Significance
Partner and localiser of BRCA2 (PALB2) is a breast cancer susceptibility gene, and the role of its product in repairing broken chromosomes has been extensively described. However, a fraction of PALB2 is also found on intact chromosomes, and it is unknown how and why PALB2 associates with undamaged chromatin. In this study, we establish that the histone binding protein MRG15 is a major interaction partner of PALB2 and plays a key role in tethering PALB2 to active genes. Failure of PALB2 to interact with MRG15 leads to the accumulation of DNA stress at active genes and chromosome instability in dividing cells. These findings shed light on why patients with PALB2 mutations often develop genome instability syndromes, such as cancer.
Keywords: PALB2, MRG15, SETD2, DNA replication, transcription
Abstract
The partner and localiser of BRCA2 (PALB2) plays important roles in the maintenance of genome integrity and protection against cancer. Although PALB2 is commonly described as a repair factor recruited to sites of DNA breaks, recent studies provide evidence that PALB2 also associates with unperturbed chromatin. Here, we investigated the previously poorly described role of chromatin-associated PALB2 in undamaged cells. We found that PALB2 associates with active genes through its major binding partner, MRG15, which recognizes histone H3 trimethylated at lysine 36 (H3K36me3) by the SETD2 methyltransferase. Missense mutations that ablate PALB2 binding to MRG15 confer elevated sensitivity to the topoisomerase inhibitor camptothecin (CPT) and increased levels of aberrant metaphase chromosomes and DNA stress in gene bodies, which were suppressed by preventing DNA replication. Remarkably, the level of PALB2 at genic regions was frequently decreased, rather than increased, upon CPT treatment. We propose that the steady-state presence of PALB2 at active genes, mediated through the SETD2/H3K36me3/MRG15 axis, ensures an immediate response to DNA stress and therefore effective protection of these regions during DNA replication. This study provides a conceptual advance in demonstrating that the constitutive chromatin association of repair factors plays a key role in the maintenance of genome stability and furthers our understanding of why PALB2 defects lead to human genome instability syndromes.
Inherited mutations in the partner and localiser of BRCA2 (PALB2) gene predispose to breast and pancreatic cancer (1–3) and also congenital malformations, growth retardation, and early childhood cancer in a rare subgroup of Fanconi anemia (FA-N) patients (4, 5). The best characterized function of PALB2 is to physically link the two main breast cancer susceptibility gene products, BRCA1 and BRCA2, at sites of DNA damage (6–8), thus playing a pivotal role in the repair of DNA double-strand breaks (DSBs) by homologous recombination (HR). BRCA1 facilitates DNA-end resection to produce single-stranded (ss) DNA (9) and concomitantly attracts PALB2, together with BRCA2 and RAD51, to DSB sites (6–8). In turn, BRCA2 promotes the loading of the RAD51 recombinase onto ssDNA (10–12), which is critical for the strand invasion and exchange phase of HR (13). This mechanism of repair factor recruitment is important for the timely activation of HR at sites of DNA damage, and its perturbation in individuals with impaired PALB2 function is presumed to cause human pathologies.
Although PALB2 is commonly described as a repair factor recruited to sites of DNA breaks, we and others have provided evidence that PALB2 also associates with chromatin in the absence of DNA damage (14–17). A recent genome-wide analysis of PALB2 chromatin occupancy revealed enrichment at highly active genes, with PALB2 occupying the entire body of these genes (18). PALB2 was shown to support the transcription of a subset of NF-κB– and retinoic acid-responsive genes. However, PALB2 does not behave as a general transcription coactivator, and the physiological significance of its association with active genes remains unclear.
Here, we provide direct evidence that PALB2 associates with active genes through its major binding partner MRG15 (MORF-related gene on chromosome 15). Cells expressing a PALB2 variant, harboring missense mutations that hinder MRG15 binding, exhibit elevated sensitivity to the topoisomerase I (TOP1) inhibitor camptothecin (CPT) and increased levels of aberrant metaphase chromosomes and DNA stress in gene bodies, which were suppressed by preventing DNA replication. Collectively, our findings suggest that steady-state chromatin association of PALB2 contributes to protecting active genes from stress arising during DNA replication and give insights into the way in which PALB2 defects elicit human genome instability syndromes.
Results and Discussion
PALB2 Associates with Active Genes Through the SETD2/H3K36me3/MRG15 Axis.
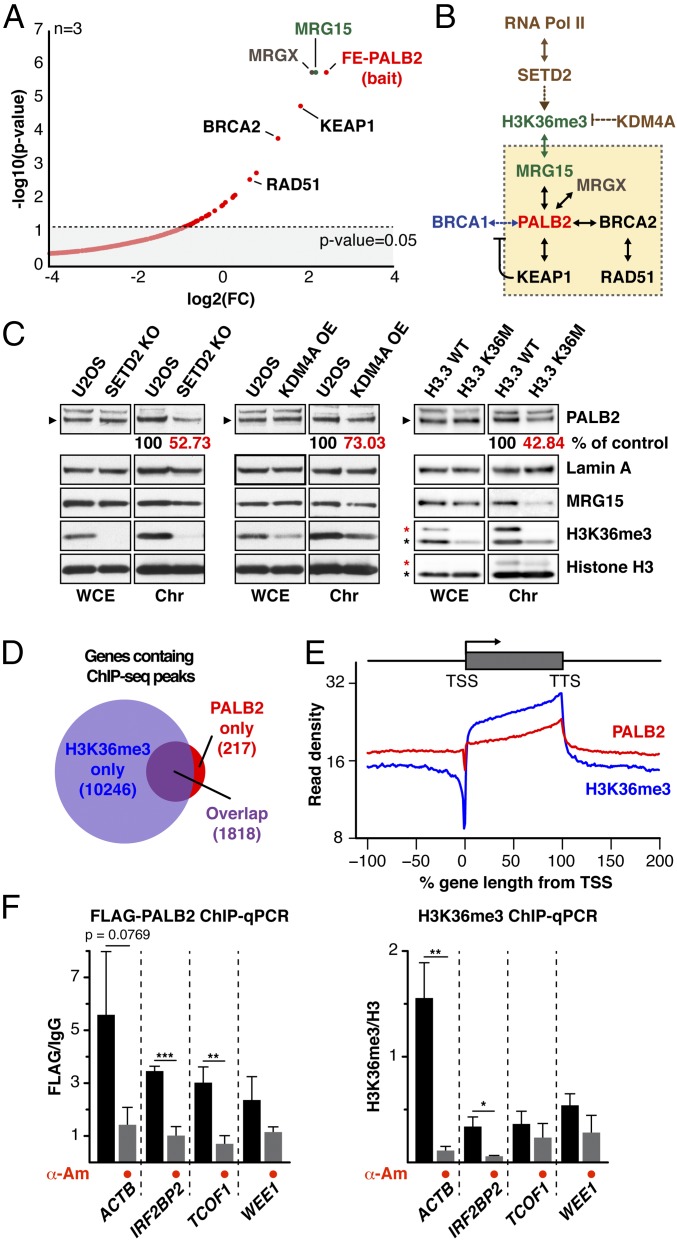
To gain insights into the mechanism of PALB2 association with undamaged chromatin, PALB2 binding partners in exponentially growing cells were identified by quantitative mass spectrometry (MS). This analysis revealed MRG15 and the product of another MORF-related gene on chromosome X (MRGX) as the two major binding partners of PALB2 (Fig. 1A and Dataset S1). Significantly, depletion of MRG15, but not MRGX or BRCA1, impaired PALB2 and BRCA2 chromatin association (Fig. S1 A–C; see also chromatin association of the PALB2 variant ablating BRCA1 binding in Fig. 2), suggesting a specific role for MRG15 in the recruitment of the PALB2–BRCA2 complex to unperturbed chromatin.
