95 is expressed at a concentration of ∼85 fmol/μg in forebrain PSDs, making it one of the most abundant adaptor/scaffold-type proteins.4 Knockdown of PSD-95 decreases AMPAR levels in specific membrane patches.121 KO of PSD-95 increases protein expression of a synaptic tyrosine phosphatase, striatal-enriched protein tyrosine phosphatase (STEP61), causing a decrease in synaptic GluN2B protein at the membrane.122 Moreover, PSD-95 KO animals have decreased spine density in the striatum and increased spine density in the hippocampus.123 PSD-95 KO animals also have specific physiological and behavioral abnormalities,124 including enhanced LTP, abrogated LTD, and impaired spatial memory deficits in aversive behaviors.125,126
Figure 5.
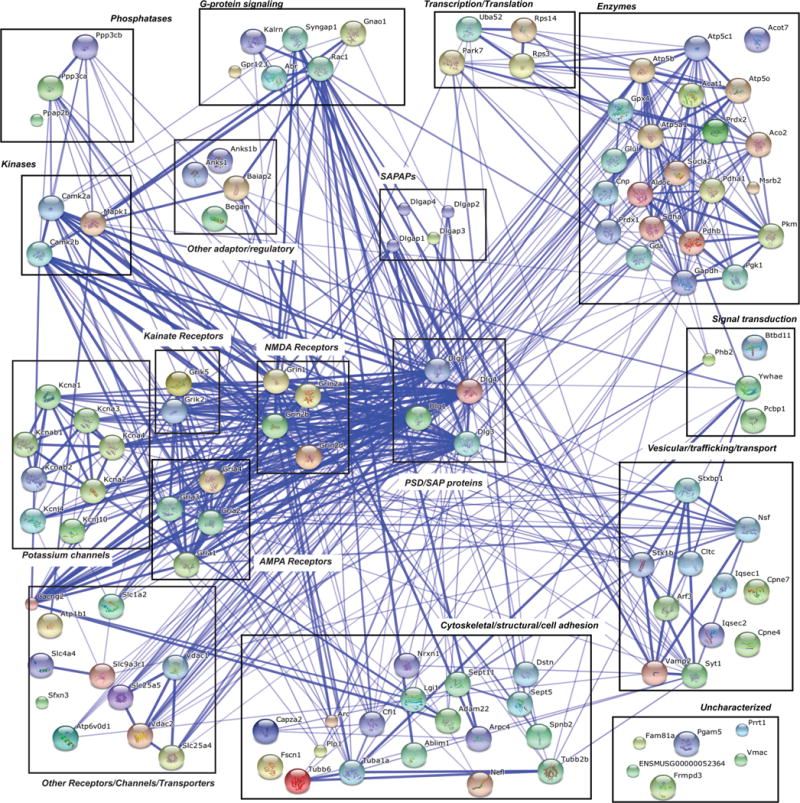
Interconnectivity of the PSD-95 interactome. PSD-95 interacting proteins identified using a tandem-affinity purification approach127 were input into the string database (http://string-db.org/) to visualize interactions between the identified components. 118 proteins were evaluated. We set parameters to only detect interactions that were found by multiple sources (see text). The thickness of the line corresponds to the confidence of interaction (thin lines, >0.4; medium lines, >0.7; thick lines, >0.9).31
While PSD-95 KO mice have many deficits, overall they are viable and do not have as severe of a neurological phenotype as may be thought given PSD-95 abundance and ability to recruit and stabilize proteins at the PSD. This minimal synaptic phenotype may be due to overlapping function of other adaptor/scaffolding proteins at the PSD. Specifically, 12 adaptor/regulatory proteins associated with PSD-95 in a proteomics screen, including: PSD-93, SAP-102, SAP-97, SAPAP1, SAPAP2, SAPAP3, and SAPAP4.127 While these proteins may compensate for loss of PSD-95, they are normally expressed at 6–40-fold lower levels than PSD-95 (∼2–15 fmol/μg).4 Grant and colleagues identified 118 proteins in at least three biological replicates that coprecipitated with PSD-95 that they have defined as constituents of the “core” PSD. We have plotted these proteins using the String database and the protein function categories given by Fernandez et al. (Figure 5).127 As expected there are multiple known nodes of association between the identified PSD proteins. To understand how the PSD-95 interactome compares to other synaptic protein interactomes, we can compare the NMDAR interactome (Figure 2)30 with the PSD-95 interactome (Figure 5).127 If we use the same parameters as Figure 2 and only plot associations that have been experimentally validated, a large majority of the proteins are not connected (data not shown). Therefore, we evaluated interactions that were not only experimentally validated, but also validated by textmining, databases, coexpression, neighborhood, gene fusion, and/or co-occurrence. Even adding other evidence of interactions, there were still 18 proteins that were unconnected. Given that the more recent studies use updated mass spectrometers, it is possible that these studies have probed deeper and have not been as well validated. Conversely, by detecting lower abundance interactors, it is possible that some of the interactions are nonspecific. Looking at specific proteins, one notices both overlap and unique proteins in each interactome. Examples of overlap include kinases, phosphatases, SynGAP, and cytoskeletal proteins. Conversely, some differences were observed. For instance, Shank and Homer were present in the NMDAR complex, whereas in the PSD-95 complex, Shank was only observed in a single biological replicate whereas Homer was not detected. Moreover, vesicular trafficking proteins and multiple ATPases were detected in the PSD-95 complexes, but not in the NMDAR complexes. These variances in data may be due to differences in the interactions between these molecules, but may also be due to technical advances in HPLC separations and parameters and mass spectrometers. As instrumentation and methodologies continue to improve, additional data will be generated, even from the same types of experiments. In addition to technical differences, isolation of NMDAR complexes would be predicted to include both synaptic and extrasynaptic complexes, which may have different interactions and organization. Comparing complexes from core PSD proteins such as PSD-95 with other synaptic protein complexes (like the NMDAR, which is localized both within and outside of the core PSD) will allow for the determination of the core components of the PSD as well as the identification of proteins that reside outside of the PSD that may have unique synaptic functions.
