Significance
Mechanical cues, such as changes in rigidity (resistance to deformation) of the surrounding microenvironment, regulate cell behaviors. Integrin-based rigidity sensing has been examined extensively, but studies investigating cadherin-based rigidity sensing at cell–cell contacts are lacking. Our study provides insight into mechanisms underlying E-cadherin–dependent rigidity sensing. We demonstrate that E-cadherin adhesions are acutely sensitive to a change in the rigidity of polyacrylamide (PA) gels functionalized with the extracellular domain of E-cadherin (Ecad-Fc) and that actin nucleating proteins and Rho GTPases activated upon cell adhesion to Ecad-Fc PA gels are also required for early stages in cell–cell adhesion. We hypothesize that E-cadherin rigidity sensing facilitates the transition from initial cell–cell adhesion to more stable contacts.
Keywords: E-cadherin, rigidity, formins, Cdc42, actin dynamics
Abstract
Mechanical cues are sensed and transduced by cell adhesion complexes to regulate diverse cell behaviors. Extracellular matrix (ECM) rigidity sensing by integrin adhesions has been well studied, but rigidity sensing by cadherins during cell adhesion is largely unexplored. Using mechanically tunable polyacrylamide (PA) gels functionalized with the extracellular domain of E-cadherin (Ecad-Fc), we showed that E-cadherin–dependent epithelial cell adhesion was sensitive to changes in PA gel elastic modulus that produced striking differences in cell morphology, actin organization, and membrane dynamics. Traction force microscopy (TFM) revealed that cells produced the greatest tractions at the cell periphery, where distinct types of actin-based membrane protrusions formed. Cells responded to substrate rigidity by reorganizing the distribution and size of high-traction-stress regions at the cell periphery. Differences in adhesion and protrusion dynamics were mediated by balancing the activities of specific signaling molecules. Cell adhesion to a 30-kPa Ecad-Fc PA gel required Cdc42- and formin-dependent filopodia formation, whereas adhesion to a 60-kPa Ecad-Fc PA gel induced Arp2/3-dependent lamellipodial protrusions. A quantitative 3D cell–cell adhesion assay and live cell imaging of cell–cell contact formation revealed that inhibition of Cdc42, formin, and Arp2/3 activities blocked the initiation, but not the maintenance of established cell–cell adhesions. These results indicate that the same signaling molecules activated by E-cadherin rigidity sensing on PA gels contribute to actin organization and membrane dynamics during cell–cell adhesion. We hypothesize that a transition in the stiffness of E-cadherin homotypic interactions regulates actin and membrane dynamics during initial stages of cell–cell adhesion.
Cell adhesion is essential for tissue structure and function. Cells use specialized types of adhesions to interact with the surrounding environment, including integrin-based focal adhesions at cell–extracellular matrix (cell–ECM) contacts and cadherin-based adhesions at cell–cell contacts (1). Integrins bind to the ECM and intracellular proteins that link to the actin cytoskeleton and important signaling pathways (2). Similarly, cadherins regulate cell–cell recognition and adhesion (3) and, through cytoplasmic adaptor proteins (catenins, vinculin) (4, 5), also link to the actin cytoskeleton and other proteins with signaling and scaffolding functions (6).
Initiation of cell–cell adhesion requires significant reorganization of the actin cytoskeleton and is tightly controlled by the activities of actin nucleating proteins and Rho GTPases. Adhesion is initiated when filopodia from opposing cells come into contact with one another (7, 8), and this process is regulated by Cdc42 activity (9, 10) and formin-dependent actin polymerization (11–14). Intermediate stages of cell–cell contact formation involve lateral expansion of the contact by Rac1-induced and Arp2/3-dependent lamellipodial activity (15, 16). Finally, compaction of cell–cell adhesion is driven by RhoA-induced actomyosin contraction at the distal edges of the contact (15).
Cell adhesion is also regulated by mechanical cues. This has been studied extensively in the context of cellular responses to increased ECM rigidity, which showed that integrin adhesions grow in size and are strengthened by local assembly of the actin cytoskeleton and activation of actomyosin contraction (17–19). These mechanical cues have physiological consequences, as ECM rigidity influences stem cell fate and cell differentiation (20). Cadherins are also mechanosensitive proteins (21, 22). E-cadherin is under constitutive tension at cell–cell junctions and dynamically responds to changes in tension (23). Cellular responses to exogenous tension applied directly to cadherins using functionalized beads have been studied extensively (22, 24, 25). However, few studies have investigated how cadherins sense and respond to changes in rigidity of cell–cell interactions. Using N-cadherin–functionalized substrates, a previous study showed that N-cadherin adhesions in C2 myogenic cells grew in size and exerted greater traction forces in response to increased substrate rigidity (21). However, it remains unknown whether such cellular responses to 2D substrate rigidity involve different actin nucleating proteins and Rho GTPases or correlate with stages of native cell–cell adhesions.
Here, we investigated how cells responded to changes in substrate rigidity at E-cadherin homotypic interactions within the physiological range of elastic moduli (30–60 kPa) found in vivo. Using mechanically tuned polyacrylamide (PA) gels functionalized with the extracellular domain of E-cadherin (Ecad-Fc), we found significant differences in the morphology and membrane dynamics of Madin–Darby canine kidney (MDCK) epithelial cells in response to a change in Ecad-Fc PA gel elastic modulus (30–60 kPa). In contrast, there were relatively minor alterations in the morphology and adhesion dynamics of cells adhered to 30-kPa and 60-kPa PA gels functionalized with collagen I. Traction force microscopy (TFM) showed that the distinct cell morphology and protrusion dynamics on 30-kPa and 60-kPa Ecad-Fc PA gels corresponded to differences in PA substrate strain and the distribution of high-traction-stress regions at the cell periphery. We showed that there is a delicate balance of different actin nucleating proteins and Rho GTPase activities required for these cellular responses to changes in E-cadherin substrate modulus, which also correlated with their roles in different stages of native cell–cell adhesion. These results indicate that E-cadherin–dependent rigidity sensing and local signal transduction contribute to initial cell–cell contact formation.
Results
We prepared PA gels of different elastic moduli functionalized with either the ECM protein collagen I or the correctly oriented extracellular domain of E-cadherin fused at the C terminus with human Fc (Ecad-Fc) (26); these proteins were covalently linked to the surface of the PA gel with the protein–substrate linker sulfo-SANPAH. Substrates immobilized with Ecad-Fc mimic cell–cell interactions, and cellular E-cadherin protein dynamics at the Ecad-Fc interface are similar to those found at endogenous cell–cell junctions (27, 28). Saturating amounts of Ecad-Fc were bound to substrates to ensure maximum engagement of E-cadherin in adhering cells (23).
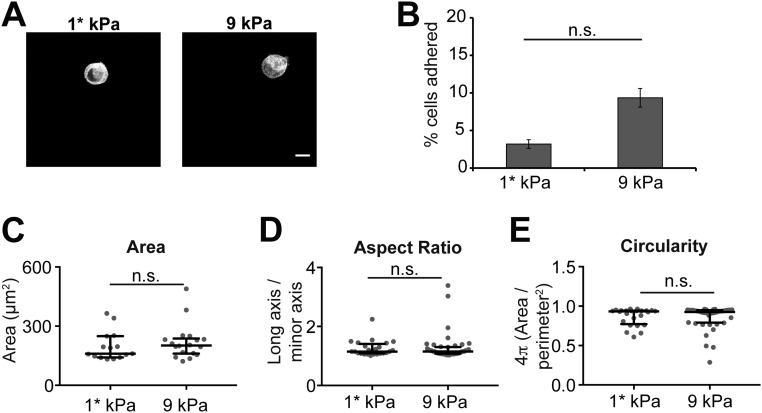
The stiffness of epithelial cell–cell contacts in vivo is unknown and reported elastic moduli of living cells vary widely. Some studies have indicated that the elastic modulus of the cell cortex is ∼1–5 kPa (29), but Ecad-Fc PA gels of ∼1 kPa and 9 kPa did not support MDCK cell adhesion or spreading (Fig. S1 and Table S1). Other work indicated that the elastic modulus of an MDCK cell monolayer is 33 kPa (30); this was derived by microscale indentation of multicellular groups with cell–cell contacts, which may be more relevant when considering the stiffness at cell–cell contacts. Therefore, we chose elastic moduli of ∼30 kPa and 60 kPa for this study, as these values are within the physiological range of tissues in vivo (0.1–100 kPa) (31, 32), are similar to values previously reported for an MDCK cell monolayer (∼33 kPa), and support MDCK adhesion and spreading (Fig. 1). Mechanically tunable PA gels were made by adjusting the total polymer content (%T) and cross-linker concentration (%C) in solution (33). The elastic moduli of the two gel formulations used in this study were verified by atomic force microscopy (AFM): 10%T, 1%C, 27.5–31.8 kPa (median: 29.07 kPa) and 10%T, 2.5% C, 52.6–67.2 kPa (median: 57.34 kPa) (Table S1). We also prepared glass coverslips (with an elastic modulus of ∼100 GPa) coated with Ecad-Fc, which have been used in most other studies (34–37).
Fig. S1.
Soft Ecad-Fc PA gels do not support MDCK cell adhesion and spreading. (A) Representative images of MDCK cells fixed and stained with phalloidin attached to Ecad-Fc PA gels with elastic moduli ∼1* kPa and 9 kPa. (B) Bar graph represents the percentage of seeded cells that adhered to 1*-kPa and 9-kPa Ecad-Fc PA gels averaged from three independent experiments. Error bars represent SEM. (C–E) Quantification of morphological features, including area (C), aspect ratio (D), and circularity (E). Cumulative data from three independent experiments are represented in a dot plot, where each dot represents one cell. The solid black line indicates the median and the upper and lower bars represent the interquartile range (n ≥ 15 cells per condition). *As indicated in Table S1, a gel formulation of 3.1%T, 3.22%C with a predicted elastic modulus of ∼1 kPa was used for these studies. However, the elastic modulus could not be confirmed by AFM due to the probe sticking to the gel.
Table S1.
AFM Measurements of polyacrylamide gels
| Gel formulation | Experiment | Elastic modulus, kPa | SD | n |
| 3.1%T, 3.22%C | 1 | * | * | * |
| 5.1%T, 2.91%C | 1 | 9.61491 | 0.2818 | 25 |
| 2 | 9.21432 | 0.6096 | 26 | |
| 10%T, 1%C | 1 | 31.75031 | 0.810869 | 25 |
| 2 | 29.32711 | 2.187512 | 22 | |
| 3 | 27.54688 | 0.871303 | 30 | |
| 10%T, 2.5%C | 1 | 52.56912 | 1.264124 | 26 |
| 2 | 67.15331 | 1.7727621 | 25 | |
| 3 | 57.29712 | 0.866444 | 29 |
Shown are AFM measurements indicating the elastic moduli of the gel formulations used in this study. For sample nomenclature, “%T” indicates the total polymer content, and “%C” indicates the cross-linker concentration used during polymerization. For each experiment, 22–30 total indentations were made in five different regions of the gel to account for any heterogeneity, which resulted in a total of 51 measurements for the “5.1%T, 2.91%C” group, 77 measurements for the “10%T, 1%C” group, and 80 measurements for the “10%T, 2.5%C” group.
The gel formulation of “3.1%T, 3.22%C” is expected to have an elastic modulus of ∼1 kPa, but we were unable to determine the elastic modulus of that sample due to the AFM probe sticking to the gel sample.
Fig. 1.
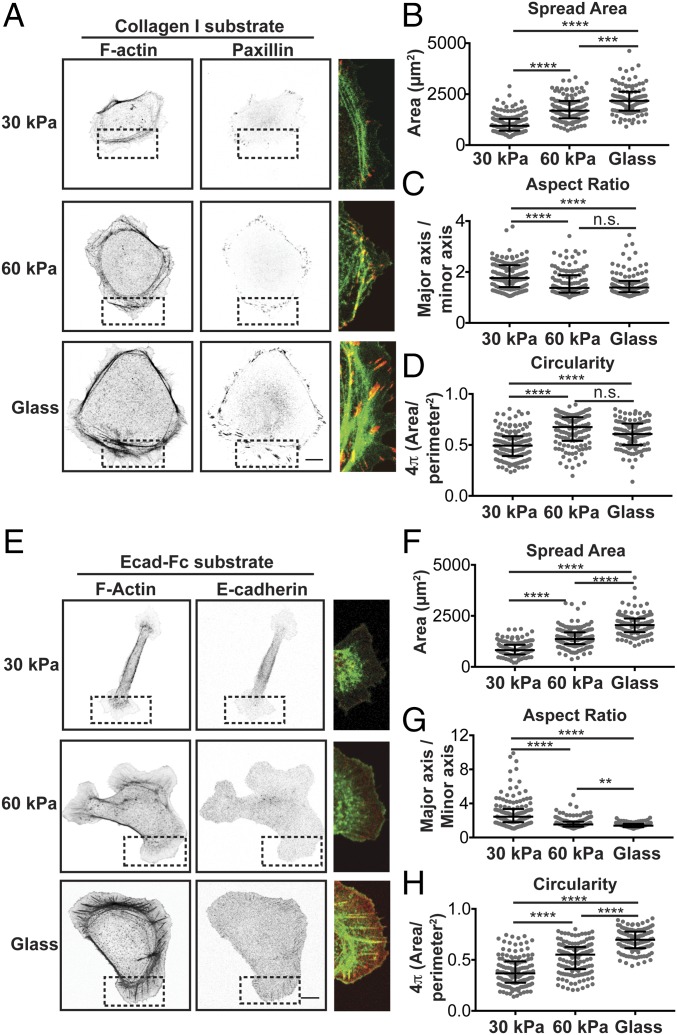
Effects of integrin– and E-cadherin–based rigidity sensing on adhesion organization and cell morphology. (A) Confocal images of MDCK cells fixed and stained for F-actin (phalloidin) (Left) and paxillin (Center) on collagen-I–coated substrates of different moduli. (A, Right) The dotted box in each image is merged and expanded (F-actin, green; paxillin, red). (Scale bar, 10 µm.) (B–D) Quantification of morphological features, including cell spread area (B), aspect ratio (C), and circularity (D). Cumulative data from three experiments are represented in a dot plot, where each dot represents one cell. The solid black line indicates the median and the upper and lower bars represented the interquartile range (n > 140 cells per condition). (E) Confocal images of MDCK cells stably expressing E-cadherin:dsRed fixed and stained for F-actin (phalloidin) on Ecad-Fc–functionalized substrates of different moduli. (E, Right) Dotted box in each image is merged and expanded (F-actin, green; E-cadherin:dsRed, red). (Scale bar, 10 µm.) (F–H) Quantification of morphological features, including cell spread area (F), aspect ratio (G), and circularity (H). Cumulative data from three independent experiments are represented in a dot plot, where each dot represents one cell. The solid black line indicates the median and upper and lower bars represent the interquartile range (n > 115 cells per condition). Statistics were determined using a Kruskal–Wallis test with Dunn’s posttest for multiple comparisons, **P < 0.001, ***P < 0.0002, ****P < 0.0001. Noncumulative averages and statistics for the three independent experiments presented in B–D and F–H are listed in Table S2.