Fig. 1.
MRG15, SETD2, and H3K36me3 are required for PALB2 chromatin association in unperturbed cells. (A) Volcano plot showing FLAG-EGFP-PALB2 (FE-PALB2)-associated proteins identified by MS. The log2 of the fold change (FC) over a FLAG-EGFP control was plotted against negative log10 P values (n = 3). (B) Schematic of the pathway linking transcription and PALB2. (C) WB analysis of PALB2 in the chromatin fraction of U2OS cells with CRISPR SETD2 knockout (Left), KDM4A overexpression (OE) (Middle), and wild type (WT) or mutant (K36M) histone variant H3.3 overexpression (Right). Asterisks indicate endogenous H3 (black) and exogenous H3.3 (red). (D) Venn diagram showing the overlap of H3K36me3-containing and PALB2-bound genes. (E) The averaged profile of H3K36me3 and PALB2 around gene loci with H3K36me3 peaks. Read density (y axis) is normalized for total number of mapped reads. (F) ChIP-qPCR quantification of FLAG-PALB2 (Left) and H3K36me3 (Right). Mean values ± SD (n = 3, with triplicate qPCR reactions). Where indicated, cells were treated with α-Am. Asterisks indicate two-tailed paired Student's t test; *P < 0.05, **P < 0.01, ***P < 0.001.
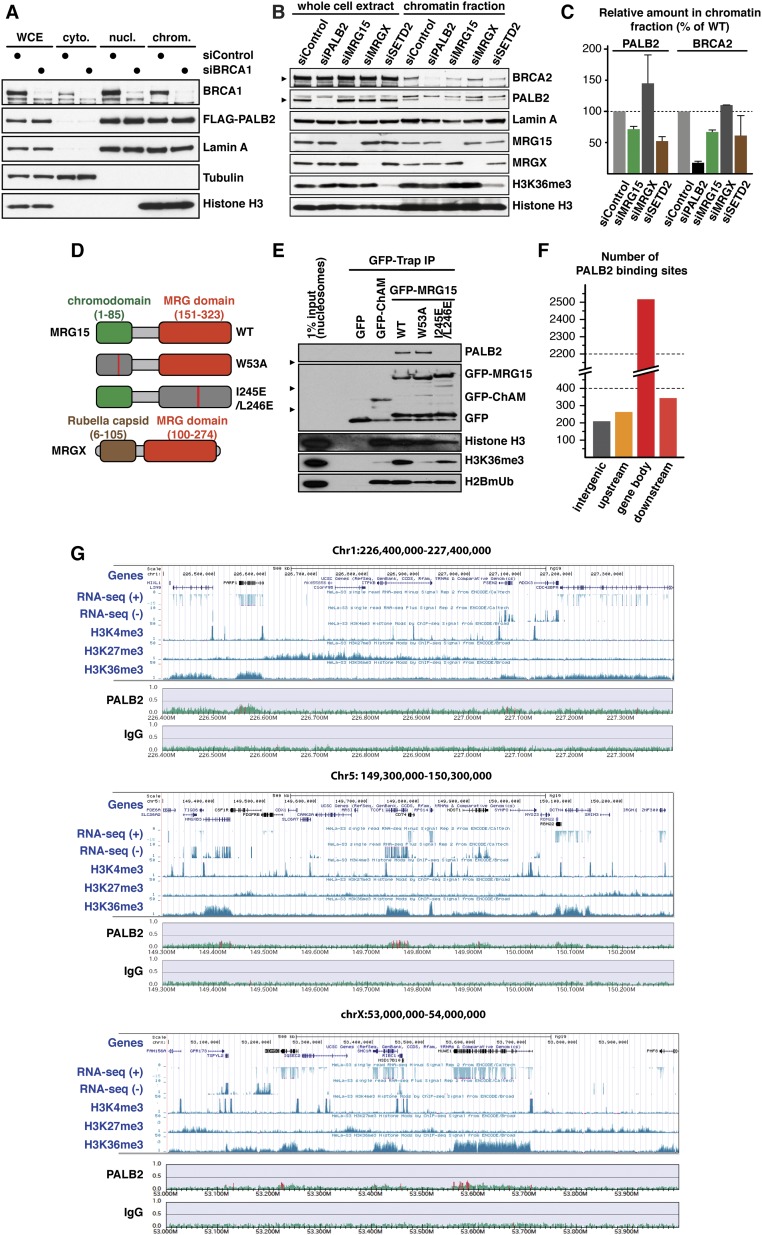
Fig. S1.