In addition to traditional isolation of PSDs using immuno or peptide isolation strategies, recent studies overexpressed a PSD-95 protein fused to an E. coli. biotinylating enzyme, BirA.128 This fusion protein allows for biotinylation of proteins near the fused PSD-95 protein and streptavidin pull-down to identify proteins in proximity to PSD-95. Following streptavidin pulldown and mass spectrometry, multiple known PSD-95 interacting proteins were identified, including NMDARs, AMPARs, and scaffolding proteins (e.g., Shank, PSD93, SAP97, SAP102).
Robust characterization of the PSD-95 interactome has identified multiple synaptic proteins and interactions, many of which have been implicated in neurological diseases, including autism spectrum disorder and psychiatric diseases.129–131 Future studies will need to identify cell-specific changes in the PSD-95 interactome under different conditions and/or disease states.
SUMMARY, EMERGING APPROACHES, AND FUTURE DIRECTIONS
Proteomics approaches have identified and characterized multiple synaptic protein complexes. These approaches are well-suited to understanding how proteins interact and how those interactions are regulated in different cell types and under pathological conditions. Moving forward, technological advances will allow for a deeper understanding of the synaptic interactome. This deeper understanding will aid in identification of novel therapeutic targets.
As mentioned above, proximity labeling approaches were used to identify proteins in proximity to PSD-95. One potential drawback to this approach is that it may not detect tertiary interacting proteins, depending upon the distance of the tertiary interacting protein from the biotinylating enzyme. Also, addition of the biotinylating enzyme may induce ectopic changes to the interaction network. Furthermore, appropriate controls must be performed to account for proteins that nonspecifcally bind to streptavidin.132 However, proximity labeling approaches like these133 allow for detection of both direct and indirect interactions, as well as interactions that tend to be more transient or that are difficult to biochemically isolate.134,135 Moreover, this approach allows for temporal and cell specific isolation of protein complexes. This proximity approach was used to biotinylate proteins that are near gephryin,128 a protein enriched at inhibitory postsynaptic densities, an area that is difficult to biochemically isolate. This approach can enhance immunoprecipitation approaches that have been used to immuno-isolate inhibitory and purkinje synapses.136,137
Another proximity labeling approach uses horseradish peroxidase (HRP) or an engineered peroxidase called APEX (or APEX2 depending on iteration) that uses hydrogen peroxide and biotin-phenol substrate to label nearby proteins.138,139 Specifically, neuronal cultures expressing HRP-tagged proteins were incubated with hydrogen peroxide and a membrane impermeable biotin phenol. This allowed for biotinylation of proteins in close proximity to the HRP-conjugated protein. Both excitatory and inhibitory synaptic cleft proteins were identified using mass spectrometry on streptavidin-isolated proteins.132 Further refinement of this approach allows the APEX2 protein to be split into two parts, allowing for identification of proteomes only where both parts of the protein are expressed. Fusing one-half of APEX2 to neurexin and one-half to neuroligin allowed for identification of specific synapse proteomes in the visual system.140 One of the drawbacks to these approaches may be toxicity of using hydrogen peroxide. However, this is not an issue with the BirA (BioID) approaches. Moving forward, one could envision using these types of approaches to further restrict expression of tagged proteins to specific cell types in addition to specific regions.