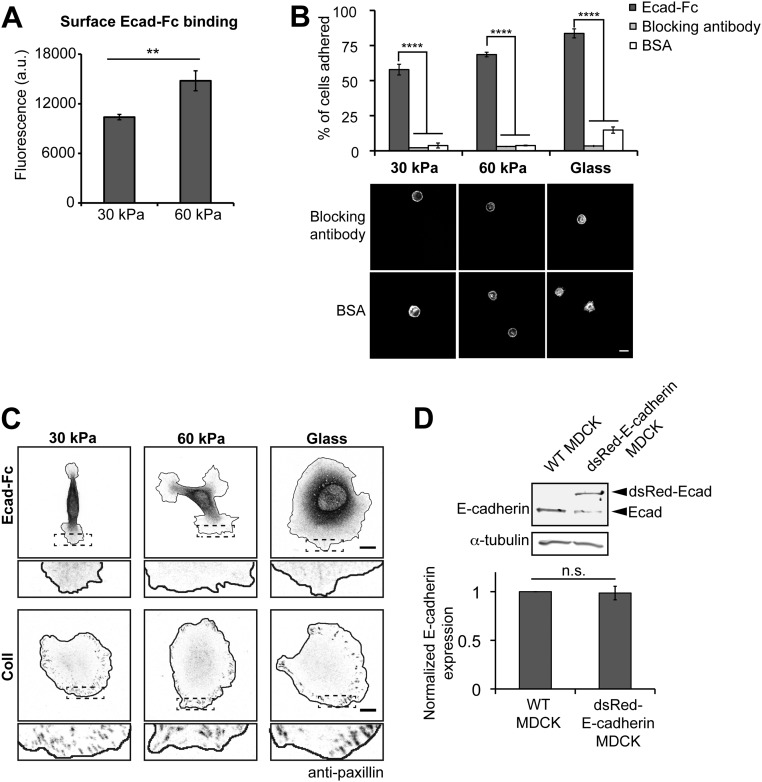
Ecad-Fc binding on the surface of the gels was quantified with SYPRO Ruby red protein stain. Whereas saturating amounts of E-cadherin were used to ensure maximal E-cadherin binding, more Ecad-Fc bound to the surface of 60-kPa PA gels compared with that to the surface of 30-kPa PA gels (Fig. S2A). The percentage of cells that attached to Ecad-Fc gels increased with increasing substrate modulus, ranging from 57% to 83% (Fig. S2B). To test the specificity of cell adhesion to Ecad-Fc substrates, we used three controls: (i) Substrates functionalized with BSA did not support cell adhesion (4–15% of cells adhered) and blocked cell spreading (Fig. S2B); (ii) blocking Ecad-Fc adhesion with an E-cadherin function-blocking antibody (rr-1) (38) inhibited cell adhesion to Ecad-Fc substrates (<5% of cells adhered), and the few cells that attached remained rounded and did not spread (Fig. S2B); and (iii) to test whether secreted ECM proteins facilitated cell adhesion to Ecad-Fc PA gels, cells were fixed and stained for the integrin-based focal adhesion protein paxillin. Paxillin staining in cells attached to and spread on Ecad-Fc PA gels was diffuse throughout the cytoplasm and focal adhesion-like clusters were not detected (Fig. S2C); in contrast, cells attached to collagen-functionalized gels showed a clear enrichment of paxillin at focal adhesions around the cell periphery (Fig. S2C). Taken together, we conclude that MDCK cell adhesion to Ecad-Fc PA gels involves specific engagement of cellular E-cadherin with Ecad-Fc to promote cell attachment and spreading.
Fig. S2.
Validation of Ecad-Fc PA gels. (A) Quantification of Ecad-Fc on the surface of functionalized gels. Fluorescence of SYPRO Ruby gel protein stain is represented in a bar graph. Data represent the fluorescence intensities averaged from three independent experiments. Error bars represent SEM. (B, Top) Bar graph indicates the average percentage of seeded cells that adhered to Ecad-Fc, Ecad-Fc + rr-1 blocking antibody, or BSA-coated substrates from three independent experiments. Error bars represent SEM. (B, Bottom) Representative images of MDCK cells fixed and stained with phalloidin adhered to rr-1 antibody blocked or BSA-coated substrates. (Scale bar, 10 µm.) (C) Representative images of MDCK cells adhered to Ecad-Fc or collagen-coated substrate stained for the focal adhesion protein paxillin. (Scale bar, 10 µm.) (D) Cell lysates from WT and E-cadherin:dsRed MDCK cells subjected to SDS/PAGE. The bar graph represents the average total E-cadherin expression (endogenous + E-cadherin:dsRed) relative to the loading control “tubulin” and normalized to the “WT MDCK” cell condition from three independent experiments. Error bars represent SEM. Statistics were determined using a Student’s t test, **P = 0.02, ****P < 0.0001.
To assess the effects of substrate rigidity on cell morphology and adhesion organization, single MDCK cells were plated on collagen-I– or Ecad-Fc–functionalized 30-kPa or 60-kPa PA gels or glass coverslips. MDCK cells were chosen for this study because cell–cell adhesion dynamics in these cells have been extensively studied and are well defined (7, 15), and single cells were examined to avoid competition with native cell–cell adhesions in larger cell aggregates. Five hours after plating, cells were fixed and processed for immunofluorescence microscopy.
First, we examined cells adhered to collagen-I–coated substrates by imaging phalloidin to mark F-actin, and paxillin to mark integrin-based focal adhesions (Fig. 1A). Regardless of substrate modulus (ECM rigidity), all cells were flat and circular with a circumferential ring of F-actin and paxillin foci just beyond the F-actin belt and around the cell periphery (Fig. 1A). The size of paxillin-positive focal adhesions appeared to increase as the ECM rigidity increased. Quantification of morphological features of cells revealed that the cell area increased with increased ECM rigidity (Fig. 1B and Table S2), in agreement with previous studies (17, 39, 40). There were also relatively small, but statistically significant changes in the aspect ratio and circularity of cells with changes in ECM rigidity (Fig. 1 C and D and Table S2).
Table S2.
Averages and statistics of collagen and Ecad-Fc spread area and morphology data
| Average ± SEM | P value | |||||
| Condition | 30 kPa | 60 kPa | Glass | 30 kPa vs. 60 kPa | 60 kPa vs. glass | 30 kPa vs. glass |
| Collagen–spread area | 1048 ± 72 | 2013 ± 124 | 2515 ± 127 | 0.002 | 0.03 | 0.0004 |
| Collagen–aspect ratio | 2.07 ± 0.15 | 1.57 ± 0.07 | 1.49 ± 0.07 | 0.04 | n.s. | 0.02 |
| Collagen–circularity | 0.50 ± 0.03 | 0.64 ± 0.02 | 0.61 ± 0.03 | 0.009 | n.s. | 0.03 |
| Ecad-Fc–spread area | 929 ± 28 | 1550 ± 48 | 2039 ± 128 | 0.0003 | 0.02 | 0.0001 |
| Ecad-Fc–aspect ratio | 2.62 ± 0.019 | 1.49 ± 0.02 | 1.46 ± 0.06 | 1.39E-06 | n.s. | 6.80E-05 |
| Ecad-Fc–circularity | 0.39 ± 0.03 | 0.59 ± 0.03 | 0.72 ± 0.03 | 0.009 | 0.02 | 0.0009 |
Shown is analysis corresponding with the data presented in Fig. 1. Values presented are averages and SEM calculated from three independent experiments. Statistics were determined using a Student’s t test.
Next, we examined MDCK cells stably expressing E-cadherin:dsRed adhered to Ecad-Fc substrates of different moduli; the total level of E-cadherin expression in these cells was similar to that in control MDCK cells (Fig. S2D). Confocal imaging of cells 5 h after plating revealed that cell spread area and morphology differed significantly between cells adhered to Ecad-Fc substrates of different rigidity (Fig. 1 E–H). On Ecad-Fc–functionalized glass, cells were very flat and round, with large focal adhesion-like cadherin plaques at the end of thick radial F-actin bundles around the periphery (Fig. 1E), similar to observations in previous studies (34–37). These prominent F-actin and E-cadherin structures were very different from the organization of these proteins in cells adhered to Ecad-Fc PA gels. On a 30-kPa Ecad-Fc PA gel, cells appeared to have very few F-actin bundles, and E-cadherin was generally diffuse with weak staining in small protrusions at either end of the cell (Fig. 1E). Cells adhered to a 60-kPa Ecad-Fc PA gel had a few small F-actin bundles and an enrichment of F-actin at the edge of broad protrusions, whereas E-cadherin was generally diffuse and did not appear to be organized into prominent puncta or plaques (Fig. 1E).
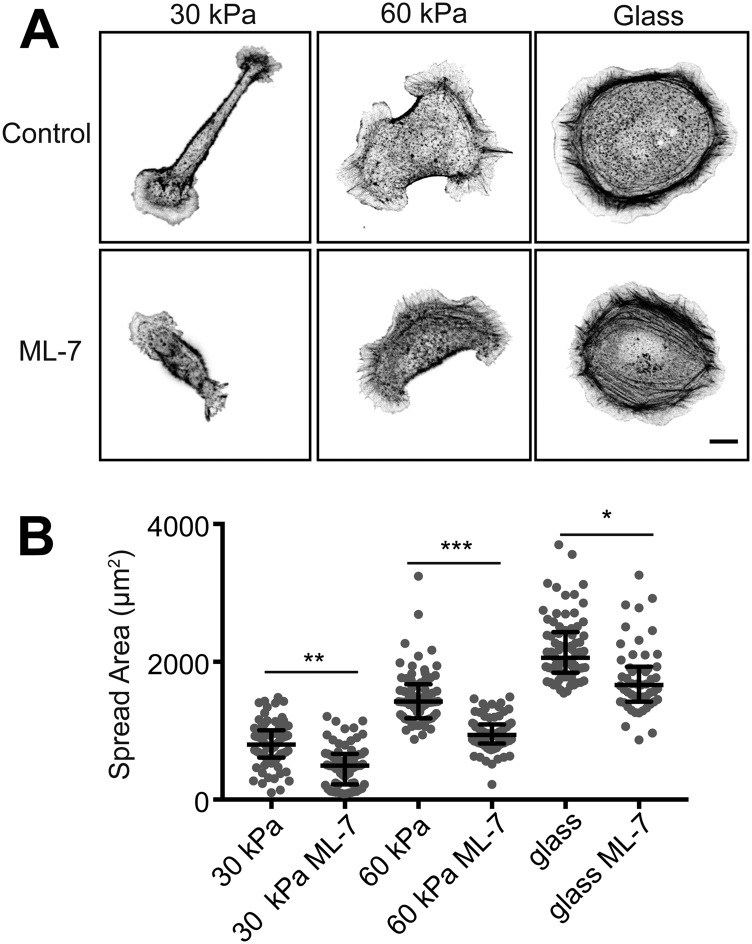
In contrast to cells adhered to collagen-I substrates, cell morphologies were very different, depending on the Ecad-Fc substrate rigidity. Cells adhered to a 30-kPa Ecad-Fc PA gel were narrow and elongated with a large, flat membrane lamellipodium with small, thin membrane protrusions at either end of the cell body (Fig. 1F and Table S2). Cells adhered to a 60-kPa Ecad-Fc PA gel were generally flatter and more circular, with a larger spread area and many broad, lamellipodia-like protrusions (Fig. 1F). Finally, cells adhering to Ecad-Fc–functionalized glass had the largest spread area and few protrusions beyond the prominent cortical F-actin ring (Fig. 1F and Table S2). Cell spreading on 30-kPa and 60-kPa Ecad-Fc PA gels and Ecad-Fc–functionalized glass was similarly dependent on myosin II, as treatment with ML-7 resulted in a 30% decrease in spread area but did not result in complete rounding of cells (Fig. S3). The elongated cell morphology on a 30-kPa Ecad-Fc gel had a significantly higher aspect ratio than that of cells adhered to a 60-kPa Ecad-Fc PA gel or glass (Fig. 1G and Table S2); as expected, there were corresponding increases in cell circularity with increasing Ecad-Fc rigidity (Fig. 1H and Table S2). Together, these results indicate that increasing the ECM (collagen) rigidity had relatively small effects on overall cell morphology or the organization of F-actin and adhesive structures. In contrast, increasing the Ecad-Fc substrate rigidity significantly affected cell morphology and the organization of F-actin and E-cadherin.
Fig. S3.
Cell spreading on Ecad-Fc substrates is partially dependent on actomyosin contractility. (A) Representative images of control and ML-7–treated (25 µM) cells fixed and stained with phalloidin. (Scale bar, 10 µm.) (B) Quantification of spread area. Cumulative data from three independent experiments are represented in a dot plot, where each dot represents one cell. The solid black line indicates the median and the upper and lower bars represent the interquartile range (n ≥ 50 cells per condition). Statistics were performed using a Mann–Whitney test to compare the “control” and “ML-7” conditions for each substrate stiffness.