(Related to Fig. 1) (A) WB analysis of chromatin-associated FLAG-PALB2 following treatment of U2OS cells with siRNA targeting BRCA1. (B) WB showing chromatin-associated PALB2 in HT-1080 cells treated with siRNA targeting indicated genes. Arrowheads denote specific bands. (C) Quantification of PALB2 and BRCA2 levels in the chromatin fraction of HT-1080 cells treated with the indicated siRNA. Mean values ± SD (n = 2). (D) Schematic diagram of the MRG15 and MRGX proteins. The chromodomain (green), MRG domain (red), rubella capsid-like domain (brown), and point mutations (red bars) are depicted. (E) Nucleosome pull-down assay using affinity-purified GFP-fusion of ChAM or MRG15 variants and partially purified human nucleosomes. The presence in the pull-down samples of histone H3, H3K36me3, and monoubiquitinated H2B (H2BmUB) was examined by WB. (F) Genome-wide PALB2 occupancy in S-phase HeLa cells was determined by ChIP-seq, and PALB2 peaks were categorized as upstream (≤5 kb upstream of gene bodies), downstream (≤5 kb downstream of gene bodies), and intergenic. (G) Snapshots of PALB2 ChIP-seq aligned with the expression levels of genes (Caltech RNA-seq; HeLa-S3 whole cells) and histone modifications (Broad HeLa-S3; H3K4me3, H3K27me3, H3K36me3) at the UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) assembly.
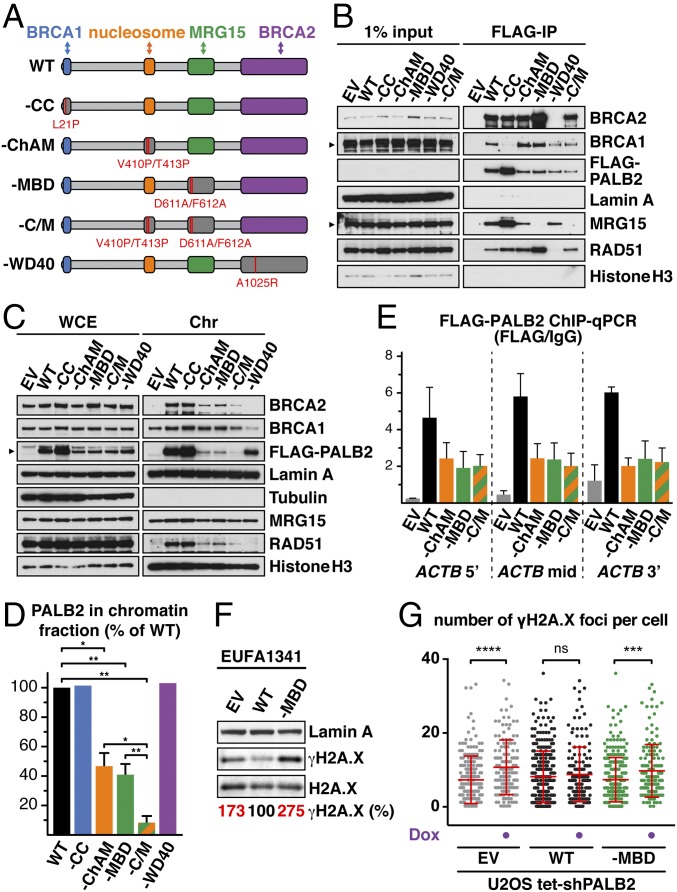
Fig. 2.
Ablation of PALB2 binding to MRG15 confers an increased level of DNA stress in unperturbed cells. (A) Schematic of PALB2 variants depicting point mutations (red bars) disrupting each domain. (B) Anti-FLAG IPs from EUFA1341 cells stably expressing FLAG-PALB2 variants. Lamin A was used as a negative control. (C) WB of indicated proteins in the chromatin fraction of EUFA1341 cells stably expressing FLAG-PALB2 variants. (D) The levels of PALB2 in the chromatin fraction of cells expressing FLAG-PALB2 variants were quantified and, following normalization against their respective levels in whole-cell extract, expressed as % of WT. Mean values ± SD (n = 3). (E) ChIP-qPCR quantification of FLAG-PALB2 variants at the ACTB gene. (F) WB showing the levels of γH2A.X in untreated EUFA1341 cells complemented with FLAG-PALB2 variants. (G) The number of γH2A.X foci in U2OS Flp-In T-REx cells stably expressing FLAG-PALB2 variants. Where indicated, cells were treated with 2 μg/mL doxycycline (Dox) for 5 d to induce shRNA targeting endogenous PALB2. Dots represent individual cells and bars mean values ± SD. Statistical significance was determined using two-tailed paired Student’s t test (D) or Mann–Whitney U test (G). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. EV, empty vector; ns, nonsignificant.
In addition to an MRG domain, which binds various transcriptional regulators and chromatin-remodeling factors, MRG15 contains an N-terminal chromodomain that binds K36 trimethylated histone H3 (H3K36me3; Fig. S1 D and E) (19, 20). This prompted us to investigate whether H3K36me3 was required for MRG15-mediated PALB2 chromatin association. In mammalian cells, H3K36me3 is catalyzed by the lysine methyltransferase SET domain containing 2 (SETD2) (21), which facilitates transcription by associating with RNA polymerase II (Pol II) (22, 23) and therefore acts as a general epigenetic mark of active genes (Fig. 1B). Strikingly, knockout of SETD2, which is also implicated in HR (24–26), resulted in a concomitant reduction of chromatin-associated PALB2 and H3K36me3 levels (Fig. 1C and Fig. S1 B and C). Similarly, reduced PALB2 chromatin association was found in cells overexpressing lysine demethylase 4A (KDM4A), which counteracts SETD2 (27), or a missense mutant histone variant H3.3 nonmethylatable at K36 (H3.3 K36M), which is incorporated at transcriptionally active loci independently of replication (28, 29) (Fig. 1C). Additionally, our independent genome-wide PALB2 ChIP-sequencing (ChIP-seq) analysis highlighted an enrichment of PALB2 occupancy at H3K36me3-containing genes (Fig. 1 D and E and Fig. S1 F and G). ChIP-qPCR quantification of PALB2 occupancy at ACTB, TCOF1, WEE1, and IRF2BP2, which were identified as PALB2-bound genes by ChIP-seq analysis both in this study and by Gardini et al. (18), showed a reduced PALB2 level at these loci following inhibition of Pol II-mediated transcription with α-amanitin (α-Am), concomitant with reduced levels of H3K36me3 and chromatin-associated Pol II (Fig. 1F and Fig. S2A). These observations demonstrate that active transcription is required for PALB2 binding to these genes. Notably, gene set enrichment analysis (GSEA) revealed a higher tendency of PALB2 to bind to periodically expressed genes (Fig. S2B), which are associated with a high level of H3K36me3 (30). Conversely, we found little correlation between the level of gene expression and PALB2 association (Fig. S2 C–G). Together, we conclude that PALB2 associates with active genes, with a preference for periodic genes, via the SETD2/H3K36me3/MRG15 axis. It is plausible that PALB2 has higher affinity for newly marked H3K36me3, which periodically changes during the cell cycle, potentially through a mechanism involving PALB2 chromatin-association motif (ChAM) (14) and/or other PALB2 binding partners.