While proximity labeling approaches will allow for the identification of proteins in specific compartments such as the PSD, newer technological advances will improve identification of synaptic proteins as well. A standard workflow (see Abstract graphic) for isolation of synaptic protein complexes includes immunoprecipitation of the complex, followed by separation of the complex and identification of the complex components using mass spectrometry. Standard separation techniques include gel-based separation. However, advances in separation technologies, such as MudPIT, will allow for identification of lower-abundance proteins. This approach couples strong cation exchange followed by reversed phase HPLC to enhance the separation of peptides.141 In addition to detecting more proteins by increasing detection of lower-abundance proteins and peptides, we will be able to identify post-translational modifications (PTMs) that occur at low levels. Coupling MudPIT with other technological advances such as improvements in mass spectrometers as well as in ionization approaches (e.g., electron transfer dissociation (ETD)) will be useful in detecting more PTMs by enhancing coverage and more faithfully preserving PTMs such as phosphorylation.142 Moreover, top-down mass spectrometry approaches to analyze intact proteins will be useful in identifying specific “proteoforms” of proteins.108,143 Specifically, this approach could map all of the PTMS on a single protein molecule. Furthermore, using multiple enzymes to digest complex mixtures along with the above newer technologies and chromatagrophy improvements has allowed for near complete examination of the yeast proteome.144–146 One would predict that utilizing these approaches would allow for identification of a near-complete or complete interactome. These approaches will be critical for identification of low abundance, endogenous synaptic interactomes. However, with improved detection capabilities and the high abundance of certain synaptic proteins, especially in fractionated samples, appropriate controls including KO-based strategies are important in validating specific interactions.147
Moving forward, improvements in mass spectrometry instrumentation and data analysis tools will allow us to further identify protein interactions that underlie normal and pathological synaptic communication. In addition to improvements in instrumentation, label-based approaches are being developed to allow for relative or absolute quantification of proteins using stable-isotope labeling of amino acids in culture (SILAC) or isobaric mass tags (e.g., iTRAQ and TMT).148 These approaches have been used to quantify tens to hundreds of proteins concurrently149 and have been used to analyze relative PSD protein abundance and abundance of phosphorylation sites.150 Targeted, label-free quantitative techniques such as selected reaction monitoring (SRM) and Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra (SWATH-MS) can be used to analyze tens to hundreds (SRM) or tens of thousands (SWATH-MS) of proteins at one time.151 Specifically, SRM methods have been designed to quantify ∼100 rat PSD proteins.152 In addition, newer label free quantitation approaches such as isoQuant20 or MaxQuant software and delayed normalization and maximal peptide ratio extraction153 are useful. MaxQuant has been utilized to quantify levels of over 13 000 protein groups isolated from different structures and different cell types in the brain.154
Taken together, advanced methodologies along with improved hardware will enhance our ability to quantitatively probe the depths of global synaptic protein abundance, PTMs, and protein interaction networks under normal and pathological conditions in specific brain regions and cell types. In addition to novel approaches and methodologies, performing multiple biological and technical replicates is also important in enhancing the full complement of detected proteins. Obtaining a complete and specific synaptic interactome and understanding physiological and pathological changes that regulate this interactome will be critical for identifying novel druggable pathways to treat myriad different neurological disorders.
Acknowledgments
I would like to thank Drs. Andy Hudmon and Emily Anderson-Baucum (Indiana University School of Medicine), Dr. Terunaga Nakagawa (Vanderbilt University), Dr. Mark Dell’Acqua (University of Colorado Anschutz Medical Campus), and members of the Baucum Laboratory for critical review of the manuscript.
Funding
A.J.B. was supported by K01-NS073700 and R21-DA041876 Department of Biology, IUPUI.
Footnotes
ORCID
Anthony J. Baucum II: 0000-0002-4756-9865
Notes
The author declares no competing financial interest.
References
- 1.Pontes AH, de Sousa MV. Mass Spectrometry-Based Approaches to Understand the Molecular Basis of Memory. Front Chem. 2016;4:40. doi: 10.3389/fchem.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erondu NE, Kennedy MB. Regional distribution of type II Ca2+/calmodulin-dependent protein kinase in rat brain. J Neurosci. 1985;5(12):3270–7. doi: 10.1523/JNEUROSCI.05-12-03270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell JW. CaMKII: claiming center stage in postsynaptic function and organization. Neuron. 2014;81(2):249–65. doi: 10.1016/j.neuron.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5(6):1158–70. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Dosemeci A, Makusky AJ, Jankowska-Stephens E, Yang X, Slotta DJ, Markey SP. Composition of the synaptic PSD-95 complex. Mol Cell Proteomics. 2007;6(10):1749–60. doi: 10.1074/mcp.M700040-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dosemeci A, Tao-Cheng JH, Vinade L, Jaffe H. Preparation of postsynaptic density fraction from hippocampal slices and proteomic analysis. Biochem Biophys Res Commun. 2006;339(2):687–94. doi: 10.1016/j.bbrc.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 7.Li K, Hornshaw MP, van Minnen J, Smalla KH, Gundelfinger ED, Smit AB. Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J Proteome Res. 2005;4(3):725–33. doi: 10.1021/pr049802+. [DOI] [PubMed] [Google Scholar]