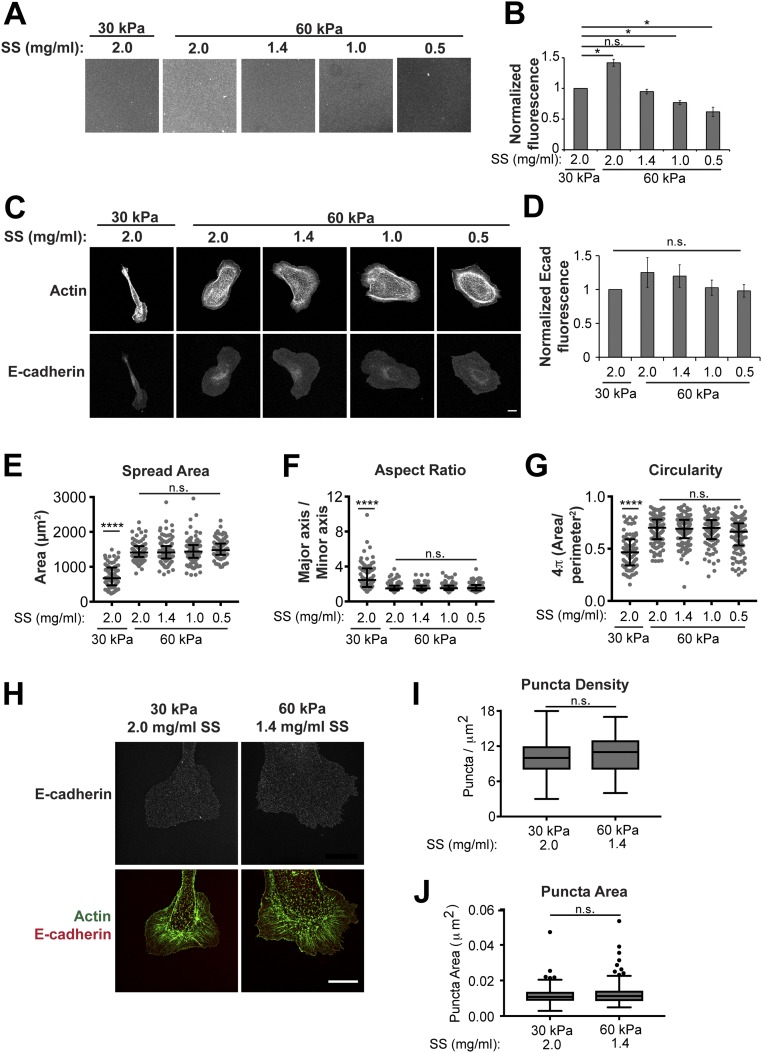
A caveat to this conclusion is that more Ecad-Fc ligand bound to the surface of the 60-kPa PA gel compared with a 30-kPa PA gel (Fig. S2A), and thus differences in cell morphology might be due to differences in ligand density, rather than gel modulus. Previous studies demonstrated that protein tethering to the surface of PA gels can be modulated by varying the concentration of the substrate–protein cross-linker sulfo-SANPAH (SS) (41). Therefore, a range of SS concentrations (0.5–2.0 mg/mL) were used to functionalize a 60-kPa PA gel, and Ecad-Fc ligand binding was measured using an anti–E-cadherin antibody (Fig. S4A). This allowed us to establish 60-kPa PA gel conditions in which Ecad-Fc ligand density was equal to (1.4 mg/mL SS) or lower than that of a 30-kPa gel (0.5 mg/mL and 1.0 mg/mL SS) (Fig. S4 A and B). We measured levels of cellular E-cadherin fluorescence at the cell–substrate interface on 30-kPa PA gels and 60-kPa PA gels with different levels of Ecad-Fc ligand density. Whereas there was a slight trend in lower levels of cellular E-cadherin fluorescence with lower Ecad-Fc ligand density (Fig. S4 C and D), the difference in cellular E-cadherin levels between all 30-kPa and 60-kPa Ecad-Fc PA gels was not statistically significant. Importantly, the amount of cell spreading and overall morphology on a 60-kPa PA gel did not change as a result of decreasing Ecad-Fc ligand density, and cells adhered to different 60-kPa PA gel Ecad-Fc ligand densities had a spread area and morphology that were statistically different from those of cells adhered to a 30-kPa gel (Fig. S4 E–G). These data indicate that differences in cell spreading and morphology on 30-kPa and 60-kPa Ecad-Fc PA gels are due to substrate rigidity and not differences in Ecad-Fc ligand density, and that the similar levels of cellular E-cadherin on these Ecad-Fc substrates likely reflect saturated binding over this range of ligand densities.
Fig. S4.
Adjusting Ecad-Fc ligand density does not affect cell spreading, morphology, or E-cadherin organization on 60-kPa PA gels. (A) Representative images of 30-kPa and 60-kPa Ecad-Fc PA gels prepared with varying concentrations of SS stained with an anti–E-cadherin antibody. (Scale bar, 50 µm.) (B) Quantification of anti–E-cadherin antibody staining on the surface of functionalized gels. Data represent the fluorescence intensities normalized to the “30 kPa, 2.0 mg/mL” condition averaged from three independent experiments. Error bars represent SEM. Statistics were determined using a Student’s t test, *P < 0.05. (C) Representative images of E-cadherin:dsRed MDCK cells on 30-kPa and 60-kPa PA gels functionalized with varying concentrations of SS. Cells were fixed and stained with phalloidin. (Scale bar, 10 µm.) (D) Average cellular E-cadherin fluorescence intensity on 30-kPa and 60-kPa PA Ecad-Fc gels functionalized with varying concentrations of SS normalized to the 30 kPa, 2.0 mg/mL condition from three independent experiments. Error bars represent SEM. Statistics were determined using a Student’s t test. (E–G) Quantification of morphological features, including cell spread area (E), aspect ratio (F), and circularity (G). Cumulative data from three independent experiments are represented in a dot plot, where each dot represents one cell. The solid black line indicates the median and upper and lower bars represent the interquartile range (n > 80 cells per condition). Statistics were determined using a Kruskal–Wallis test with Dunn’s posttest for multiple comparisons, ****P < 0.0001. (H) Representative SIM superresolution images of cells stably expressing E-cadherin:dsRed and stained for actin (phalloidin) on 30-kPa and 60-kPa PA Ecad-Fc gels functionalized with varying concentrations of SS. (Scale bar, 10 µm.) (I) Quantification of E-cadherin puncta per square micrometer represented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box represents the median, and whiskers outside the box extend to the highest and lowest value within the 1.5× interquartile range. Data shown are cumulative from three independent experiments. (J) Quantification of E-cadherin puncta size represented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box represents the median, and whiskers outside the box extend to the highest and lowest value within the 1.5× interquartile range. Data shown are cumulative from three independent experiments. For quantification (I and J), five 1-µm2 regions within cell protrusions were analyzed per cell and at least 10 cells per condition were quantified. Statistics were determined using a Kruskal–Wallis test with Dunn’s posttest for multiple comparisons.
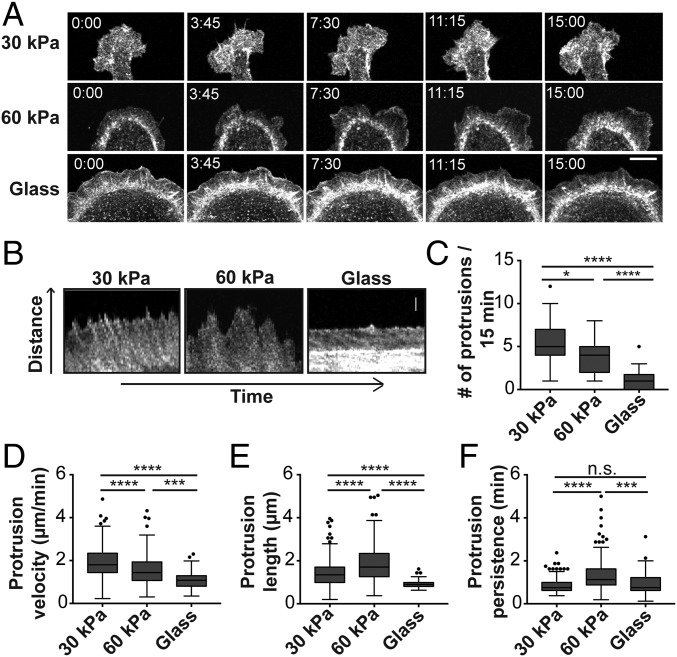
Given the striking difference in cell spreading and plasma membrane protrusions between Ecad-Fc–functionalized 30-kPa and 60-kPa PA gels (Fig. 1E), we measured plasma membrane dynamics in MDCK cells expressing GFP-LifeAct. Cells adhered to a 30-kPa Ecad-Fc PA gel had a large, flat, and rather stable membrane lamellipodium at both ends of the elongated cell body, from which dynamic filopodia-like extensions rapidly extended and retracted (Fig. 2A and Movie S1); few, if any, lamellipodia or filopodia were detected along the sides of these elongated cells. In contrast, cells adhered to a 60-kPa Ecad-Fc PA gel had large, dynamic lamellipodia that ruffled around the entire cell periphery (Fig. 2A and Movie S2); few filopodia were detected in these cells, compared with cells on a 30-kPa Ecad-Fc PA gel. Cells adhered to Ecad-Fc–functionalized glass were flat and circular, and the plasma membrane at the periphery was relatively undynamic and extended very few protrusions (Fig. 2A and Movie S3). Kymograph analysis of live cell images (Fig. 2B) showed that the number (Fig. 2C) and velocity (Fig. 2D) of membrane protrusions decreased in cells adhered to Ecad-Fc substrates of increasing rigidities; in general, cells adhered to 30-kPa and glass Ecad-Fc substrates extended shorter and less persistent protrusions, compared with cells adhered to the 60-kPa PA gel (Fig. 2 E and F).
Fig. 2.
Ecad-Fc substrate rigidity influences actin dynamics in adhering cells. (A) Time-lapse images of MDCK cells expressing GFP-LifeAct adhered to Ecad-Fc–functionalized surfaces of varying rigidities. (Scale bar, 10 µm.) (B) Representative kymograph images of membrane protrusions from MDCK cells expressing GFP-LifeAct adhered to Ecad-Fc substrates. (Scale bar, 2 µm.) (C–F) Quantification of number of protrusions (C), protrusion velocity (D), protrusion length (E), and protrusion persistence (F) over a 15-min interval. Four regions from at least 10 different cells were analyzed for each substrate, resulting in quantification of 209 protrusions (30 kPa), 143 protrusions (60 kPa), and 43 protrusions (glass). Results are represented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box represents the median, and whiskers outside the box extend to the highest and lowest value within the 1.5× interquartile range. Statistics were determined using a Kruskal–Wallis test with Dunn’s posttest for multiple comparisons, *P < 0.05, ***P < 0.0002, ****P < 0.0001.
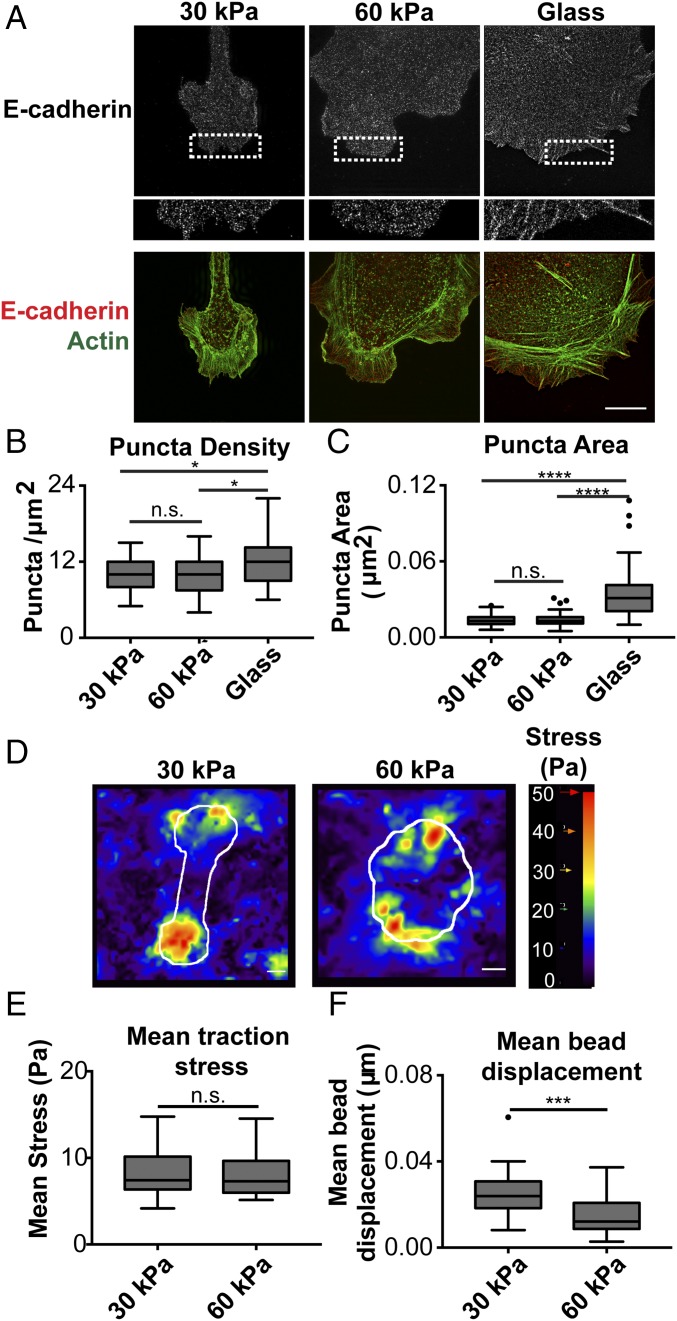
Next, we sought to determine whether differences in protrusion dynamics were mediated by differences in clustering of cellular E-cadherin. MDCK cells stably expressing E-cadherin:dsRed plated on Ecad-Fc gels were fixed and processed for structured illumination microscopy (SIM) 5 h after seeding. Similar to confocal image analysis (Fig. 1E), SIM analysis showed that E-cadherin localization on 30-kPa and 60-kPa Ecad-Fc PA gels was generally diffuse throughout the cell body and at membrane protrusions, but at higher resolution with SIM, small puncta were now visible (Fig. 3A). The level of E-cadherin in and the size of these puncta were similar in cells on 30-kPa and 60-kPa Ecad-Fc PA gels (Fig. 3 B and C). Because Ecad-Fc ligand density on the surface of the gel could contribute to E-cadherin clustering, we also measured E-cadherin puncta on 30-kPa and 60-kPa PA gels with matched Ecad-Fc ligand density and found little or no difference in puncta density and size (Fig. S4 H–J). In contrast, focal adhesion-like E-cadherin plaques were present around the periphery of cells on Ecad-Fc–functionalized glass, and the level of E-cadherin and size of these clusters were significantly greater than the small E-cadherin puncta on 30-kPa and 60-kPa Ecad-Fc PA gels (Fig. 3 B and C). These data indicate that differences in E-cadherin organization and clustering are not responsible for the differences in cell shape or membrane protrusion dynamics on 30-kPa and 60-kPa Ecad-Fc PA gels.
Fig. 3.