Fig. S2.
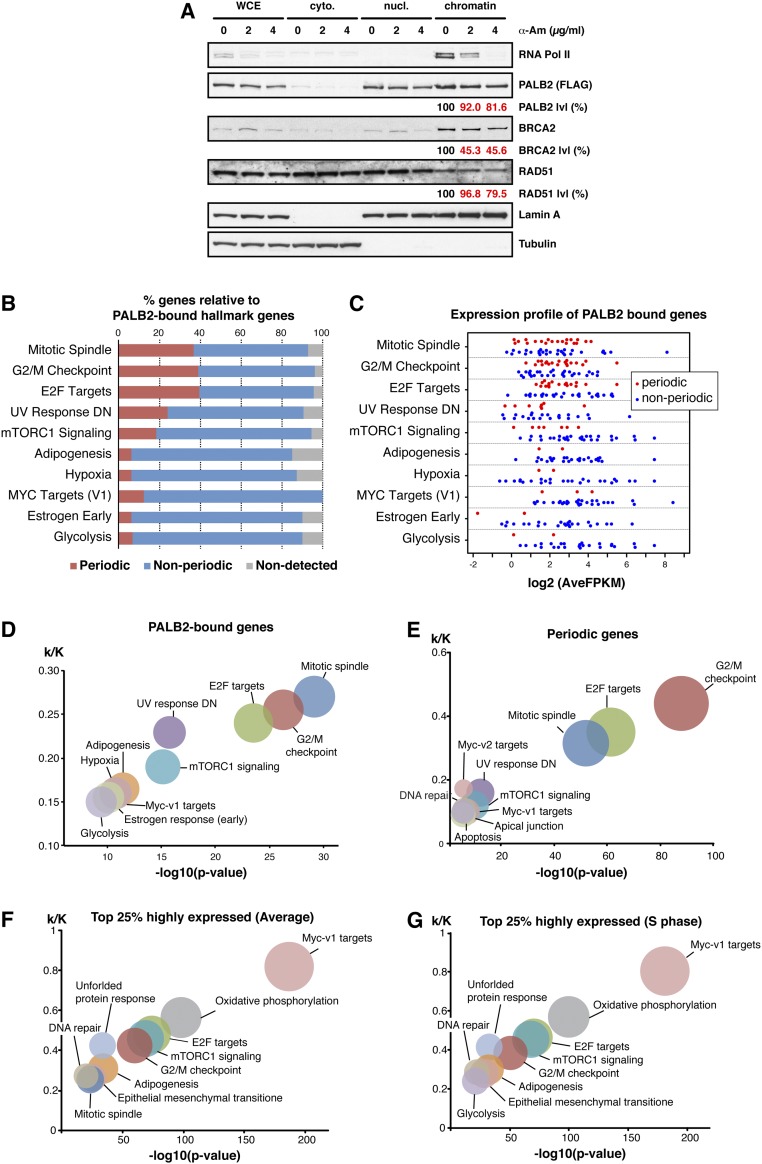
(Related to Fig. 1) (A) WB analysis of chromatin-associated FLAG-PALB2 WT in EUFA1341 cells following 17 h of treatment with the indicated concentrations of α-Am. The levels of FLAG-PALB2, BRCA2, and Rad51 in the chromatin fraction were quantified and, following normalization against their respective levels in whole-cell extract, expressed as % of untreated cells. (B) The 2,050 genes containing PALB2-binding sites were categorized by GSEA, and the proportion of periodic and nonperiodic genes, as identified by Dominguez et al. (30), is shown in each category. (C) A strip-plot showing the average expression level of PALB2-bound genes during the cell cycle. Red and blue dots, respectively, indicate periodic and nonperiodic genes. (D) As in B, but the ratio of k (number of genes containing PALB2-binding sites) against K (number of genes consisting of a given hallmark gene set) was plotted against negative log10 P values. The size of each circle indicates the respective k value. (E) GSEA of 1,183 periodic genes as identified by Dominguez et al. (30). (F) The GSEA of the top 25% highly expressed genes (2,204 genes), as detected by RNA-seq of synchronized HeLa cells. The level of average gene expression was calculated as mean of FPKM (fragments per kilobase of transcript per million mapped reads) at each time point during the cell cycle. (G) The GSEA of the top 25% highly expressed genes in S phase (2,204 genes), as in F but at 3 h after the thymidine release.
PALB2 Association with Active Genes Is Necessary to Maintain Genome Integrity.