- 8.Husi H. NMDA receptors, neural pathways, and protein interaction databases. Int Rev Neurobiol. 2004;61:49–77. doi: 10.1016/S0074-7742(04)61003-8. [DOI] [PubMed] [Google Scholar]
- 9.Husi H, Grant SG. Proteomics of the nervous system. Trends Neurosci. 2001;24(5):259–66. doi: 10.1016/s0166-2236(00)01792-6. [DOI] [PubMed] [Google Scholar]
- 10.Husi H, Grant SG. Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J Neurochem. 2001;77(1):281–91. doi: 10.1046/j.1471-4159.2001.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- 11.Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20(11):4069–80. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–47. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- 13.Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3(9):857–71. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279(2):987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 15.Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279(20):21003–11. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- 16.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88(3):759–68. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 17.Li KW, Jimenez CR. Synapse proteomics: current status and quantitative applications. Expert Rev Proteomics. 2008;5(2):353–60. doi: 10.1586/14789450.5.2.353. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara Y. Quantification of postsynaptic density proteins: glutamate receptor subunits and scaffolding proteins. Hippocampus. 2012;22(5):942–53. doi: 10.1002/hipo.20950. [DOI] [PubMed] [Google Scholar]
- 19.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 20.Distler U, Schmeisser MJ, Pelosi A, Reim D, Kuharev J, Weiczner R, Baumgart J, Boeckers TM, Nitsch R, Vogt J, Tenzer S. In-depth protein profiling of the postsynaptic density from mouse hippocampus using data-independent acquisition proteomics. Proteomics. 2014;14(21–22):2607–13. doi: 10.1002/pmic.201300520. [DOI] [PubMed] [Google Scholar]
- 21.Phillips GR, Florens L, Tanaka H, Khaing ZZ, Fidler L, Yates JR, 3rd, Colman DR. Proteomic comparison of two fractions derived from the transsynaptic scaffold. J Neurosci Res. 2005;81(6):762–75. doi: 10.1002/jnr.20614. [DOI] [PubMed] [Google Scholar]
- 22.Sanz-Clemente A, Nicoll RA, Roche KW. Diversity in NMDA receptor composition: many regulators, many consequences. Neuroscientist. 2013;19(1):62–75. doi: 10.1177/1073858411435129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang LR, Liu JP, Zhang N, Wang YJ, Gao XL, Wu Y. Different expression of NR2B and PSD-95 in rat hippocampal subregions during postnatal development. Microsc Res Tech. 2009;72(7):517–24. doi: 10.1002/jemt.20708. [DOI] [PubMed] [Google Scholar]
- 24.Law AJ, Weickert CS, Webster MJ, Herman MM, Kleinman JE, Harrison PJ. Expression of NMDA receptor NR1, NR2A and NR2B subunit mRNAs during development of the human hippocampal formation. European journal of neuroscience. 2003;18(5):1197–205. doi: 10.1046/j.1460-9568.2003.02850.x. [DOI] [PubMed] [Google Scholar]
- 25.Rauner C, Kohr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem. 2011;286(9):7558–66. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tovar KR, McGinley MJ, Westbrook GL. Triheteromeric NMDA receptors at hippocampal synapses. J Neurosci. 2013;33(21):9150–60. doi: 10.1523/JNEUROSCI.0829-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13(2):325–38. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 28.Akashi K, Kakizaki T, Kamiya H, Fukaya M, Yamasaki M, Abe M, Natsume R, Watanabe M, Sakimura K. NMDA receptor GluN2B (GluR epsilon 2/NR2B) subunit is crucial for channel function, postsynaptic macromolecular organization, and actin cytoskeleton at hippocampal CA3 synapses. J Neurosci. 2009;29(35):10869–82. doi: 10.1523/JNEUROSCI.5531-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakimura K, Kutsuwada T, Ito I, Manabe T, Takayama C, Kushiya E, Yagi T, Aizawa S, Inoue Y, Sugiyama H, et al. Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature. 1995;373(6510):151–5. doi: 10.1038/373151a0. [DOI] [PubMed] [Google Scholar]
- 30.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–9. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(D1):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BS, Roche KW. Regulation of NMDA receptors by phosphorylation. Neuropharmacology. 2007;53(3):362–8. doi: 10.1016/j.neuropharm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sim AT, Scott JD. Targeting of PKA, PKC and protein phosphatases to cellular microdomains. Cell Calcium. 1999;26(5):209–17. doi: 10.1054/ceca.1999.0072. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Rao W, Zhang C, Zhang C, Liu MD, Han F, Yao LB, Han H, Luo P, Su N, Fei Z. Scaffolding protein Homer1a protects against NMDA-induced neuronal injury. Cell Death Dis. 2015;6:e1843. doi: 10.1038/cddis.2015.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cousins SL, Papadakis M, Rutter AR, Stephenson FA. Differential interaction of NMDA receptor subtypes with the post-synaptic density-95 family of membrane associated guanylate kinase proteins. J Neurochem. 2008;104(4):903–13. doi: 10.1111/j.1471-4159.2007.05067.x. [DOI] [PubMed] [Google Scholar]