Cells adhered to 30-kPa and 60-kPa Ecad-Fc PA gels exhibit similar cadherin clustering and mean traction stress, but the distribution of stress and strain differs. (A) Representative SIM images of cells stably expressing E-cadherin:dsRed and stained for actin (phalloidin). (A, Lower) Dotted box in the E-cadherin image is expanded. (Scale bar, 10 µm.) (B) Quantification of E-cadherin puncta per square micrometer represented in a box and whisker format, in which the ends of the box mark the upper and lower quartiles, the horizontal line in the box represents the median, and whiskers outside the box extend to the highest and lowest value within the 1.5× interquartile range. Data shown are cumulative from three independent experiments. (C) Quantification of E-cadherin puncta size represented in a box and whisker format. Data shown are cumulative from three independent experiments. For quantification (B and C), five 1-µm2 regions within cell protrusions were analyzed per cell and at least 10 cells per condition were quantified. Statistics were determined using a Kruskal–Wallis test with Dunn’s posttest for multiple comparisons, *P < 0.05, ****P < 0.0001. (D) Heat maps of the traction stress (magnitude shown) produced by cells adhered to Ecad-Fc PA gels, with cell boundary as overlay in white. (Scale bar, 10 µm.) (E) Mean traction stress (measured within cell boundaries) produced by cells on 30-kPa and 60-kPa Ecad-Fc PA gels. Results are represented in box and whisker format. n = 27 cells (30 kPa) and 19 cells (60 kPa). Statistics were determined using a Mann–Whitney test. (F) Mean bead displacements within 30-kPa and 60-kPa Ecad-Fc PA gels resulting from cell tractions. n = 27 (30 kPa) and 19 (60 kPa). Statistics were determined using a Mann–Whitney test. ***P = 0.0005.
To test the possibility that differences in substrate deformation and traction stress on 30-kPa and 60-kPa Ecad-Fc PA gels may contribute to differences in protrusion dynamics, we performed TFM by tracking the displacement of fluorescent beads embedded in Ecad-Fc PA gels (42). Heat maps of traction stress magnitude in cells on 30-kPa and 60-kPa Ecad-Fc PA gels showed that regions of highest stress were located at the cell periphery, where membrane protrusions were found. Cells with elongated morphologies on 30-kPa Ecad-Fc PA gels produced regions with the greatest stress only at either end of the cell, whereas cells on 60-kPa Ecad-Fc PA gels had the high-stress regions distributed all around the periphery (Fig. 3D). As a result, the distribution of high-traction-stress regions differed in cells on 30-kPa and 60-kPa Ecad-Fc PA gels. Cells produced fewer regions of high stress on 30-kPa Ecad-Fc PA gels but these regions were larger in area. Cells generally produced a greater number of high-stress regions on 60-kPa Ecad-Fc PA gels, but these regions were smaller in area (Fig. 3D). The mean traction stress magnitude within the cell boundary was remarkably similar on 30-kPa and 60-kPa Ecad-Fc PA gels and differences were not statistically significant (Fig. 3E). However, we measured a significantly larger substrate strain (using mean bead displacement, Fig. 3F) within cell boundaries on 30-kPa PA gels than on 60-kPa PA gels. Taken together, these data indicate that high-stress regions are located at the cell periphery near membrane protrusions, and the localized areas of high traction stress and strain exerted by cells on the substrate are greater in cells adhering to 30-kPa than to 60-kPa Ecad-Fc PA gels.
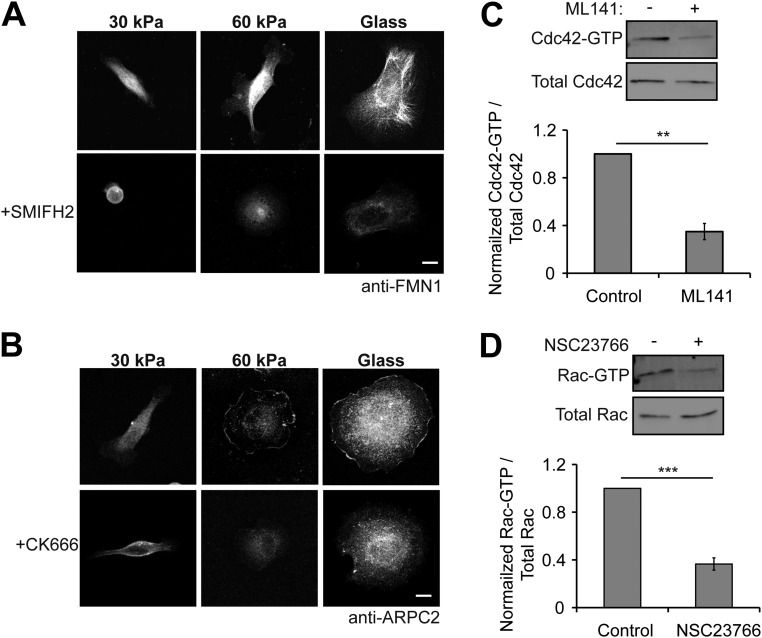
We hypothesized that differences in cadherin rigidity sensing on 30-kPa and 60-kPa Ecad-Fc PA gels involved the activation of different signaling pathways that regulate actin dynamics and contribute to cell shape and membrane protrusion dynamics. We could not use siRNA knockdown of protein expression or dominant-negative mutants, as constitutive knockdown or inhibition of proteins of interest is known to affect cell–cell adhesion and thus would disrupt initial cell attachment and spreading on PA gel substrates. Therefore, we used small molecule inhibitors that could be added and then washed out at specific times after cell attachment; target protein disruption by a given inhibitor was validated by immunofluorescence microscopy (formins, Arp2/3) or activity pull-down assays (Cdc42, Rac1) (Fig. S5).
Fig. S5.
Validation of small molecule inhibitors. (A) MDCK cells adhered to Ecad-Fc substrates and stained for the formin protein FMN1 in the presence or absence of the small molecule inhibitor SMIFH2. (Scale bar, 10 µm.) (B) MDCK cells adhered to Ecad-Fc substrates and stained for the Arp2/3 subunit ARPC2 in the presence or absence of the Arp 2/3 inhibitor CK666. (Scale bar, 10 µm.) (C) GTP-bound Cdc42 isolated from control and ML141-treated cells and analyzed by SDS/PAGE. Bar graph indicates the average from three independent experiments. Activity levels were normalized by setting the control condition to “1.” Error bars represent SEM. (D) GTP-bound Rac isolated from control and NSC23766-treated cells and analyzed by SDS/PAGE. Bar graph indicates the average from three independent experiments. Activity levels were normalized by setting the control condition to 1. Error bars represent SEM.
First, we sought to determine whether differences in actin dynamics were mediated by distinct actin nucleating proteins. MDCK cells stably expressing E-cadherin:dsRed were plated on a 30-kPa or 60-kPa Ecad-Fc PA gel and imaged by fluorescence microscopy and differential interference contrast (DIC) microscopy. Cells adhered to Ecad-Fc–functionalized glass were not examined in this assay because they appeared to be relatively undynamic and extended very few actin-based protrusions (Fig. 2 and Movie S3). DIC (Fig. S6) and fluorescence images (Fig. 4) were collected for 15 min, and baseline protrusive area and plasma membrane activity were measured from fluorescence images; DIC contrast was poor due to light interference from the PA gel. Small molecule inhibitors were added to inhibit formins (20 µM SMIFH2) and Arp2/3 (100 µM CK666) activity. These concentrations have been shown previously to disrupt formin and Arp2/3 activities in MDCK cells without inducing cytotoxic or off-target effects (14, 43–45). Importantly, the effects of these inhibitor concentrations are reversible so that recovery of normal function can be monitored. Small molecule inhibitors were added, cells were imaged for 60 min, and then the inhibitor was washed out and imaging was continued for a further 60 min.
Fig. S6.
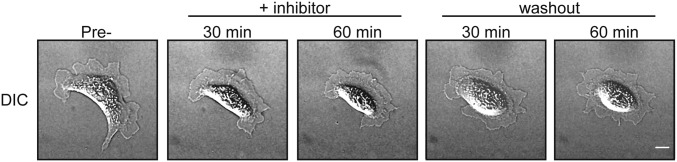
DIC time-lapse images of MDCK cell adhered to an Ecad-Fc PA gel. Image corresponds to the “60-kPa PA gel, NSC23766” fluorescent images shown in Fig. 3. DIC images were collected during all experiments, but were not used for analysis due to poor contrast caused by the PA gel. (Scale bar, 10 µm.)
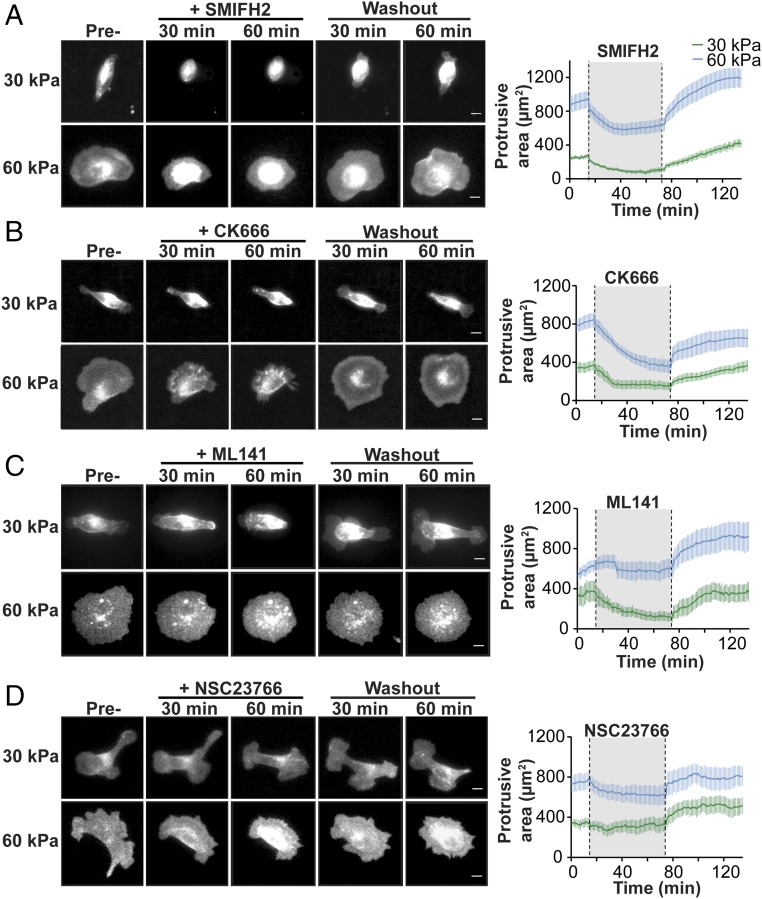
Fig. 4.
Cell adhesion to a 30-kPa and 60-kPa Ecad-Fc PA gel requires the activities of distinct signaling molecules. (A–D) Representative montage of MDCK cells stably expressing E-cadherin:dsRed adhered to a 30-kPa or 60-kPa Ecad-Fc PA gel. Each panel is representative of the effects on cell protrusive area following the addition and washout of different inhibitors: (A) pan-formin inhibitor, SMIFH2; (B) Arp2/3 inhibitor, CK666; (C) Cdc42 inhibitor, ML141; and (D) Rac inhibitor, NSC23766. (Scale bar, 10 µm.) (A–D, Right) Line graphs represent the mean protrusive area for each condition throughout the time course of the experiment. For each line graph, the mean protrusive area of cells on a 30-kPa Ecad-Fc PA gel and on a 60-kPa Ecad-Fc PA gel is indicated by the green and blue lines, respectively. The gray region between the dotted lines indicates the period in which the inhibitor was present. n ≥ 10 cells per condition from at least three independent experiments; error bars represent SEM.
Formins induce the polymerization of long, parallel actin filament bundles found in filopodia (46, 47) that are similar to the dynamic plasma membrane protrusions observed in cells adhered to a 30-kPa Ecad-Fc PA gel (Fig. 2A). Addition of the pan-formin inhibitor SMIFH2 disrupted formin localization (Fig. S5A) and caused cells on a 30-kPa Ecad-Fc PA gel to rapidly lose adhesion to the substrate and round up; after SMIFH2 washout, the cells regained an elongated shape and reextended a large, flat plasma membrane lamellipodium with filopodia-like protrusions at both ends of the cell (Fig. 4A and Movie S4). Addition of SMIFH2 to cells adhered to a 60-kPa Ecad-Fc PA gel resulted in a smaller decrease in spread area and a reduction in the size of protrusions extending from the cell body; however, cells remained well spread on this substrate compared with complete cell rounding on the 30-kPa PA gel in the presence of SMIFH2 (Fig. 4A and Movie S5). Washout of SMIFH2 resulted in cell respreading on the 60-kPa Ecad-Fc PA gel concomitant with the reappearance of large lamellipodia around the cell periphery (Fig. 4A).
Next, we tested the role of Arp2/3, which nucleates branched actin networks commonly found in lamellipodia (48, 49) that are similar to plasma membrane protrusions observed in cells adhered to a 60-kPa Ecad-Fc PA gel (Fig. 2A). The small molecule inhibitor CK666 disrupted Arp2/3 localization (Fig. S5B) and decreased plasma membrane protrusive area and activity in cells adhered to a 30-kPa Ecad-Fc PA gel, although cells did not lose adhesion to the same extent as they did in the presence of SMIFH2. Inhibitor washout resulted in the reextension of protrusions at the distal ends of the elongated cell body (Fig. 4B and Movie S6). Inhibition of Arp2/3 in cells adhered to a 60-kPa Ecad-Fc PA gel resulted in a decrease in the cell spread area and retraction of large lamellipodia; washout of CK666 resulted in cell respreading concomitant with the reappearance of large lamellipodia (Fig. 4B and Movie S7).
Overall, these results indicate that formins are required for cell adhesion, spreading, and membrane protrusive activity on a 30-kPa Ecad-Fc PA gel. Formins do not appear to be required for cell adhesion to a 60-kPa Ecad-Fc PA gel, but contribute to cell spreading and protrusive area. Arp2/3 activity does not appear to be required for cell adhesion to either a 30-kPa or a 60-kPa Ecad-Fc PA gel, but contributes to the cell spread area and formation of lamellipodia independent of substrate rigidity.