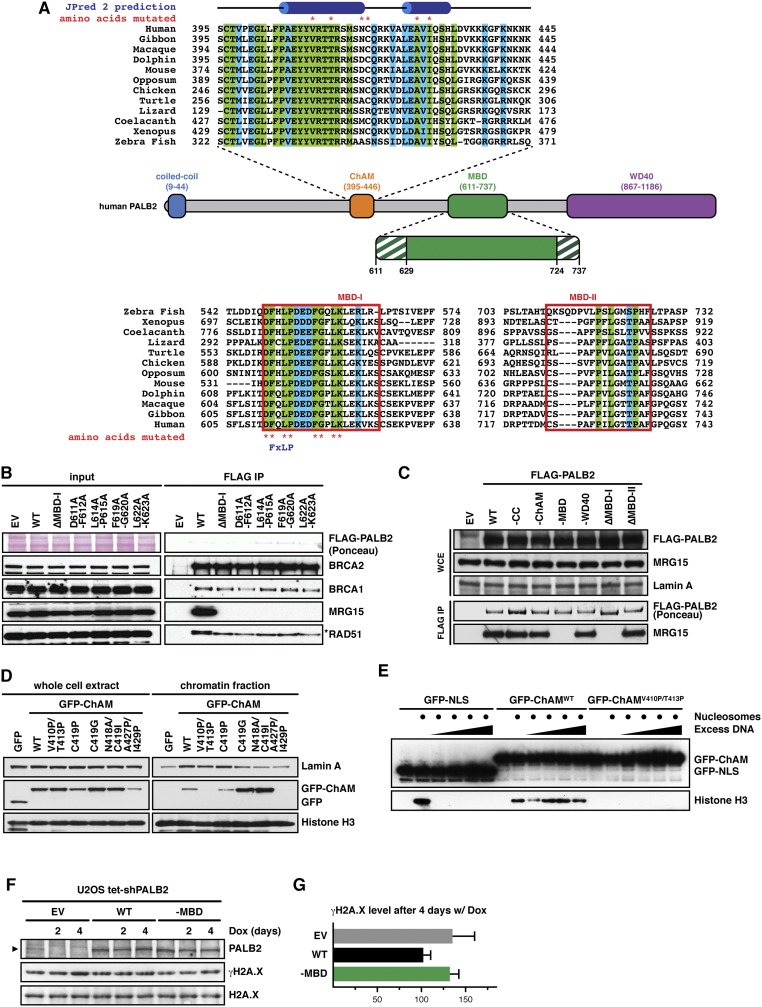
To further examine the physiological role of PALB2 at gene bodies, we first characterized missense mutations blocking PALB2 binding to MRG15. PALB2 MRG15-binding domain (MBD) (31) contains two highly conserved regions: MBD-I (residues 611–629), which includes a MRG-binding FxLP motif (32), and MBD-II (residues 724–737; Fig. S3A). Notably, deletion of MBD I or missense mutations within the FxLP motif and other MBD-I conserved residues, such as D611A/F612A (designated as PALB2–MBD), hindered PALB2 binding to MRG15 (Fig. S3 B and C). PALB2 binding was also blocked by missense mutations of MRG15 at positions 245 and 246 (I245E/L246E), which lie outside the hydrophobic FxLP-binding pocket within the MRG domain (32) (Fig. S1 D and E). These findings indicate that the PALB2 interaction with MRG15 involves a noncanonical interface.
Fig. S3.
(Related to Fig. 2) (A) Schematic diagram of the PALB2 protein depicting the position of the coiled-coil domain (blue), the ChAM (orange), the MBD (green), and the WD40-repeat domain (purple). Protein sequence alignments for ChAM and conserved regions MBD-I and MBD-II are also presented. Identical and conserved residues are, respectively, highlighted in green and blue. Asterisks indicate residues mutated in this study. ChAM helical secondary structure as predicted by JPred 2 and the FxLP motif within MBD-I are indicated at the top and bottom, respectively. (B) FLAG-PALB2 variants with a MBD-I deletion or indicated point mutations were expressed in HEK293T cells and immunoprecipitated from whole-cell lysates. WB detection of the indicated proteins was performed to assess co-IP. (C) The indicated FLAG-PALB2 variants were expressed in HEK293T cells and immunoprecipitated from whole-cell lysates. WB detection of MRG15 was performed to assess co-IP. (D) GFP-ChAM peptides, with the indicated mutations, were transiently expressed in HEK293T cells and their chromatin association was determined following chemical cell fractionation. Lamin A and histone H3 are markers for extraction of nuclear and chromatin-associated proteins, respectively. (E) Pull-down assay using affinity-purified GFP-ChAM and recombinant mononucleosomes. Salmon sperm DNA was included to outcompete nonspecific interactions. (F) WB showing the levels of γH2A.X in U2OS Flp-In T-REx cells stably expressing FLAG-PALB2 variants, following down-regulation of endogenous PALB2 using shRNA induced by 2 µg/mL doxycycline (Dox). Arrowhead indicates PALB2-specific band. (G) Levels of γH2A.X on day 4 were quantified and expressed as % of untreated. Mean values ± SD (n = 3). EV, empty vector.
To consolidate our analysis, we introduced missense mutations ablating other known functional domains of PALB2 (Fig. 2A). Through systematic mutagenesis of conserved residues (Fig. S3 A, D, and E), we identified mutations impairing ChAM and generated single (PALB2–ChAM) and double ChAM/MBD (PALB2–C/M) mutants. Mutations disrupting PALB2 interaction with BRCA1 (PALB2–CC) and BRCA2 (PALB2–WD40) (7, 33) were also introduced, and each PALB2 variant was stably expressed in PALB2-defective EUFA1341 fibroblasts (5). As expected, BRCA1, MRG15, and BRCA2 were found in protein complexes containing either PALB2WT or PALB2–ChAM but were respectively absent from those containing PALB2–CC, PALB2–MBD, and PALB2–WD40 (Fig. 2B). Furthermore, cell fractionation analyses established that PALB2–CC and PALB2–WD40 associate proficiently with chromatin, whereas PALB2–ChAM and PALB2–MBD, along with BRCA2 and RAD51, showed pronounced reduction of chromatin association, which was further decreased in PALB2–C/M (Fig. 2 C and D). Importantly, however, our ChIP-qPCR analysis showed comparable levels of PALB2–ChAM, PALB2–MBD, and PALB2–C/M within the ACTB gene body (Fig. 2E), suggesting that ChAM and MRG15 cooperatively recruit PALB2 to active genes.