- 36.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23(3):569–82. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 37.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5(10):771–81. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 38.Sheng M, Kim E. The postsynaptic organization of synapses. Cold Spring Harbor Perspect Biol. 2011;3(12):a005678. doi: 10.1101/cshperspect.a005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruneau EG, Esteban JA, Akaaboune M. Receptor-associated proteins and synaptic plasticity. FASEB J. 2009;23(3):679–88. doi: 10.1096/fj.08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan X, Jin WY, Wang YT. The NMDA receptor complex: a multifunctional machine at the glutamatergic synapse. Front Cell Neurosci. 2014;8:160. doi: 10.3389/fncel.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaka R, Thornton C, Vagts AJ, Phamluong K, Bonci A, Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc Natl Acad Sci U S A. 2002;99(8):5710–5. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46(5):745–60. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20(5):895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- 44.Kim JH, Lee HK, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci. 2003;23(4):1119–24. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O’Carroll CM, Martin SJ, Morris RG, O’Dell TJ, Grant SG. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22(22):9721–32. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halpain S, Greengard P. Activation of NMDA receptors induces rapid dephosphorylation of the cytoskeletal protein MAP2. Neuron. 1990;5(3):237–46. doi: 10.1016/0896-6273(90)90161-8. [DOI] [PubMed] [Google Scholar]
- 47.Rosenmund C, Westbrook GL. Rundown of N-methyl-D-aspartate channels during whole-cell recording in rat hippocampal neurons: role of Ca2+ and ATP. J Physiol. 1993;470:705–29. doi: 10.1113/jphysiol.1993.sp019884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei S, Czerwinska E, Czerwinski W, Walsh MP, MacDonald JF. Regulation of NMDA receptor activity by F-actin and myosin light chain kinase. J Neurosci. 2001;21(21):8464–72. doi: 10.1523/JNEUROSCI.21-21-08464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farinelli M, Heitz FD, Grewe BF, Tyagarajan SK, Helmchen F, Mansuy IM. Selective regulation of NR2B by protein phosphatase-1 for the control of the NMDA receptor in neuroprotection. PLoS One. 2012;7(3):e34047. doi: 10.1371/journal.pone.0034047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao GY, Wagner DA, Hsu MH, Leonard JP. Evidence for direct protein kinase-C mediated modulation of N-methyl-D-aspartate receptor current. Mol Pharmacol. 2001;59(5):960–4. doi: 10.1124/mol.59.5.960. [DOI] [PubMed] [Google Scholar]
- 51.Sessoms-Sikes S, Honse Y, Lovinger DM, Colbran RJ. CaMKIIalpha enhances the desensitization of NR2B-containing NMDA receptors by an autophosphorylation-dependent mechanism. Mol Cell Neurosci. 2005;29(1):139–47. doi: 10.1016/j.mcn.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Tavalin SJ, Colbran RJ. CaMKII-mediated phosphorylation of GluN2B regulates recombinant NMDA receptor currents in a chloride-dependent manner. Mol Cell Neurosci. 2017;79:45. doi: 10.1016/j.mcn.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung HJ, Huang YH, Lau LF, Huganir RL. Regulation of the NMDA receptor complex and trafficking by activity-dependent phosphorylation of the NR2B subunit PDZ ligand. J Neurosci. 2004;24(45):10248–59. doi: 10.1523/JNEUROSCI.0546-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanz-Clemente A, Matta JA, Isaac JT, Roche KW. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron. 2010;67(6):984–96. doi: 10.1016/j.neuron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghafari M, Hoger H, Keihan Falsafi S, Russo-Schlaff N, Pollak A, Lubec G. Mass spectrometrical identification of hippocampal NMDA receptor subunits NR1, NR2A-D and five novel phosphorylation sites on NR2A and NR2B. J Proteome Res. 2012;11(3):1891–6. doi: 10.1021/pr201099u. [DOI] [PubMed] [Google Scholar]
- 56.Zamzow DR, Elias V, Shumaker M, Larson C, Magnusson KR. An increase in the association of GluN2B containing NMDA receptors with membrane scaffolding proteins was related to memory declines during aging. J Neurosci. 2013;33(30):12300–5. doi: 10.1523/JNEUROSCI.0312-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wills TA, Baucum AJ, 2nd, Holleran KM, Chen Y, Pasek JG, Delpire E, Tabb DL, Colbran RJ, Winder DG. Chronic intermittent alcohol disrupts the GluN2B-associated proteome and specifically regulates group I mGlu receptor-dependent long-term depression. Addict Biol. 2015 doi: 10.1111/adb.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30(1):227–40. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 59.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1995;92(12):5734–8. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14(2):433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 61.Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11(10):682–96. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsons MP, Raymond LA. Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron. 2014;82(2):279–93. doi: 10.1016/j.neuron.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 63.Papouin T, Oliet SH. Organization, control and function of extrasynaptic NMDA receptors. Philos Trans R Soc B. 2014;369(1654):20130601. doi: 10.1098/rstb.2013.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudmon A, Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 65.Lou LL, Schulman H. Distinct autophosphorylation sites sequentially produce autonomy and inhibition of the multifunctional Ca2+/calmodulin-dependent protein kinase. J Neurosci. 1989;9(6):2020–32. doi: 10.1523/JNEUROSCI.09-06-02020.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baucum AJ, Shonesy BC, Rose KL, Colbran RJ. Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem Neurosci. 2015;6:615–31. doi: 10.1021/cn500337u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colbran RJ. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem J. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science (Washington, DC, U S) 1998;279(5352):870–3. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 69.Glazewski S, Giese KP, Silva A, Fox K. The role of alpha-CaMKII autophosphorylation in neocortical experience-dependent plasticity. Nat Neurosci. 2000;3(9):911–8. doi: 10.1038/78820. [DOI] [PubMed] [Google Scholar]