We next examined the role of Rho GTPases as downstream transducers of E-cadherin rigidity sensing and regulators of actin-based membrane protrusion dynamics. We focused on Cdc42 activity, which is largely associated with filopodia formation (and early stages of cell–cell adhesion) (7, 9, 10), and Rac1 activity, which is associated with lamellipodial activity (and intermediate stages of cell–cell adhesion) (15, 16, 50). We did not investigate the role of RhoA because its activity is important in late stages of cell–cell adhesion and compaction (15).
Addition of 20 µM ML141, a small molecule inhibitor of Cdc42 activity (51, 52), resulted in a 65% decrease in Cdc42 activity (Fig. S5C) and a significant decrease in cell protrusive area and spreading on a 30-kPa Ecad-Fc PA gel; washout of ML141 resulted in the reestablishment of an elongated cell shape and the reappearance of large, flat lamellipodia with filopodia-like protrusions at both ends of the cell (Fig. 4C and Movie S8). In contrast, Cdc42 inhibition appeared to have little or no effect on cells adhered to a 60-kPa Ecad-Fc PA gel (Fig. 4C and Movie S9). We next tested the role of Rac1 activity, using the small molecule inhibitor NSC23766 (100 µM), which was shown previously to inhibit Rac activity in MDCK cells (53, 54) and resulted in a 63% reduction of Rac activity (Fig. S5D). We observed little or no effect of NSC23766 on cell adhesion or protrusive area on a 30-kPa Ecad-Fc PA gel (Fig. 4D and Movie S10) and only a modest decrease in cell protrusive area on a 60-kPa Ecad-Fc PA gel (Fig. 4D and Movie S11). Taken together, we conclude that different Rho GTPases contribute to membrane dynamics and cell spreading/adhesion on Ecad-Fc gels of specific moduli: Cdc42, but not Rac1 activity is required for cell adhesion and spreading on a 30-kPa Ecad-Fc PA gel; Rac1, but not Cdc42 activity plays a modest role in cell spreading on a 60-kPa Ecad-Fc PA gel.
For comparison, we also tested whether formin, Arp2/3, Cdc42, or Rac1 activities were required for cell adhesion and spreading on 30-kPa or 60-kPa collagen-I (ECM) PA gels. Significantly, addition of SMIFH2, CK666, ML141, or NSC23766 reduced the protrusion area of cells on both 30-kPa and 60-kPa collagen-I PA gels to a similar extent (Fig. S7), indicating that all of these activities are required for integrin-based cell adhesions regardless of ECM rigidity.
Fig. S7.
Effects of inhibiting actin nucleating proteins and Rho GTPases in cells on collagen-I–coated 30-kPa and 60-kPa PA gels. Line graphs illustrate the effects on protrusive cell area by (A) SMIFH2, (B) CK666, (C) ML141, and (D) NSC23766. The solid line indicates the mean protrusive area for each condition and the error bars indicate SEM (n ≥ 10 cells per condition from at least three independent experiments).
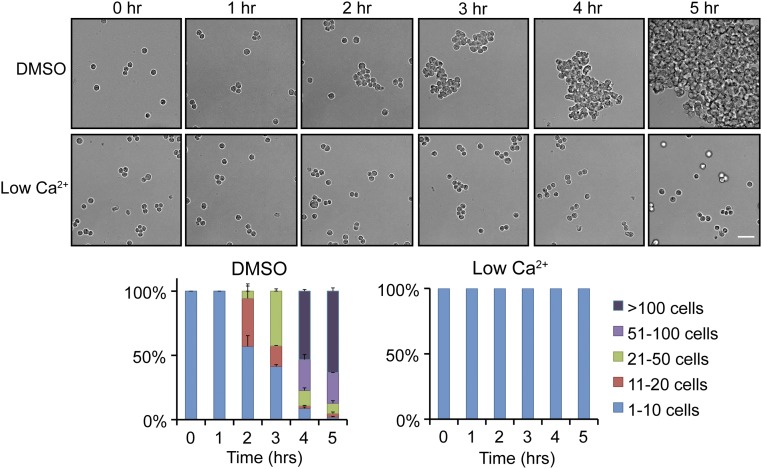
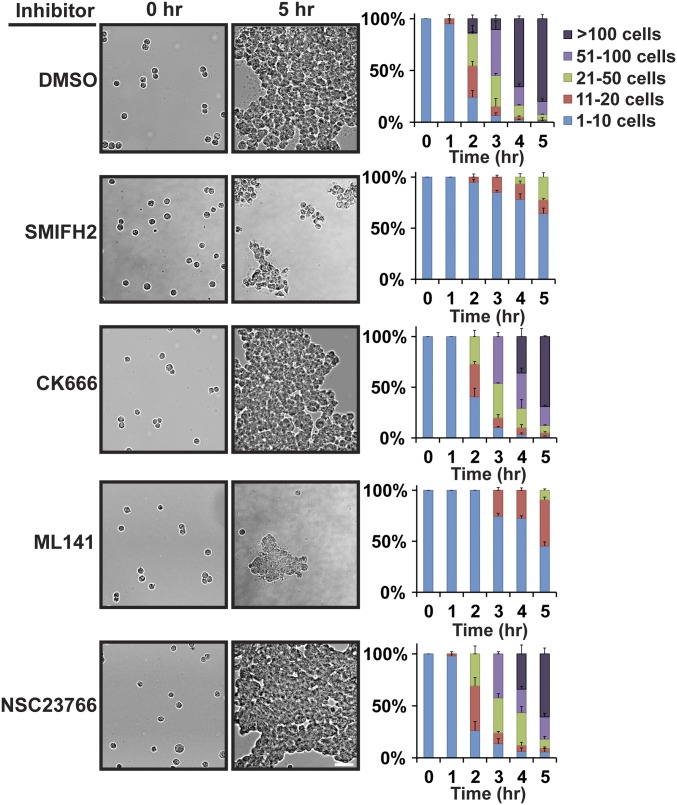
To substantiate our signal transduction results in a more biologically relevant system, we extended our reductionist approach of single-cell adhesion on Ecad-Fc PA gels to native cell–cell adhesion between MDCK cells in 2D and 3D. We first measured cadherin-based cell–cell adhesion, using a hanging-drop assay, in which MDCK cells form large, 3D aggregates in a Ca2+- (Fig. S8) and cadherin-dependent manner (55) in suspension in the absence of integrin-dependent ECM adhesion or cell migration (Fig. 5). Inhibitors of formin, Arp2/3, Cdc42, or Rac1 activity were added to a suspension of single cells, and cell aggregate formation was quantified for up to 5 h, when large 100+ cell aggregates had formed in the control (DMSO). Addition of CK666 or NSC23766 had little or no effect on the kinetics of cell aggregation compared with the control (Fig. 5). In contrast, ∼75% of cell aggregates in the presence of SMIFH2 and 90% of cell aggregates in the presence of ML141 comprised less than 20 cells after 5 h (Fig. 5). These results indicate that Cdc42 and formins, but not Rac1 or Arp2/3 activities are required for initial cell–cell adhesion.
Fig. S8.
Formation of large aggregates in a hanging-drop assay is calcium dependent. (Top) Bright-field images showing aggregation of MDCK cells at 0- to 5-h time points in “normal” (1.8 mM) and low Ca2+ (5 µM) media. (Scale bar, 50 µm.) (Bottom) Quantification of hanging-drop assays in normal and low Ca2+ media. Cells were binned into cluster classes: 1–10 cells, 11–20 cells, 21–50 cells, 51–100 cells, or >100 cells. The percentage of cells in each cluster size for each time point is indicated. The data shown are averages from three independent experiments and error bars represent SEM.
Fig. 5.
Formin or Cdc42 inhibition disrupts cell–cell adhesion in a 3D hanging-drop assay. (Left) Bright-field images showing aggregation of MDCK cells at 0-h and 5-h time points in the presence of the indicated inhibitors. (Scale bar, 50 µm.) (Right) Quantification of hanging-drop assays in the presence of the indicated inhibitors. Cells were binned into cluster classes: 1–10 cells, 11–20 cells, 21–50 cells, 51–100 cells, or >100 cells. The percentage of cells in each cluster size for each time point is indicated. The data shown are averaged from three independent experiments and errors bars represent SEM.
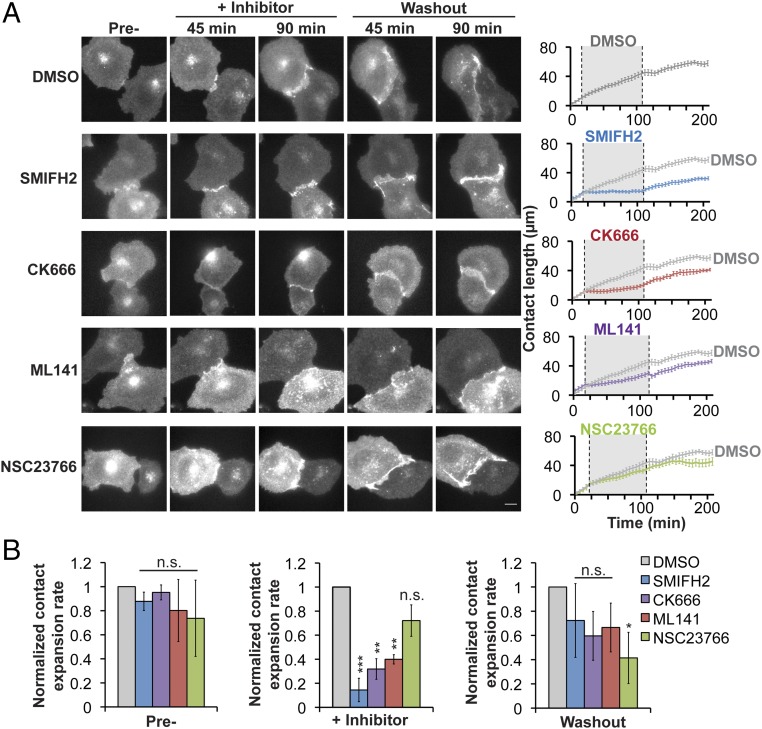
To examine the formation of individual cell–cell adhesions in detail, we used live cell imaging of initial adhesion between pairs of MDCK cells stably expressing E-cadherin:dsRed. MDCK cell–cell contact was initiated when filopodia from opposing cells came into contact with one another (Fig. 6, DMSO) (7). The contact between cells then spread laterally by lamellipodial protrusions that increased membrane interactions between neighboring cells along the expanding cell–cell contact until compaction (Fig. 6, DMSO) (15). As cells initiated contacts, SMIFH2, CK666, ML141, or NSC23766 was added to the media and the expansion of the cell–cell contact was imaged for 90 min, and then the inhibitor was washed out and imaging continued for an additional 90 min (Fig. 6A, images). The rate of cell–cell contact expansion 15 min before, during, and after inhibitor addition was measured from time-lapse movies and plotted; note that the rate of contact expansion in the control (DMSO) is replotted for comparison with each inhibitor (Fig. 6B, bar graphs).
Fig. 6.
Disruption of cell–cell contact expansion between pairs of MDCK cells, using small molecule inhibitors. (A, Left) Images of pairs of MDCK cells stably expressing E-cadherin:dsRed during the formation and expansion of cell–cell contacts. (Scale bar, 10 µm.) (A, Right) Quantification of cell–cell contact length over time. The solid line is representative of the mean contact length for each condition; the DMSO control line is copied in each of the graphs of the inhibitors as a reference. The gray region indicates the period during which the inhibitor was present (n ≥ 10 contacts per condition from at least three independent experiments; error bars indicate SEM). (B) Bar graphs for the expansion rate for cell–cell adhesion for each condition during pretreatment, addition of the inhibitor, and washout period, normalized to the rate in the DMSO control for each time point (value of 1). Statistics were performed using a Student’s t test. *P < 0.05, **P < 0.001, ***P < 0.0001.
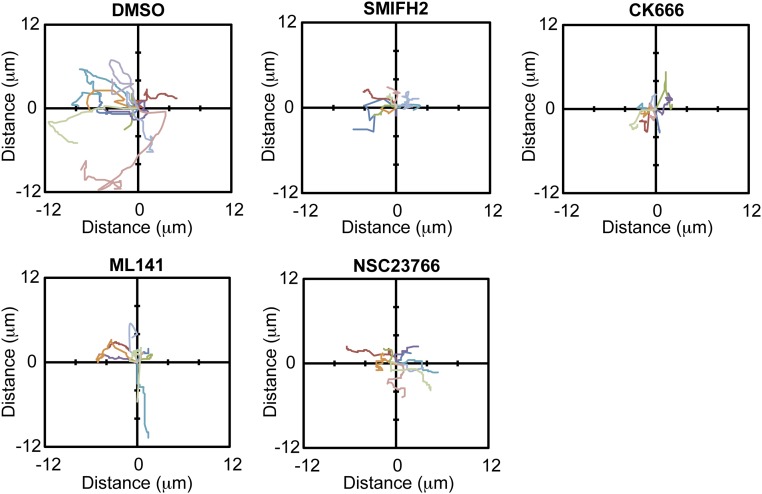
The contact expansion rate before inhibitor addition varied slightly across conditions, depending on the time at which initial contact between pairs of cells was established during the 15-min interval before inhibitor addition (Fig. 6B, “Pre-”). However, once contact was established, the rate of contact expansion was the same in all conditions. In the presence of the formin inhibitor SMIFH2, existing cell–cell adhesions that had formed before the addition of the inhibitor remained, but did not grow in length; however, when the inhibitor was washed out, contact expansion resumed at a rate similar to the control (DMSO) (Fig. 6 A and B). Treatment with CK666 or ML141 also reduced the rate of initial contact expansion, which was reversed upon inhibitor washout (Fig. 6 A and B). Addition of NSC23766 had little or no effect on any stage in early cell–cell adhesion (Fig. 6 A and B). We also measured the center of mass motion during cell–cell adhesion (Fig. S9). Although we detected a decrease in the center of mass motion in the presence of any of the inhibitors, the effect was similar for all inhibitors, suggesting that changes in cell contact expansion rate were not directly due a migration defect but likely involved other activities such as changes in membrane protrusion dynamics.
Fig. S9.
Center of mass motion during contact formation. Center of mass motion of MDCK cells forming contacts was tracked using FIJI and plotted in a smoothed-line scatter plot to display the trajectory for each condition. Each colored line represents a different cell. At least nine cells were tracked per condition.