While generating EUFA1341 cells complemented with PALB2 variants, we noticed that PALB2–MBD cells grew more slowly than those expressing PALB2WT. Indeed, an increased level of Ser139 phosphorylated histone variant H2A.X (γH2A.X), a marker of DNA stress, was detectable in cells expressing PALB2–MBD compared with PALB2WT (Fig. 2F). Similarly, increased levels of γH2AX and γH2AX foci were detectable in U2OS cells stably expressing exogenous PALB2–MBD following the down-regulation of endogenous PALB2 by shRNA (Fig. 2G and Fig. S3 F and G).
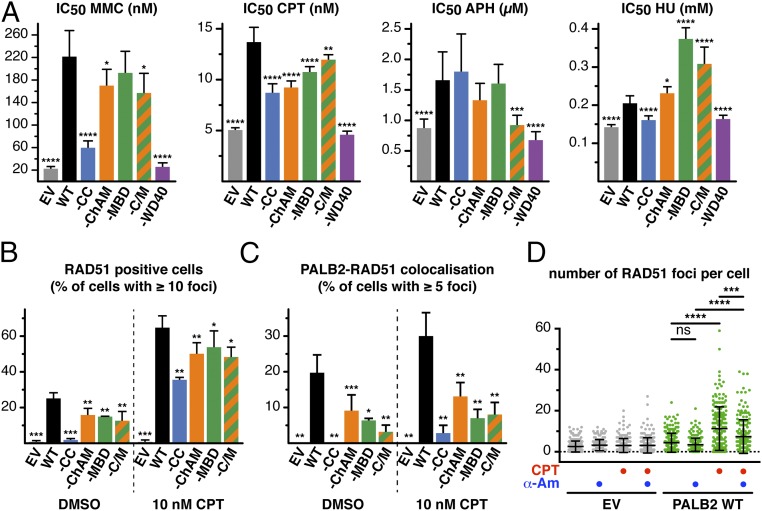
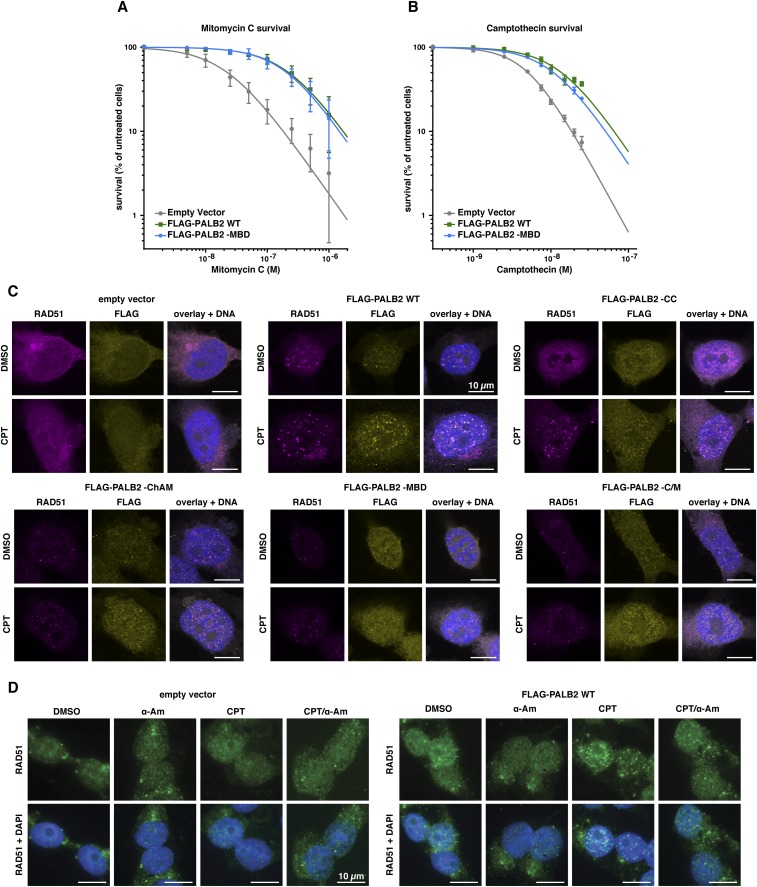
To identify the nature of the DNA stress detected in PALB2–MBD cells, we assessed the survival of cells expressing PALB2 variants following treatment with the crosslinking agent mitomycin C (MMC), the TOP1 inhibitor CPT, the DNA polymerase inhibitor aphidicolin (APH), or the ribonucleotide reductase inhibitor hydroxyurea (HU; Fig. 3A). Each variant conferred a distinct drug sensitivity signature, with PALB2–WD40 most closely matching the phenotype of PALB2-defective cells, exhibiting hypersensitivity to all four drugs. Importantly, cells expressing PALB2–MBD displayed a specific sensitivity to CPT (Fig. 3A and Fig. S4 A and B). Furthermore, compared with cells expressing PALB2WT, a modest but significant impairment of RAD51 foci formation was detectable in PALB2–MBD cells (Fig. 3B). In particular, we noticed a marked impairment of RAD51 colocalization with PALB2–MBD in CPT-induced foci (Fig. 3C) and a reduced fluorescence intensity of CPT-induced RAD51 foci (Fig. S4C). CPT-induced RAD51 foci formation was additionally impaired upon transcription inhibition (Fig. 3D and Fig. S4D), indicating that RAD51 recruitment is dependent on active transcription.
Fig. 3.
Ablation of PALB2 binding to MRG15 confers hypersensitivity to CPT. (A) The IC50 for MMC, CPT, APH, and HU were determined by WST-1 assay. n = 3, with two technical replicates. Error bars indicate 95% CI. (B) Quantification of RAD51-positive cells (% of cells with ≥10 foci per nucleus). Mean values ± SD (n ≥ 3, >210 nuclei scored per repeat). (C) Quantification of RAD51 and PALB2 colocalization (% of total cells with ≥5 overlapping foci per nucleus). Mean values ± SD (n ≥ 3, >210 nuclei scored per repeat). (D) Quantification of the number of RAD51 foci per cell. Where indicated, cells were treated with CPT or α-Am. Dots represent individual cells and bars mean values ± SD. Statistical significance was determined using the extra sum-of-squares f test (A), two-tailed paired Student’s t test (B and C), or Mann–Whitney U test (D). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, nonsignificant.
Fig. S4.