- 70.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science (Washington, DC, U S) 1992;257(5067):206–11. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 71.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science (Washington, DC, U S) 1992;257(5067):201–6. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 72.Coultrap SJ, Freund RK, O’Leary H, Sanderson JL, Roche KW, Dell’Acqua ML, Bayer KU. Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Rep. 2014;6(3):431–7. doi: 10.1016/j.celrep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller SG, Kennedy MB. Distinct forebrain and cerebellar isozymes of type II Ca2+/calmodulin-dependent protein kinase associate differently with the postsynaptic density fraction. J Biol Chem. 1985;260(15):9039–46. [PubMed] [Google Scholar]
- 74.van Woerden GM, Hoebeek FE, Gao Z, Nagaraja RY, Hoogenraad CC, Kushner SA, Hansel C, De Zeeuw CI, Elgersma Y. betaCaMKII controls the direction of plasticity at parallel fiber-Purkinje cell synapses. Nat Neurosci. 2009;12(7):823–5. doi: 10.1038/nn.2329. [DOI] [PubMed] [Google Scholar]
- 75.Chen X, Vinade L, Leapman RD, Petersen JD, Nakagawa T, Phillips TM, Sheng M, Reese TS. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci U S A. 2005;102(32):11551–6. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suzuki T, Okumura-Noji K, Tanaka R, Tada T. Rapid translocation of cytosolic Ca2+/calmodulin-dependent protein kinase II into postsynaptic density after decapitation. J Neurochem. 1994;63(4):1529–37. doi: 10.1046/j.1471-4159.1994.63041529.x. [DOI] [PubMed] [Google Scholar]
- 77.Dosemeci A, Reese TS, Petersen J, Tao-Cheng JH. A novel particulate form of Ca(2+)/calmodulin-dependent [correction of Ca(2+)/CaMKII-dependent] protein kinase II in neurons. J Neurosci. 2000;20(9):3076–84. doi: 10.1523/JNEUROSCI.20-09-03076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen K, Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science (Washington, DC, U S) 1999;284(5411):162–6. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- 79.Shen K, Teruel MN, Connor JH, Shenolikar S, Meyer T. Molecular memory by reversible translocation of calcium/calmodulin-dependent protein kinase II. Nat Neurosci. 2000;3(9):881–6. doi: 10.1038/78783. [DOI] [PubMed] [Google Scholar]
- 80.Sharma K, Fong DK, Craig AM. Postsynaptic protein mobility in dendritic spines: long-term regulation by synaptic NMDA receptor activation. Mol Cell Neurosci. 2006;31(4):702–12. doi: 10.1016/j.mcn.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Barcomb K, Buard I, Coultrap SJ, Kulbe JR, O’Leary H, Benke TA, Bayer KU. Autonomous CaMKII requires further stimulation by Ca2+/calmodulin for enhancing synaptic strength. FASEB J. 2014;28(8):3810–9. doi: 10.1096/fj.14-250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pi HJ, Otmakhov N, Lemelin D, De Koninck P, Lisman J. Autonomous CaMKII can promote either long-term potentiation or long-term depression, depending on the state of T305/T306 phosphorylation. J Neurosci. 2010;30(26):8704–9. doi: 10.1523/JNEUROSCI.0133-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pi HJ, Otmakhov N, El Gaamouch F, Lemelin D, De Koninck P, Lisman J. CaMKII control of spine size and synaptic strength: role of phosphorylation states and nonenzymatic action. Proc Natl Acad Sci U S A. 2010;107(32):14437–42. doi: 10.1073/pnas.1009268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gustin RM, Shonesy BC, Robinson SL, Rentz TJ, Baucum AJ, 2nd, Jalan-Sakrikar N, Winder DG, Stanwood GD, Colbran RJ. Loss of Thr286 phosphorylation disrupts synaptic CaMKIIalpha targeting, NMDAR activity and behavior in preadolescent mice. Mol Cell Neurosci. 2011;47(4):286–92. doi: 10.1016/j.mcn.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411(6839):801–5. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- 86.Bayer KU, LeBel E, McDonald GL, O’Leary H, Schulman H, De Koninck P. Transition from reversible to persistent binding of CaMKII to postsynaptic sites and NR2B. J Neurosci. 2006;26(4):1164–74. doi: 10.1523/JNEUROSCI.3116-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robison AJ, Bass MA, Jiao Y, MacMillan LB, Carmody LC, Bartlett RK, Colbran RJ. Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2. J Biol Chem. 2005;280(42):35329–36. doi: 10.1074/jbc.M502191200. [DOI] [PubMed] [Google Scholar]
- 88.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science (Washington, DC, U S) 1992;256(5060):1199–202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 89.Sawallisch C, Berhorster K, Disanza A, Mantoani S, Kintscher M, Stoenica L, Dityatev A, Sieber S, Kindler S, Morellini F, Schweizer M, Boeckers TM, Korte M, Scita G, Kreienkamp HJ. The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J Biol Chem. 2009;284(14):9225–36. doi: 10.1074/jbc.M808425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou J, Jones DR, Duong DM, Levey AI, Lah JJ, Peng J. Proteomic analysis of postsynaptic density in Alzheimer’s disease. Clin Chim Acta. 2013;420:62–8. doi: 10.1016/j.cca.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140(4):567–78. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 92.Migues PV, Lehmann IT, Fluechter L, Cammarota M, Gurd JW, Sim AT, Dickson PW, Rostas JA. Phosphorylation of CaMKII at Thr253 occurs in vivo and enhances binding to isolated postsynaptic densities. J Neurochem. 2006;98(1):289–99. doi: 10.1111/j.1471-4159.2006.03876.x. [DOI] [PubMed] [Google Scholar]
- 93.Skelding KA, Suzuki T, Gordon S, Xue J, Verrills NM, Dickson PW, Rostas JA. Regulation of CaMKII by phospho-Thr253 or phospho-Thr286 sensitive targeting alters cellular function. Cell Signalling. 2010;22(5):759–69. doi: 10.1016/j.cellsig.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 94.Dosemeci A, Gollop N, Jaffe H. Identification of a major autophosphorylation site on postsynaptic density-associated Ca2+/calmodulin-dependent protein kinase. J Biol Chem. 1994;269(50):31330–3. [PubMed] [Google Scholar]
- 95.Miller SG, Patton BL, Kennedy MB. Sequences of autophosphorylation sites in neuronal type II CaM kinase that control Ca2(+)-independent activity. Neuron. 1988;1(7):593–604. doi: 10.1016/0896-6273(88)90109-2. [DOI] [PubMed] [Google Scholar]