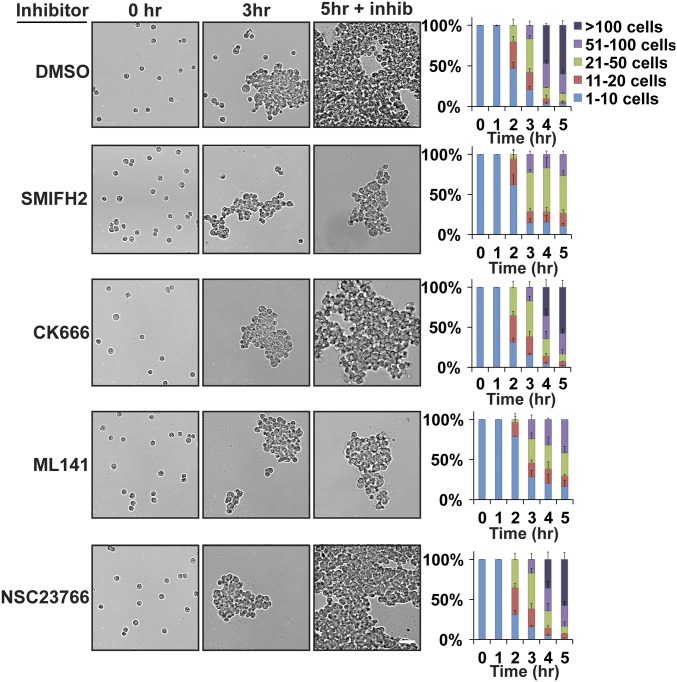
Cdc42 and formins appeared to be required for the initial, but not later stages of cell–cell adhesion, as indicated by live-cell imaging (Fig. 6) and cell aggregation in 3D (Fig. 5). If this was the case, we predicted that addition of Cdc42 or formin inhibitors to the 3D cell adhesion assay when intermediate-sized cell aggregates had formed would inhibit further cell aggregation, but not affect existing (mature) adhesions. Cells were allowed to form intermediate-sized aggregates for 3 h in 3D, at which time actin and Rho GTPase inhibitors were added for 2 h (Fig. 7). Cells treated with CK666 or NSC23766 continued to form large aggregates at rates similar to the DMSO control. In contrast, addition of SMIFH2 or ML141 blocked further enlargement of the aggregates beyond the size achieved at 3 h. These results indicate that initiation of new cell–cell interactions requires Cdc42 and formin activities, whereas the maintenance of mature adhesions does not.
Fig. 7.
Formin or Cdc42 inhibition disrupts the formation of new adhesions, but not the maintenance of existing adhesions. (Left) Bright-field images showing MDCK cell aggregation at times before (0 h, 3 h) and after (5 h) addition of inhibitors at 3 h. (Scale bar, 50 µm.) (Right) Quantification of hanging-drop assay in the presence of the indicated inhibitors. Cells were binned into cluster classes: 1–10 cells, 11–20 cells, 21–50 cells, 51–100 cells, or >100 cells. The percentage of cells in each cluster size for each time point is indicated. The data shown are averaged from three independent experiments and errors bars represent SEM.
Discussion
The effects of ECM rigidity on integrin-based cell adhesions have been studied extensively (20, 56, 57), but comparable studies on cadherin-based cell–cell adhesion are lacking. This is due in part to the difficulty in gaining direct access to cadherin–cadherin interactions to measure their stiffness and contribution to the rigidity sensed by adhering cells. Previous work showed that cells adhering to a more rigid (95-kPa) N-cadherin–coated PA gel were flat and well spread with large N-cadherin adhesion plaques (21), similar to cells adhered to Ecad-Fc–coated glass coverslips (∼100 GPa) (Fig. 1E) (34–37). In contrast, cells adhered to a softer (10-kPa) N-cadherin–coated PA gel had a reduced spread area and smaller N-cadherin adhesions (21), similar to small E-cadherin clusters in cells adhering to 2D-supported membranes functionalized with the E-cadherin extracellular domain (58), 30-kPa and 60-kPa Ecad-Fc PA gels (Figs. 1E and 3A), and native cell–cell junctions (59). We could not measure adhesion to 9-kPa PA gels functionalized with Ecad-Fc as MDCK cells attached poorly and did not spread (Fig. S1 and Table S1). This difference might be due to differences in cell type, cell adhesion to Ncad-Fc and Ecad-Fc, the PA gel formulations used, or the type of mechanical characterization used to derive the elastic modulus of the substrates (micro- vs. macroscale indentation). Nevertheless, these studies indicate that cell–cell adhesion is sensitive to the stiffness and/or mobility of cadherin-based homotypic interactions. However, the mechanisms underlying stiffness-dependent differences in cadherin adhesions and the relationship, if any, between those differences and native cell–cell adhesions were unknown. Our experimental design sought to address these knowledge gaps.
Our results showed that cell behavior and morphology appeared to be very sensitive to relatively small changes in Ecad-Fc substrate rigidity, whereas ECM rigidity sensing within the same range of elastic moduli did not result in significant differences in cell behavior or morphology. Furthermore, inhibition of actin nucleating proteins or Rho GTPases affected adhesion similarly on all ECM substrates regardless of substrate rigidity, whereas adhesion to Ecad-Fc gels of different moduli required the activity of different actin nucleating proteins and Rho GTPases.
Adhesion to Ecad-Fc substrates of different moduli, independent of ligand density, had a profound effect on cell morphology, membrane dynamics, and actin organization. These differences did not appear to be due to different cellular E-cadherin organization imaged by superresolution microscopy, which revealed that E-cadherin was organized into small puncta of similar size and density on 30-kPa and 60-kPa Ecad-Fc PA gels, independent of Ecad-Fc ligand density. E-cadherin puncta had an average area of 0.014 µm2, with an estimated diameter of 120–130 nm. This estimation approaches the spatial resolution limit of our microscope and, therefore, may overestimate the diameter of E-cadherin clusters. The size of these puncta is similar to that of E-cadherin clusters at cell–cell contacts in A431D cells (112 nm), but larger than that of E-cadherin clusters in EphA4 cells (60 nm) (60). It is possible that E-cadherin cluster size differs in distinct cell types and at different stages of cell–cell adhesion (61), but additional studies at high resolution are required to track E-cadherin puncta size and organization over time.
TFM studies revealed nearly identical mean traction stress magnitude within cell boundaries on 30-kPa and 60-kPa Ecad-Fc PA gels (Fig. 3E). The number and area of these high-stress regions differed in cells on 30-kPa or 60-kPa Ecad-Fc PA gels, but the mean traction stress within these regions was similar. Whereas it is not possible to perform SIM and TFM experiments simultaneously with our current experimental setup, we were able to extrapolate meaningful, comparative data from both. We calculated the average force per E-cadherin punctum within the high-stress regions based on the area and mean stress magnitude within high-stress regions from our TFM studies and the density of E-cadherin puncta from SIM. We estimated the same mean force of ∼2 pN per E-cadherin punctum in cells on both the 30-kPa and 60-kPa PA gels (comparing the median value of our calculations for both substrates). This estimate assumes that the E-cadherin density in regions of high traction stress is the same as those measured by SIM, although we do not know the number of “active” E-cadherin molecules bearing force within each punctum. Nevertheless, this result is consistent with previous work showing that E-cadherin molecular tension remains constant between pairs of cells, regardless of differences in cell shape, traction stress, or cell–cell force (62).
We showed that differences in adhesion and cell spreading on Ecad-Fc PA gels required a delicate balance of distinct Rho GTPases (Table S3). Cdc42 activity is required for cell adhesion and spreading on a 30-kPa Ecad-Fc PA gel, but not on a more rigid 60-kPa Ecad-Fc PA gel (Fig. 4C). Moreover, Cdc42 activity was required for the initiation of new cell–cell adhesions in 3D aggregates (Figs. 5 and 7) and contact expansion of initial contacts between pairs of cells (Fig. 6), but not for the maintenance of mature cell–cell adhesions in either assay (Figs. 6 and 7). Previous studies of the role of E-cadherin engagement and Cdc42 activity have generated conflicting results (9, 10, 63, 64) but, based on our results, it is possible that these differences could be due to the mechanical properties of cell–cell adhesions in different cell types.
Table S3.
Summary of inhibitor effects in each assay
| Protrusive area | ||||
| Inhibitor | 30 kPa | 60 kPa | Hanging drop | Contact expansion |
| SMIFH2 | +++ | + | +++ | +++ |
| CK666 | + | ++ | — | ++ |
| ML141 | ++ | — | +++ | ++ |
| NSC23766 | — | — | — | — |
The degree of the effect of each inhibitor for each assay is indicated using a +/− system, where “+” indicates an effect and “−” indicates no effect. Degree of effect is indicated by the numbers of +, where “+++” is the greatest effect.
Rac1 inhibition had little or no effect on cells adhered to a 30-kPa Ecad-Fc PA gel and only a modest effect on cells adhered to a 60-kPa Ecad-Fc PA gel (Fig. 4D). This was surprising, because studies have indicated that Rac1 is essential for cell–cell adhesion (10, 13, 15, 36). However, Rac1 may play a role in maintaining cadherin adhesions at stiffer elastic moduli that may be representative of cell–cell adhesions that are more mature than those studied here.
Our results also revealed that the role of different actin nucleating protein activities is influenced by the rigidity of the Ecad-Fc substrate. In general, inhibition of formin or Arp2/3 activities caused more cell rounding (Fig. 4) than inhibition of actomyosin activity (Fig. S3), indicating that maintenance of cell adhesion requires active actin and membrane dynamics and that myosin-dependent forces on stable actin structures may play a secondary role on these PA substrates. In particular, formin activity, which polymerizes long, parallel actin bundles found in filopodia (46, 47), appeared to be essential for cell adhesion and dynamic filopodia-like protrusions on a 30-kPa Ecad-Fc PA gel, but not on a 60-kPa Ecad-Fc PA gel. These results are consistent with previous studies that demonstrated a role for the diaphanous formin mDia1 (11, 65, 66) and formin-like proteins FMNL2 and FMNL3 (12, 13) in the recruitment of actin and E-cadherin and the formation of filopodia at initial cell–cell contacts. At present, we do not know which formin(s) is (are) regulated by the stiffness of E-cadherin interactions. However, formin1 binds directly to the E-cadherin complex through α-catenin (67), and it is possible that forces at E-cadherin adhesions activate α-catenin binding to formins via force-dependent changes in α-catenin conformation (68), perhaps similar to that required for binding to F-actin (5). In this context, FMNL3 recruitment to junctions is tension dependent and regulated by Cdc42 (14). Our results indicate that the requirement for Cdc42 and formin activity becomes less important as substrate modulus increases, which could be related to the roles of mDia1 and FMNL3 in stabilizing early junctions and decreasing E-cadherin mobility (14).
Inhibition of Arp2/3 affected cell adhesion on 30-kPa and 60-kPa Ecad-Fc PA gels (Fig. 4B). In both cases, cells adhered to the substrate with lamellipodia, albeit with very different organizations—a large, flat membrane lamellipodium at either end of the cell body adhered to the 30-kPa PA gel, and large circular lamellipodia in flat, well-spread cells adhered to the 60-kPa Ecad-Fc PA gel. Arp2/3 inhibition in cells adhered to the 30-kPa Ecad-Fc PA gel could affect formin-dependent actin polymerization during filopodia initiation (69, 70). Similarly, Arp2/3 inhibition disrupted native cell–cell contact formation between pairs of cells, which also required Cdc42 and formin activities (Fig. 6). However, Arp2/3 inhibition did not affect formation of 3D cell aggregates in the hanging-drop assay (Figs. 5 and 7). These apparently contradictory results could be due to differences in E-cadherin adhesion in 2D vs. 3D. In 3D suspension, cells are rounded and aggregate by minimally spreading on neighboring cells, which would not necessarily require extensive Arp2/3-dependent lamellipodia activity (Figs. 5 and 7). In contrast, cells in 2D are flat and spread out and migrate with protrusive lamellipodia to make contact with each other, where upon expansion of the cell–cell contact area by lamellipodia is required, which would require Arp2/3 activity (Fig. 6). In line with this idea, inhibition of Arp2/3 (as well as formins, Cdc42, and Rac) resulted in a reduction in center of mass motion during cell–cell contact expansion, suggesting decreased cell migration.
We do not know the stiffness of E-cadherin interactions during native cell–cell adhesion. However, insights can be extrapolated from our TFM results and a comparison of cell behaviors and the requirements of different Rho GTPases and actin nucleators in our reductionist assay using Ecad-Fc PA gels and our 2D and 3D native cell–cell adhesion assays. Our TFM data indicate that cells may sense and react to differences in effective E-cadherin stiffness by changing the distribution of traction stress and strain on the substrate. Cells adhered to a 30-kPa Ecad-Fc PA gel exerted a higher strain on the gel than cells adhered to a 60-kPa Ecad-Fc gel (Fig. 3F), and high-traction-stress regions were often located at the distal edges of elongated cells on 30-kPa Ecad-Fc PA gels where dynamic filopodial protrusions originate (Fig. 3D). Previous work has shown that fibroblasts extend filopodia as a rigidity-sensing mechanism and levels of strain on the resulting substrate influence cellular behavior (71). Our data indicate that a similar mechanism may exist during cell–cell contact formation. We showed that Cdc42 and formin activities are the dominant signaling molecules that regulate cell adhesion on a 30-kPa Ecad-Fc PA gel during initial cell–cell adhesion in 2D and 3D. Therefore, it is possible that Cdc42- and formin-dependent actin polymerization supports high levels of strain exerted by opposing cells during early stages of cell–cell adhesion (8, 67). During intermediate stages of cell–cell contact, when the area of the contact increases and E-cadherin is stabilized at the junction, strain and stress at the contact would redistribute as the contact expands (similar to the changes in stress distribution observed in 30-kPa and 60-kPa Ecad-Fc PA gels, Fig. 3D). Our data indicate that this transition would result in a change in actin-based protrusions that are less dependent on formin and Cdc42 activities and more dependent on Arp2/3 actin polymerization and lamellipodia as the contact expands (15). Taken together, we suggest that different stages of cell–cell adhesion may involve a transition in E-cadherin rigidity sensing at the contact that results in reorganization of stress between opposing cells and corresponding changes in the downstream signaling pathways involved in actin reorganization and plasma membrane dynamics. Direct evidence of this transition will need the development of methods to assay cell–cell adhesion stiffness directly.