(Related to Fig. 3) Survival curves for EUFA1341 cells complemented with FLAG-PALB2 WT or -MBD mutant following treatment with indicated concentrations of MMC (A) or CPT (B). Mean values ± SD (n = 3). (C) EUFA1341 cells complemented with FLAG-PALB2 variants were stained for RAD51 and FLAG-PALB2 foci following 17 h of treatment with DMSO or 10 nM CPT. For each cell line used in this study, representative pictures of RAD51 (purple) and FLAG-PALB2 (yellow) foci are shown. (D) Representative pictures of RAD51 foci (green) in EUFA1341 cells complemented with WT FLAG-PALB2. Where indicated, cells were treated for 17 h with DMSO, 4 µg/mL α-Am, or 10 nM CPT. (Scale bars, 10 μm.)
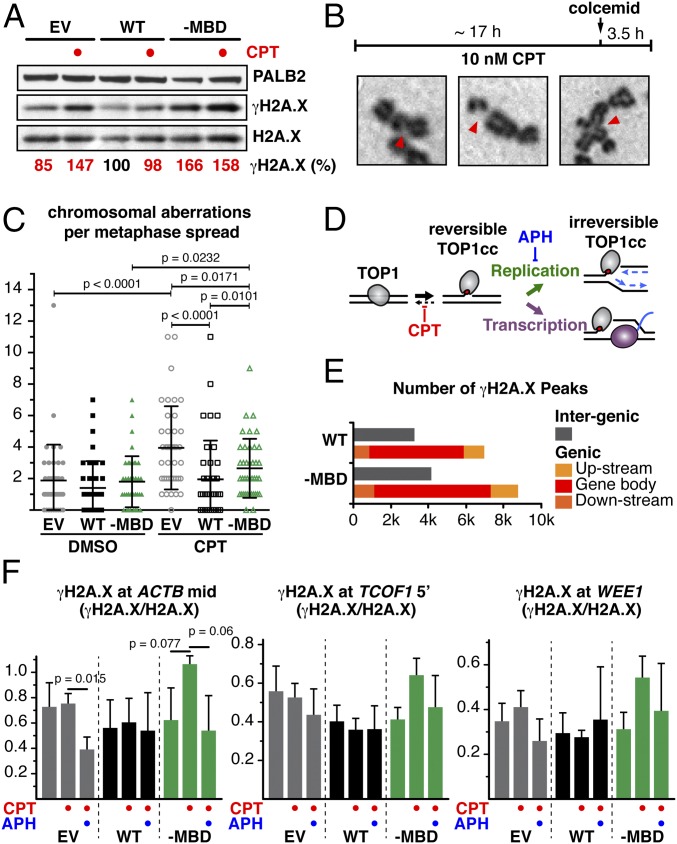
The impact of CPT treatment on genome stability was further evaluated by examining metaphase chromosome spreads. Strikingly, exposure of PALB2–MBD cells to 10 nM CPT, a dose that typically causes mild replicative stress without inducing substantial DNA breaks (34) (Fig. 4A), triggered a significant increase in the number of chromosomal aberrations (Fig. 4 B and C). In contrast, and as expected, the chromosomal aberration frequency remained unchanged in PALB2WT cells. These results reflect the reduced cellular survival of PALB2–MBD cells upon CPT treatment, further emphasizing the role of MRG15-mediated PALB2 chromatin association in preventing genome instability.
Fig. 4.
PALB2 interaction with MRG15 protects active genes during DNA replication. (A) WB showing γH2A.X levels in EUFA1341 cells expressing FLAG-PALB2 variants. (B) Workflow of metaphase chromosome spread preparation and examples of chromosomal aberrations. (C) Aberrant chromosomes per metaphase cell. Dots represent individual cells and bars mean values ± SD (n = 40). P values are for Mann–Whitney U test. (D) Diagram depicting the mechanism of TOP1 poisoning by CPT, stabilizing TOP1 cleavage complexes (TOP1cc) at active genes. (E) Genome-wide distribution of γH2A.X in EUFA1341 cells expressing PALB2 WT or -MBD mutant, treated with CPT, was determined by ChIP-seq. γH2A.X peaks were categorized as upstream (≤5 kb upstream of gene bodies), downstream (≤5 kb downstream of gene bodies), or intergenic. (F) ChIP-qPCR quantification of γH2A.X (γH2A.X/H2A.X ratio). Where indicated, EUFA1341 cells expressing FLAG-PALB2 variants were treated with CPT alone or in combination with APH. Mean values ± SD (n = 3, with triplicate qPCR reactions).
Steady-State Chromatin Association of PALB2 Protects Transcribed Genes from Replication-Associated Stress.