- 96.Patton BL, Miller SG, Kennedy MB. Activation of type II calcium/calmodulin-dependent protein kinase by Ca2+/calmodulin is inhibited by autophosphorylation of threonine within the calmodulin-binding domain. J Biol Chem. 1990;265(19):11204–12. [PubMed] [Google Scholar]
- 97.Martins-de-Souza D, Guest PC, Vanattou-Saifoudine N, Rahmoune H, Bahn S. Phosphoproteomic differences in major depressive disorder postmortem brains indicate effects on synaptic function. Eur Arch Psychiatry Clin Neurosci. 2012;262(8):657–66. doi: 10.1007/s00406-012-0301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics. 2006;5(5):914–22. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 99.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7(7):2845–51. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oppermann FS, Gnad F, Olsen JV, Hornberger R, Greff Z, Keri G, Mann M, Daub H. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8(7):1751–64. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herskowitz JH, Seyfried NT, Duong DM, Xia Q, Rees HD, Gearing M, Peng J, Lah JJ, Levey AI. Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration. J Proteome Res. 2010;9(12):6368–79. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baucum AJ, 2nd, Shonesy BC, Rose KL, Colbran RJ. Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem Neurosci. 2015;6(4):615–31. doi: 10.1021/cn500337u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown AM, Baucum AJ, Bass MA, Colbran RJ. Association of protein phosphatase 1 gamma 1 with spinophilin suppresses phosphatase activity in a Parkinson disease model. J Biol Chem. 2008;283(21):14286–94. doi: 10.1074/jbc.M801377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Picconi B, Gardoni F, Centonze D, Mauceri D, Cenci MA, Bernardi G, Calabresi P, Di Luca M. Abnormal Ca2+-calmodulin-dependent protein kinase II function mediates synaptic and motor deficits in experimental parkinsonism. J Neurosci. 2004;24(23):5283–91. doi: 10.1523/JNEUROSCI.1224-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, Jiang YH, Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10(3):280–2. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- 106.Wang N, Chen L, Cheng N, Zhang J, Tian T, Lu W. Active calcium/calmodulin-dependent protein kinase II (CaMKII) regulates NMDA receptor mediated postischemic long-term potentiation (i-LTP) by promoting the interaction between CaMKII and NMDA receptors in ischemia. Neural Plast. 2014;2014:827161. doi: 10.1155/2014/827161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rich MT, Abbott TB, Chung L, Gulcicek EE, Stone KL, Colangelo CM, Lam TT, Nairn AC, Taylor JR, Torregrossa MM. Phosphoproteomic Analysis Reveals a Novel Mechanism of CaMKIIalpha Regulation Inversely Induced by Cocaine Memory Extinction versus Reconsolidation. J Neurosci. 2016;36(29):7613–27. doi: 10.1523/JNEUROSCI.1108-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gregorich ZR, Ge Y. Top-down proteomics in health and disease: challenges and opportunities. Proteomics. 2014;14(10):1195–210. doi: 10.1002/pmic.201300432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kang MG, Nuriya M, Guo Y, Martindale KD, Lee DZ, Huganir RL. Proteomic analysis of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor complexes. J Biol Chem. 2012;287(34):28632–45. doi: 10.1074/jbc.M111.336644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, Klocker N, Schulte U, Fakler B. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74(4):621–33. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 111.Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR, 3rd, Nakagawa T. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 2012;1(6):590–8. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B, Schulte U. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84(1):41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 113.Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408(6815):936–43. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 114.Letts VA, Valenzuela A, Kirley JP, Sweet HO, Davisson MT, Frankel WN. Genetic and physical maps of the stargazer locus on mouse chromosome 15. Genomics. 1997;43(1):62–8. doi: 10.1006/geno.1997.4780. [DOI] [PubMed] [Google Scholar]
- 115.Noebels JL, Qiao X, Bronson RT, Spencer C, Davisson MT. Stargazer: a new neurological mutant on chromosome 15 in the mouse with prolonged cortical seizures. Epilepsy Res. 1990;7(2):129–35. doi: 10.1016/0920-1211(90)90098-g. [DOI] [PubMed] [Google Scholar]
- 116.Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA, Bredt DS. Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol. 2003;161(4):805–16. doi: 10.1083/jcb.200212116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park J, Chavez AE, Mineur YS, Morimoto-Tomita M, Lutzu S, Kim KS, Picciotto MR, Castillo PE, Tomita S. CaMKII Phosphorylation of TARPgamma-8 Is a Mediator of LTP and Learning and Memory. Neuron. 2016;92(1):75–83. doi: 10.1016/j.neuron.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McGee TP, Bats C, Farrant M, Cull-Candy SG. Auxiliary Subunit GSG1L Acts to Suppress Calcium-Permeable AMPA Receptor Function. J Neurosci. 2015;35(49):16171–9. doi: 10.1523/JNEUROSCI.2152-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Erlenhardt N, Yu H, Abiraman K, Yamasaki T, Wadiche JI, Tomita S, Bredt DS. Porcupine Controls Hippocampal AMPAR Levels, Composition, and Synaptic Transmission. Cell Rep. 2016;14(4):782–94. doi: 10.1016/j.celrep.2015.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li KW, Chen N, Smit AB. Interaction proteomics of the AMPA receptor: towards identification of receptor sub-complexes. Amino Acids. 2013;44(5):1247–51. doi: 10.1007/s00726-013-1461-9. [DOI] [PubMed] [Google Scholar]
- 121.Chen X, Nelson CD, Li X, Winters CA, Azzam R, Sousa AA, Leapman RD, Gainer H, Sheng M, Reese TS. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci. 2011;31(17):6329–38. doi: 10.1523/JNEUROSCI.5968-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Won S, Incontro S, Nicoll RA, Roche KW. PSD-95 stabilizes NMDA receptors by inducing the degradation of STEP61. Proc Natl Acad Sci U S A. 2016;113:E4736. doi: 10.1073/pnas.1609702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vickers CA, Stephens B, Bowen J, Arbuthnott GW, Grant SG, Ingham CA. Neurone specific regulation of dendritic spines in vivo by post synaptic density 95 protein (PSD-95) Brain Res. 2006;1090(1):89–98. doi: 10.1016/j.brainres.2006.03.075. [DOI] [PubMed] [Google Scholar]