Materials and Methods
Full materials and methods are available in SI Materials and Methods. Briefly, PA gels of varying elastic moduli were made by adjusting the amounts of acrylamide and bis-acrylamide in the prepolymer gel solution (33). Gels were functionalized by UV activation of 0.5–2.0 mg/mL SS (72) and subsequent incubation with collagen I or protein A/G and Ecad-Fc. MDCK type II G cells (73) and MDCK type II G cells stably expressing E-cadherin:dsRed (74) were used in this study. MDCK cells expressing GFP-LifeAct were imaged 24 h after transfection. All inhibitors were diluted in DMSO and used at concentrations previously shown to inhibit activities without inducing cytotoxic or off-target effects in MDCK cells: 20 µM SMIFH2 (14), 100 µM CK666 (45), 20 µM ML-141 (52), and 100 µM NSC-23766 (54). Three-dimensional hanging-drop assays used single-cell suspensions in droplets hanging from an inverted 35-mm Petri dish lid. Samples were triturated before fixation and 20 random images were collected for each time point. Image analysis was performed using ImageJ and statistical analyses were performed using a Student’s t test, Mann–Whitney test, or Kruskal–Wallis test in GraphPad Prism Software.
SI Materials and Methods
Cell Culture, Transfections, and Inhibitors.
MDCK G type II cells (73) and MDCK G type II cells stably expressing E-cadherin:dsRed (74) were grown in DMEM with 1 g/L sodium bicarbonate, 10% FBS (Atlas Biologicals), penicillin, streptomycin, and kanamycin. Cells were transiently transfected with GFP-LifeAct using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and imaged 24 h after transfection. The following inhibitors were used in this study: SMIFH2 (20 µM; EMD Millipore), CK666 (100 µM; EMD Millipore), ML141 (20 µM; Tocris Bioscience), and NSC23766 (100 µM; Tocris Bioscience). All inhibitors were diluted in DMSO, and DMSO alone was used as a control.
Generation and Preparation of Ecad-Fc.
Ecad-Fc was purified from culture media of HEK 293 cells expressing canine Ecad-Fc as previously described (26). Briefly, HEK293 cells were grown in DMEM with 10% FBS and 1% penicillin/streptomycin at 37 °C. Two days priors to media collection, cells were switched to serum-free DMEM to remove serum IgG. Media were collected and filtered, and Ecad-Fc was purified over a protein-A column (Thermo-Pierce). The purity of the Ecad-Fc was confirmed by SDS/PAGE, and functionality was confirmed by calcium-dependent aggregation of Ecad-Fc–coated beads.
Preparation and Functionalization of PA Gels.
PA gels were prepared on coverslips or MatTek dishes treated with 3-(trimethoxysilyl)propyl methacrylate (bind silane; Sigma). Briefly, glass coverslips were incubated with a 0.3% bind silane solution for 5 min. The silane solution was removed, and the glass coverslips were washed with 95% ethanol three times and air dried. PA gel solutions were prepared using 0.5 g/mL acrylamide and 0.025 g/mL bis-acrylamide solutions. The modulus of the PA was tuned by varying the amount of bis-acrylamide cross-linker and total acrylamide monomers (33). For this study, we used four gel formulations: 3.1%T, 3.22%C; 5.1%T, 2.91%C; 10%T, 1%C; and 10%T, 2.5%C, where the total polymer content and cross-linker concentration are calculated as follows:
| [S1] |
| [S2] |
The gel precursor solution (acrylamide, bis-acrylamide, and water) was degassed in a vacuum desiccator for 1 h before gelation. To initiate gelation, 5 μL of 10% (wt/vol) ammonium persulfate (APS; Sigma-Aldrich) was added to ∼995 μL of gel precursor solution followed by 0.5 μL of N,N,N′,N′-Tetramethylethylenediamine accelerator (TEMED; Sigma-Aldrich). The solutions were mixed by gentle pipetting before dispersing on the activated coverslips. A total of 20 μL of gel precursor solution was pipetted onto the activated coverslip and a second coverslip was sandwiched on top of the droplet to disperse the polymerizing gel over the activated coverslip. Gels were left to polymerize at room temperature for 30 min. Afterward, the gels were flooded with 1× PBS, pH 7.4, and the top coverslip was gently lifted from the gel surface, using forceps. Gels were ∼70–80 µm in thickness, as measured by confocal microscopy.
PA gels were functionalized by incubating with 2 mg/mL SS (ThermoFisher) (72). For some experiments (Fig. S4), lower concentrations of SS (0.5 mg/mL, 1.0 mg/mL, or 1.4 mg/mL) were used to adjust the level of Ecad-Fc binding on the surface of a 60-kPa PA gel. The gel was exposed to UV light to photoactivate the cross-linker and then quickly washed with PBS. For Ecad-Fc functionalization, the gels were incubated with 300 µg/mL protein A/G overnight at 4 °C, followed by a 1-h incubation with 150 µg/mL Ecad-Fc (26), and, after washing three times with PBS, the gel was blocked with 1% BSA for 30 min. For collagen-I functionalization, 150 µg/mL collagen I (BD Biosciences) was added to the gel after SS treatment. For Ecad-Fc blocking antibody experiments, gels were functionalized as described above. Following Ecad-Fc incubation, gels were incubated with 300 µg/mL Fc and blocked with an anti–E-cadherin function blocking antibody (rr-1, 1:50) for 1 h at room temperature before seeding cells.
Microscale Mechanical Characterization of Gel Stiffness.
To characterize the Young’s modulus of the PA gels used in this study, AFM indentation experiments were performed to generate force–distance curves, which were then analyzed using the Hertz elastic contact model (75) as described by Denisin and Pruitt (76). AFM indentation experiments were performed within 24 h of polymerizing the hydrogels on a WITec AFM (alpha300) with gold-coated cantilevers (NanoWorld AG: Nanosensors, PPP-NCSTAuD-10). Cantilever stiffness was measured using thermomechanical noise of 8.48 N/m. After characterization, one 50-μm diameter bead (Duke Scientific; 9050) was attached to the tip of each probe, using UV epoxy glue (Loctite 352). Samples were incubated in 1× PBS within the AFM for at least 1.5 h to stabilize the system before obtaining measurements. Optical lever sensitivity measurements were performed before analyzing each gel sample by indenting the glass surface using the following parameters: feedback control with 0.5 V set point, 1% p-gain and 0.2% i-gain, and force–distance using 0.2 μm pull and 0.6 μm push at 0.2 μm/s speed. The gel surface was then approached with the following feedback threshold parameters: 0.5 V set point, 2% p-gain, and 0.2% i-gain. Force–distance curve parameters were optimized to ensure that both zero load and ample indentation data were recorded. We performed at least 5 indentations in 5 different areas (at least 25 indentations per sample) for both gel formulations, using the following force–distance parameters: 9 µm pull and 4 µm push at 3 μm/s speed.
Immunofluorescence, Antibodies, and Image Analysis.
Cells were washed with PBS, fixed in 4% paraformaldehyde for 20 min, and permeabilized with 0.2% Triton X-100 in PBS for 2 min. Cells were blocked in 10% goat serum, incubated with primary antibodies for 1 h at room temperature, washed with PBS, incubated with fluorescently labeled secondary antibodies for 1 h at room temperature, and mounted on a coverslip with Immu-Mount (ThermoFisher). For phalloidin staining, Alexa-488 phalloidin (Invitrogen) was added to the secondary antibody solution and incubated with the cells for 1 h during secondary antibody incubation. The following primary antibodies were used: Arp2 (anti-p34-Arc/ARPC2; Millipore), formin1 (anti-FMN1; Novus Biologicals), and paxillin (anti-paxillin; BD Biosciences). All primary antibodies were used at a 1:100 dilution, and fluorescently labeled secondary antibodies were used at a 1:200 dilution. For confocal imaging and structured illumination microscopy experiments, PA gels on coverslips were mounted on a larger rectangular coverslip (24 × 50 mm, 1.5 mm thickness), using Immu-Mount mounting media (Thermo Scientific). Samples were inverted and imaged through the rectangular coverslip so that cells could be imaged from the top down, without having to image through the PA gel. Images were analyzed using ImageJ software.
Superresolution SIM.
The samples were fixed, stained, and mounted as described in Immunofluorescence, Antibodies, and Image Analysis. SIM microscopy was performed using a DeltaVision OMX V4 BLAZE system (Applied Precision). The microscope was equipped with a 100×/1.42 N.A. U-PLANAPO SIM oil immersion objective (Olympus); 405-nm, 488-nm, 568-nm, and 642-nm lasers; and three EMCCD cameras. Samples were imaged using objective oil with a 1.512 refractive index. An electro-optical high-speed SI diffraction grating engine was used to generate SI patterns. Image stacks were composed of 15 images per plane (five phases and three angles). Serial sections spaced at a z distance of 0.125 μm were collected, spanning a total distance of 0.8–1.2 μm, depending on sample thickness. The resulting images were computationally reconstructed and color channels were aligned using the SoftWoRx 3.7 imaging software package (Applied Precision). Images shown are maximum projections of stacks. Quantification of E-cadherin puncta was performed in ImageJ, using thresholded images and the “Analyze particles” function.
TFM.
MDCKs stably expressing E-cadherin:dsRed were plated on 30-kPa and 60-kPa Ecad-Fc PA gels 5 h before imaging in DMEM (without phenol red). Images of the fluorescent beads (fiducial markers) within the gel were acquired in the FITC channel while the cells were contracting the gel. The E-cadherin:dsRed channel was also imaged to determine the cell outline. Imaging was performed using a wide-field epifluorescence inverted microscope (Zeiss Observer Z1; Intelligent Imaging Innovations) fitted with a stage incubator (5% CO2 in air, 37 °C). Imaging was performed using a 63× (0.75 N.A.) oil objective, filter sets for dsRed (cell outline) and FITC (beads), an X-Cite 120 LED illumination source, and a Flash4 LT CMOS camera (Hammamatsu). We then applied 100 μL of concentrated trypsin–EDTA to detach cells and imaged the same stage positions to obtain a reference relaxed position of the fluorescent beads. Experiments were performed on three different occasions and imaged two samples per gel condition on each day.
Data processing was performed as previously described by Chang et al. (77), using the ImageJ plugins developed by Tseng et al. (42). First, lateral drift was corrected between contracted and relaxed cell frames, using the template matching and slice alignment plugin, and then the particle image velocimetry (PIV) plugin was applied to quantify bead displacements, using a cross-correlation algorithm with iterative window size of 64 pixels and 32 pixels to yield displacement vectors 16 pixels apart with a final resolution of 1.65 μm. A normalized median test (noise level of 0.2 and threshold of 5.0) was applied twice to filter and replace erroneous displacement vectors with the median value from nearby vectors (78). The Fourier transform traction cytometry (FTTC) plugin was used to convert displacements into traction forces. The parameters used in the FTTC code included a pixel size of 0.103 μm, Poisson’s ratio of 0.5, and elastic modulus of the gel used (29.07 kPa or 57.34 kPa).
The appropriate regularization factor λ needed to perform the Fourier transform in the FTTC code and smooth the final traction force reconstruction was determined using the methods described by Chang et al. (77). The residual norm was analyzed as a function of λ rather than using the L-curve corner to determine λ because using the L-curve criterion yielded traction stress values that were unrealistically high, as also found by Schwarz et al. (79). The methods by Schwarz et al. (79) were followed to apply the discrepancy principle to determine λ, setting the optimum λ by approximating the residual norm in absolute values using the χ2 distribution, based on the degrees of freedom of the inverse problem and SD of measurement errors. Similar to the fibroblast cell samples examined by Chang et al. (76), each of our samples also had a different distribution of cell force vectors, cell area, and total area. Thus, we calculated the “optimal” λ by discrepancy theorem for each of our samples, found the average optimal λ, and applied this to the processing of all of the samples: 1.3E-9 for ∼30-kPa gels and 6.6E-10 for ∼60-kPa gels.
The E-cadherin:dsRed signal was used to determine the cell outline by first thresholding the image, binarizing, determining the largest “particle” in the image (the cell), importing that selection as a region of interest, and then extending the area around the cell by dilating to account for cell tractions at the cell periphery [methods similar to those in Chang et al. (76)]. We performed a single-point force calculation by the Boussinesq solution to Green’s function, d = F(1 + ν)/uEπ, where d is the distance in micrometers, F is the force at the cell boundary approximated in our case to be 100 nN, u is the resolution of our measurements taken to be 1 μm, and E is the elastic modulus of the substrate. Following this calculation, the cell area was extended by 16 pixels (1.64 μm) for ∼30-kPa gels and by 8 pixels (0.83 μm) for ∼60-kPa gels.
Mean stress within the cell boundary was “background corrected” for each sample. We determined “background noise” for each sample by measuring the stress magnitude in a 400 × 400-pixel area in the corner of each image (far from the cell). We then subtracted the background mean stress magnitude from the mean stress magnitude underneath the cell to obtain background-corrected values. The mean traction force magnitude within a cell boundary was calculated by multiplying the background-corrected mean stress by the area of the cell boundary.
Live Cell Imaging and Confocal Microscopy on PA Gels.
Live-cell imaging was performed at 37 °C in DMEM (without phenol red) with 1 g/L sodium bicarbonate, 25 mM Hepes, 10% FBS, penicillin, streptomycin, and kanamycin. Images were acquired using an inverted wide-field epifluorescence microscope (Zeiss Observer Z1; Intelligent Imaging Innovations) fitted with a stage incubator (5% CO2 in air, 37 °C). Imaging was performed using a 63× (0.75 N.A.) or 100× (1.4 N.A.) objective, a filter set for dsRed, an X-Cite 120 Led Illumination source, and a Flash4 LT CMOS camera (Hammamatsu) or an ImageEM X2 EM-CCD camera (Hammamatsu). Images were processed using ImageJ software. Images were background subtracted and contrast was adjusted for presentation in figures only (Figs. 1 A–E, 3A, 4 A–D, and 6A). GFP-LifeAct cells and fixed samples were imaged on a Leica TCS SP8 confocal microscope (Leica Application Suite; Leica Microsystems) fitted with a stage incubator (37 °C) used for live imaging of cells expressing GFP-LifeAct. Imaging was performed using a 63× oil objective (1.4 N.A.) with 1.5× zoom magnification and a white-light laser. Excitation and emission paths were set to correspond to fluorophores of interest (dsRed, Alexa-488, EGFP, or Alexa-647).
Contact Expansion Studies.
MatTek dishes were coated with 150 µg/mL collagen I, and 180,000 MDCK cells expressing E-cadherin:dsRed were plated. Thirty minutes after plating, pairs of cells forming cell–cell contacts were imaged (1 frame per minute) for 15 min (described above), at which time the media in the MatTek dish were replaced with fresh media containing DMSO or the indicated inhibitor. After 90 min, the media containing the inhibitor were removed and the cells were washed with 5 vol of fresh media and imaged for an additional 90 min. Center of mass motion was tracked in FIJI, using the manual tracking plugin.
Hanging-Drop Assay.
MDCK cells were plated at low density, and cells were trypsinized, centrifuged, and resuspended at a density of 2.5 × 105 cells/mL in media containing 1.8 mM calcium. For “low calcium” experiments, cells were resuspended in media containing 5 µM calcium. Twenty-microliter drops of the cell suspension were placed on 35-mm culture dish lids, which were inverted on top of the dish containing medium and incubated for different times at 37 °C. At each time point, drops were triturated 15 times through a 20-µL pipette tip, and 4 µL of 16% PFA was added. The entire sample was mounted on a slide and observed at 40× magnification. Twenty random images were taken throughout the slide and scored by cell cluster size. For some experiments (Fig. 5), the indicated inhibitors were added at the time of plating the 20-µL droplets on the lid and cells were given a second dose of the inhibitor at the 2.5-h time point. When testing the maintenance of cell clusters (Fig. 7), cells were plated in droplets in the absence of inhibitors and were dosed with the indicated inhibitors at the 3-h time point.
Measuring Ecad-Fc Binding on the Surface of Gels.
SYPRO Ruby red protein gel stain.
Ecad-Fc binding on the surface of functionalized PA gels was quantified using SYPRO Ruby protein gel stain (Molecular Probes). Gels were fixed for 60 min on an orbital shaker in a solution containing 50% methanol and 7% acetic acid. Gels were stained in SYPRO Ruby gel stain overnight on an orbital shaker. Following staining, gels were washed in a solution containing 10% methanol and 7% acetic acid for 30 min, followed by two 5-min washes in ultrapure water. Gels were imaged with a Leica TCS SP8 confocal microscope (Leica Application Suite; Leica Microsystems). Fluorescence intensity at the surface of the gel was quantified in ImageJ.
Anti–E-cadherin antibody staining.
Ecad-Fc binding on the surface of functionalized PA gels was quantified using the anti–E-cadherin antibody rr-1 (Developmental Studies Hybridoma Bank, University of Iowa) (38). Gels were functionalized as described in Preparation and Functionalization of PA Gels. Following Ecad-Fc incubation, gels were blocked with 1% goat serum and purified human Fc and were stained with an anti–E-cadherin antibody (rr-1, 1:100) and fluorescent secondary antibody (1:200). Images were acquired using a Leica SP8 scanning confocal microscope (Leica Application Suite; Leica Microsystems). Fluorescence intensity at the surface of the gel was quantified in ImageJ.
Rac/Cdc42 Pulldowns and Western Blotting.
Rac and Cdc42 pulldowns were performed using a pulldown activation assay biochem kit (Cytoskeleton, Inc.) according to the manufacturer’s protocol. Briefly, cells were washed with ice-cold PBS and lysed in 50 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 M NaCl, and 2% IGEPAL. Samples were centrifuged at 4 °C for 1 min, and supernatants were incubated with 20 µg GST-PBD glutathione affinity beads for 1 h at 4 °C. Bead pellets were washed in 25 mM Tris, pH 7.5, 30 mM MgCl2, and 40 mM NaCl; resuspended in 2× Laemmli sample buffer; and subjected to SDS/PAGE. Samples were blotted for Cdc42 (Cytoskeleton, Inc.) or Rac1 (Cytoskeleton, Inc.), and Western blots were scanned with an Odyssey imager and software (LI-COR Biosciences).
For Western blot of E-cadherin expression, WT MDCKs and MDCKs stably expressing E-cadherin:dsRed were lysed in 2× Laemmli sample buffer and subjected to SDS/PAGE. Samples were blotted for E-cadherin (Cell Signaling) or α-tubulin (Santa Cruz) and scanned with an Odyssey imager and software (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Dr. Jens Moeller for initial assistance in establishing PA gel protocols and Jon Mulholland and Cell Sciences Imaging Facility for assistance with SIM experiments. This work was supported by NIH Public Health Service Grant T32 CA09151 awarded by the National Cancer Institute (to C.C.), NIH Grants R35GM118064-01 (to W.J.N.) and 1R21HL13099301 (to B.L.P.), and National Science Foundation (NSF) Grant Engineering New Technologies Based on Multicellular and Inter-kingdom Signaling Awards (EFRI MIKS) 1136790 (to W.J.N. and B.L.P.). A.K.D. is supported by graduate research fellowships from the NSF and the Stanford Office of the Vice Provost for Graduate Education. The SIM superresolution experiments were supported in part by Award 1S10OD01227601 from the National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618676114/-/DCSupplemental.
References
- 1.Collins C, Nelson WJ. Running with neighbors: Coordinating cell migration and cell–cell adhesion. Curr Opin Cell Biol. 2015;36:62–70. doi: 10.1016/j.ceb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gumbiner BM. Regulation of cadherin adhesive activity. J Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley CD, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidel-Bar R. Cadherin adhesome at a glance. J Cell Sci. 2013;126:373–378. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
- 7.McNeill H, Ryan TA, Smith SJ, Nelson WJ. Spatial and temporal dissection of immediate and early events following cadherin-mediated epithelial cell adhesion. J Cell Biol. 1993;120:1217–1226. doi: 10.1083/jcb.120.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 9.Kim SH, Li Z, Sacks DB. E-cadherin-mediated cell-cell attachment activates Cdc42. J Biol Chem. 2000;275:36999–37005. doi: 10.1074/jbc.M003430200. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda S, et al. Regulation of cell-cell adhesion of MDCK cells by Cdc42 and Rac1 small GTPases. Biochem Biophys Res Commun. 1997;240:430–435. doi: 10.1006/bbrc.1997.7675. [DOI] [PubMed] [Google Scholar]
- 11.Carramusa L, Ballestrem C, Zilberman Y, Bershadsky AD. Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J Cell Sci. 2007;120:3870–3882. doi: 10.1242/jcs.014365. [DOI] [PubMed] [Google Scholar]
- 12.Gauvin TJ, Young LE, Higgs HN. The formin FMNL3 assembles plasma membrane protrusions that participate in cell-cell adhesion. Mol Biol Cell. 2015;26:467–477. doi: 10.1091/mbc.E14-07-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grikscheit K, Frank T, Wang Y, Grosse R. Junctional actin assembly is mediated by Formin-like 2 downstream of Rac1. J Cell Biol. 2015;209:367–376. doi: 10.1083/jcb.201412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao MV, Zaidel-Bar R. Formin-mediated actin polymerization at cell-cell junctions stabilizes E-cadherin and maintains monolayer integrity during wound repair. Mol Biol Cell. 2016;27:2844–2856. doi: 10.1091/mbc.E16-06-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J Cell Biol. 2007;178:517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki D, Oikawa T, Takenawa T. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J Cell Sci. 2007;120:86–100. doi: 10.1242/jcs.03311. [DOI] [PubMed] [Google Scholar]
- 17.Califano JP, Reinhart-King CA. Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact. Cell Mol Bioeng. 2010;3:68–75. doi: 10.1007/s12195-010-0102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trichet L, et al. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA. 2012;109:6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolas A, Geiger B, Safran SA. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc Natl Acad Sci USA. 2004;101:12520–12525. doi: 10.1073/pnas.0403539101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 21.Ladoux B, et al. Strength dependence of cadherin-mediated adhesions. Biophys J. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci USA. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barry AK, Wang N, Leckband DE. Local VE-cadherin mechanotransduction triggers long-ranged remodeling of endothelial monolayers. J Cell Sci. 2015;128:1341–1351. doi: 10.1242/jcs.159954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drees F, Reilein A, Nelson WJ. Cell-adhesion assays: Fabrication of an E-cadherin substratum and isolation of lateral and basal membrane patches. Methods Mol Biol. 2005;294:303–320. doi: 10.1385/1-59259-860-9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borghi N, Lowndes M, Maruthamuthu V, Gardel ML, Nelson WJ. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc Natl Acad Sci USA. 2010;107:13324–13329. doi: 10.1073/pnas.1002662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen DJ, Gloerich M, Nelson WJ. Epithelial self-healing is recapitulated by a 3D biomimetic E-cadherin junction. Proc Natl Acad Sci USA. 2016;113:14698–14703. doi: 10.1073/pnas.1612208113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. Atomic force microscopy probing of cell elasticity. Micron. 2007;38:824–833. doi: 10.1016/j.micron.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Schulze KD, et al. Elastic modulus and hydraulic permeability of MDCK monolayers. J Biomech. 2017;53:210–213. doi: 10.1016/j.jbiomech.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: The important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemir S, West JL. Synthetic materials in the study of cell response to substrate rigidity. Ann Biomed Eng. 2010;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 33.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;47:10.16.1–10.16.16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 34.Gavard J, et al. Lamellipodium extension and cadherin adhesion: Two cell responses to cadherin activation relying on distinct signalling pathways. J Cell Sci. 2004;117:257–270. doi: 10.1242/jcs.00857. [DOI] [PubMed] [Google Scholar]
- 35.Guo Z, et al. E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci Signal. 2014;7:rs7. doi: 10.1126/scisignal.2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacs EM, Ali RG, McCormack AJ, Yap AS. E-cadherin homophilic ligation directly signals through Rac and phosphatidylinositol 3-kinase to regulate adhesive contacts. J Biol Chem. 2002;277:6708–6718. doi: 10.1074/jbc.M109640200. [DOI] [PubMed] [Google Scholar]
- 37.Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379–382. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- 38.Gumbiner B, Simons K. A functional assay for proteins involved in establishing an epithelial occluding barrier: Identification of a uvomorulin-like polypeptide. J Cell Biol. 1986;102:457–468. doi: 10.1083/jcb.102.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelham RJ, Jr, Wang YI. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tee SY, Fu J, Chen CS, Janmey PA. Cell shape and substrate rigidity both regulate cell stiffness. Biophys J. 2011;100:L25–L27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen JH, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tseng Q, et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci USA. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isogai T, van der Kammen R, Innocenti M. SMIFH2 has effects on Formins and p53 that perturb the cell cytoskeleton. Sci Rep. 2015;5:9802. doi: 10.1038/srep09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolen BJ, et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris AR, Daeden A, Charras GT. Formation of adherens junctions leads to the emergence of a tissue-level tension in epithelial monolayers. J Cell Sci. 2014;127:2507–2517. doi: 10.1242/jcs.142349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruyne D, et al. Role of formins in actin assembly: Nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 47.Mattila PK, Lappalainen P. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 48.Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF. The Arp2/3 complex branches filament barbed ends: Functional antagonism with capping proteins. Nat Cell Biol. 2000;2:385–391. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- 49.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 50.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 51.Hong L, et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J Biol Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sumida GM, Yamada S. Rho GTPases and the downstream effectors actin-related protein 2/3 (Arp2/3) complex and myosin II induce membrane fusion at self-contacts. J Biol Chem. 2015;290:3238–3247. doi: 10.1074/jbc.M114.612168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das T, et al. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol. 2015;17:276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 55.Kim JB, et al. N-Cadherin extracellular repeat 4 mediates epithelial to mesenchymal transition and increased motility. J Cell Biol. 2000;151:1193–1206. doi: 10.1083/jcb.151.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexander NR, et al. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biswas KH, et al. E-cadherin junction formation involves an active kinetic nucleation process. Proc Natl Acad Sci USA. 2015;112:10932–10937. doi: 10.1073/pnas.1513775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Näthke IS, Hinck L, Swedlow JR, Papkoff J, Nelson WJ. Defining interactions and distributions of cadherin and catenin complexes in polarized epithelial cells. J Cell Biol. 1994;125:1341–1352. doi: 10.1083/jcb.125.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y, Kanchanawong P, Zaidel-Bar R. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev Cell. 2015;32:139–154. doi: 10.1016/j.devcel.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sim JY, et al. Spatial distribution of cell-cell and cell-ECM adhesions regulates force balance while maintaining E-cadherin molecular tension in cell pairs. Mol Biol Cell. 2015;26:2456–2465. doi: 10.1091/mbc.E14-12-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erasmus J, Aresta S, Nola S, Caron E, Braga VM. Newly formed E-cadherin contacts do not activate Cdc42 or induce filopodia protrusion in human keratinocytes. Biol Cell. 2009;102:13–24. doi: 10.1042/BC20090048. [DOI] [PubMed] [Google Scholar]
- 64.Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–33308. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- 65.Homem CC, Peifer M. Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development. 2008;135:1005–1018. doi: 10.1242/dev.016337. [DOI] [PubMed] [Google Scholar]
- 66.Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735–1748. doi: 10.1128/MCB.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao M, et al. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- 69.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 70.Yang C, Svitkina T. Filopodia initiation: Focus on the Arp2/3 complex and formins. Cell Adhes Migr. 2011;5:402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong S, Guo WH, Wang YL. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc Natl Acad Sci USA. 2014;111:17176–17181. doi: 10.1073/pnas.1412285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serra-Picamal X, Conte V, Sunyer R, Muñoz JJ, Trepat X. Mapping forces and kinematics during collective cell migration. Methods Cell Biol. 2015;125:309–330. doi: 10.1016/bs.mcb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 73.Mays RW, et al. Hierarchy of mechanisms involved in generating Na/K-ATPase polarity in MDCK epithelial cells. J Cell Biol. 1995;130:1105–1115. doi: 10.1083/jcb.130.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin DC, Horkay F. Nanomechanics of polymer gels and biological tissues: A critical review of analytical approaches in the Hertzian regime and beyond. Soft Matter. 2008;4:669–682. doi: 10.1039/b714637j. [DOI] [PubMed] [Google Scholar]
- 76.Denisin AK, Pruitt BL. Tuning the range of polyacrylamide gel stiffness for mechanobiology applications. ACS Appl Mater Interfaces. 2016;8:21893–21902. doi: 10.1021/acsami.5b09344. [DOI] [PubMed] [Google Scholar]
- 77.Chang AC, et al. Single molecule force measurements in living cells reveal a minimally tensioned integrin state. ACS Nano. 2016;10:10745–10752. doi: 10.1021/acsnano.6b03314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westerweel JSF. Universal outlier detection for PIV data. Exp Fluids. 2005;39:1096–1100. [Google Scholar]
- 79.Schwarz US, et al. Calculation of forces at focal adhesions from elastic substrate data: The effect of localized force and the need for regularization. Biophys J. 2002;83:1380–1394. doi: 10.1016/S0006-3495(02)73909-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.