- 124.Gao C, Tronson NC, Radulovic J. Modulation of behavior by scaffolding proteins of the post-synaptic density. Neurobiol Learn Mem. 2013;105:3–12. doi: 10.1016/j.nlm.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O’Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396(6710):433–9. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 126.Camp MC, Feyder M, Ihne J, Palachick B, Hurd B, Karlsson RM, Noronha B, Chen YC, Coba MP, Grant SG, Holmes A. A novel role for PSD-95 in mediating ethanol intoxication, drinking and place preference. Addict Biol. 2011;16(3):428–39. doi: 10.1111/j.1369-1600.2010.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fernandez E, Collins MO, Uren RT, Kopanitsa MV, Komiyama NH, Croning MD, Zografos L, Armstrong JD, Choudhary JS, Grant SG. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353(6304):1123–9. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fossati G, Morini R, Corradini I, Antonucci F, Trepte P, Edry E, Sharma V, Papale A, Pozzi D, Defilippi P, Meier JC, Brambilla R, Turco E, Rosenblum K, Wanker EE, Ziv NE, Menna E, Matteoli M. Reduced SNAP-25 increases PSD-95 mobility and impairs spine morphogenesis. Cell Death Differ. 2015;22(9):1425–36. doi: 10.1038/cdd.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sakai Y, Shaw CA, Dawson BC, Dugas DV, Al-Mohtaseb Z, Hill DE, Zoghbi HY. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med. 2011;3(86):86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang B, Wang T, Wan H, Han L, Qin X, Zhang Y, Wang J, Yu C, Berton F, Francesconi W, Yates JR, 3rd, Vanderklish PW, Liao L. Fmr1 deficiency promotes age-dependent alterations in the cortical synaptic proteome. Proc Natl Acad Sci U S A. 2015;112(34):E4697–706. doi: 10.1073/pnas.1502258112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, Carr SA, Ting AY. Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell. 2016;166:1295–1307. doi: 10.1016/j.cell.2016.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schreiner D, Savas JN, Herzog E, Brose N, de Wit J. Synapse biology in the ’circuit-age’-paths toward molecular connectomics. Curr Opin Neurobiol. 2017;42:102–110. doi: 10.1016/j.conb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yao Z, Petschnigg J, Ketteler R, Stagljar I. Application guide for omics approaches to cell signaling. Nat Chem Biol. 2015;11(6):387–97. doi: 10.1038/nchembio.1809. [DOI] [PubMed] [Google Scholar]
- 135.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196(6):801–10. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heller EA, Zhang W, Selimi F, Earnheart JC, Slimak MA, Santos-Torres J, Ibanez-Tallon I, Aoki C, Chait BT, Heintz N. The biochemical anatomy of cortical inhibitory synapses. PLoS One. 2012;7(6):e39572. doi: 10.1371/journal.pone.0039572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 2009;7(4):e1000083. doi: 10.1371/journal.pbio.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc. 2016;11(3):456–75. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339(6125):1328–31. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, Ting AY. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat Biotechnol. 2016;34(7):774–80. doi: 10.1038/nbt.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 142.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Toby TK, Fornelli L, Kelleher NL. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu Rev Anal Chem. 2016;9(1):499–519. doi: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, Coon JJ. The one hour yeast proteome. Mol Cell Proteomics. 2014;13(1):339–47. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shishkova E, Hebert AS, Coon JJ. Now, More Than Ever, Proteomics Needs Better Chromatography. Cell Syst. 2016;3(4):321–324. doi: 10.1016/j.cels.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9(3):1323–9. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baucum AJ, 2nd, Jalan-Sakrikar N, Jiao Y, Gustin RM, Carmody LC, Tabb DL, Ham AJ, Colbran RJ. Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Mol Cell Proteomics. 2010;9(6):1243–59. doi: 10.1074/mcp.M900387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rauniyar N, Yates JR., 3rd Isobaric labeling-based relative quantification in shotgun proteomics. J Proteome Res. 2014;13(12):5293–309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schmidt C, Gronborg M, Deckert J, Bessonov S, Conrad T, Luhrmann R, Urlaub H. Mass spectrometry-based relative quantification of proteins in precatalytic and catalytically active spliceosomes by metabolic labeling (SILAC), chemical labeling (iTRAQ), and label-free spectral count. RNA. 2014;20(3):406–20. doi: 10.1261/rna.041244.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2007;7(4):684–96. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 151.Rosenberger G, Koh CC, Guo T, Rost HL, Kouvonen P, Collins BC, Heusel M, Liu Y, Caron E, Vichalkovski A, Faini M, Schubert OT, Faridi P, Ebhardt HA, Matondo M, Lam H, Bader SL, Campbell DS, Deutsch EW, Moritz RL, Tate S, Aebersold R. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Colangelo CM, Ivosev G, Chung L, Abbott T, Shifman M, Sakaue F, Cox D, Kitchen RR, Burton L, Tate SA, Gulcicek E, Bonner R, Rinehart J, Nairn AC, Williams KR. Development of a highly automated and multiplexed targeted proteome pipeline and assay for 112 rat brain synaptic proteins. Proteomics. 2015;15(7):1202–14. doi: 10.1002/pmic.201400353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–26. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, Rossner MJ, Mann M, Simons M. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18(12):1819–31. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]