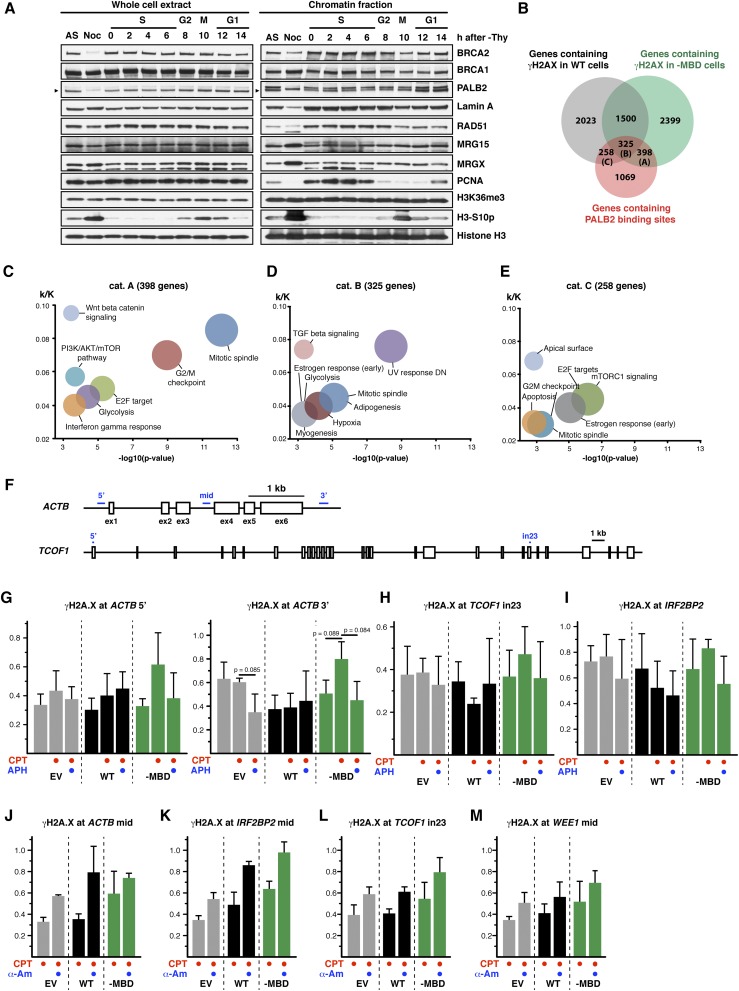
At the molecular level, CPT stabilizes the TOP1 cleavage complex, and its toxicity mainly arises from topologically constrained cellular events such as replication or transcription or conflicts between these processes (Fig. 4D) (35). Our analysis of chromatin-associated proteins in synchronized HeLa cells revealed no significant enrichment of PALB2, MRG15, or H3K36me3 on S-phase chromatin, contrasting with BRCA1 and RAD51 (Fig. S5A). Similarly, PALB2 has not been found to be enriched at stalled replication forks following exposure to HU or CPT in previous studies combining iPOND and proteomics (36, 37). These observations indicate that PALB2 association with chromatin is not induced by general stress associated with DNA replication.
Fig. S5.
(Related to Fig. 4) (A) HeLa Kyoto cells were synchronized by double thymidine block and released for the indicated time. The chromatin association of PALB2, BRCA2, BRCA1, RAD51, MRG15, and MRGX and the level of H3K36me3 were examined following fractionation of a synchronized cell population. PCNA and H3-S10p are markers for S phase and mitosis, respectively. Arrowheads indicate the specific band. (B) Venn diagram showing genes containing γH2A.X peaks in WT–expressing cells or in -MBD–expressing cells and PALB2-bound genes. (C–E) GSEA of PALB2-bound genes containing γH2A.X peaks only in -MBD–expressing cells (cat. A) (C), in both WT and -MBD expression cells (cat. B) (D), or only in WT-expressing cells (cat. C) (E). The ratio of k (number of genes containing PALB2-binding sites) against K (number of genes consisting of a given hallmark gene set) was plotted against negative log10 P values. The size of each circle indicates respective k value. (F) Schematic diagram of the beta actin (ACTB) and TCOF1 genes. Boxes and blue bars indicate exons and pairs of primers, respectively. (G–I) γH2A.X levels at the ACTB (G), TCOF1 (H), and IRF2BP2 (I) loci were analyzed by ChIP-qPCR. Where indicated, EUFA1341 cells stably expressing FLAG-PALB2 WT or -MBD mutant were treated for 17 h with 10 nM CPT alone or in combination with 0.5 µM APH. γH2A.X intensity is shown as the γH2A.X/H2A.X ChIP-qPCR signal ratio. Mean values ± SD (n = 3, with triplicate qPCR reactions). P values are for the two-tailed paired Student’s t test. (J–M) Same as G–I, except γH2A.X levels at the ACTB (J), IRF2BP2 (K), TCOF1 (L), and WEE1 (M) loci were analyzed by ChIP-qPCR following 17 h of treatment with 10 nM CPT alone or in combination with 4 µg/mL α-Am. Mean values ± SD (n = 3, with triplicate qPCR reactions).
Given the preferential localization of PALB2 to active genes, we hypothesized that MRG15-mediated PALB2 chromatin association has a unique function in the protection of genic regions. Indeed, ChIP-seq analysis of CPT-treated cells identified twice as many γH2A.X peaks in genic regions compared with intergenic regions (Fig. 4E), confirming that genic regions are more vulnerable to DNA stress associated with TOP1 defects (38). PALB2–MBD cells exhibited an overall increase in the number of γH2A.X peaks compared with PALB2WT cells, with 398 PALB2-bound genes with γH2A.X peaks detected only in PALB2–MBD cells (Fig. S5B, cat. A). The GSEA of these genes revealed a strong enrichment for genes involved in the mitotic spindle and G2/M checkpoint (Fig. S5C), which is similar to the top two categories identified in the GSEA for all PALB2-bound genes (Fig. S2D). Conversely, the GSEA for PALB2-bound genes with γH2A.X peaks in both PALB2–MBD and PALB2WT cells (Fig. S5B, cat. B) or only in PALB2WT cells (Fig. S5B, cat. C) showed an enrichment for different categories—namely, damage- and growth factor-responsive genes (Fig. S5 D and E). These observations support the notion that MRG15-mediated PALB2 chromatin association has a distinctive role in protecting periodic genes.
Additional ChIP-qPCR quantification of γH2A.X at PALB2-bound genes revealed that cells expressing PALB2–MBD triggered H2A.X hyperphosphorylation upon CPT treatment, something that was not seen with PALB2-defective cells (Fig. 4F and Fig. S5 F–I). This may reflect the partial complementation of EUFA1341 cells by PALB2–MBD, mediating a DNA stress response independently of MRG15, such as through BRCA1 interaction. Most importantly, the level of CPT-induced γH2A.X was reduced by DNA replication inhibition with APH, demonstrating that PALB2 in complex with MRG15 suppresses stress arising from DNA replication. In contrast, the level of γH2A.X was intensified by transcription inhibition with α-Am in all cell lines (Fig. S5 J–M). Taken together, these observations suggest that, although actively transcribed regions are subject to stress during DNA replication, transcription also helps to protect active genes from DNA stress. It is worth noting, however, that because α-Am is a slow-acting Pol II inhibitor, transcription may have remained active in the early phase of the treatment, resulting in the observed increase of γH2A.X through the conflict of transcription and replication, which was triggered by immediate CPT-induced stress. In this scenario, gradual reduction of PALB2 chromatin association (Fig. 1F) and overall inhibition of gene expression might have contributed to an accumulation of DNA stress at active genes.
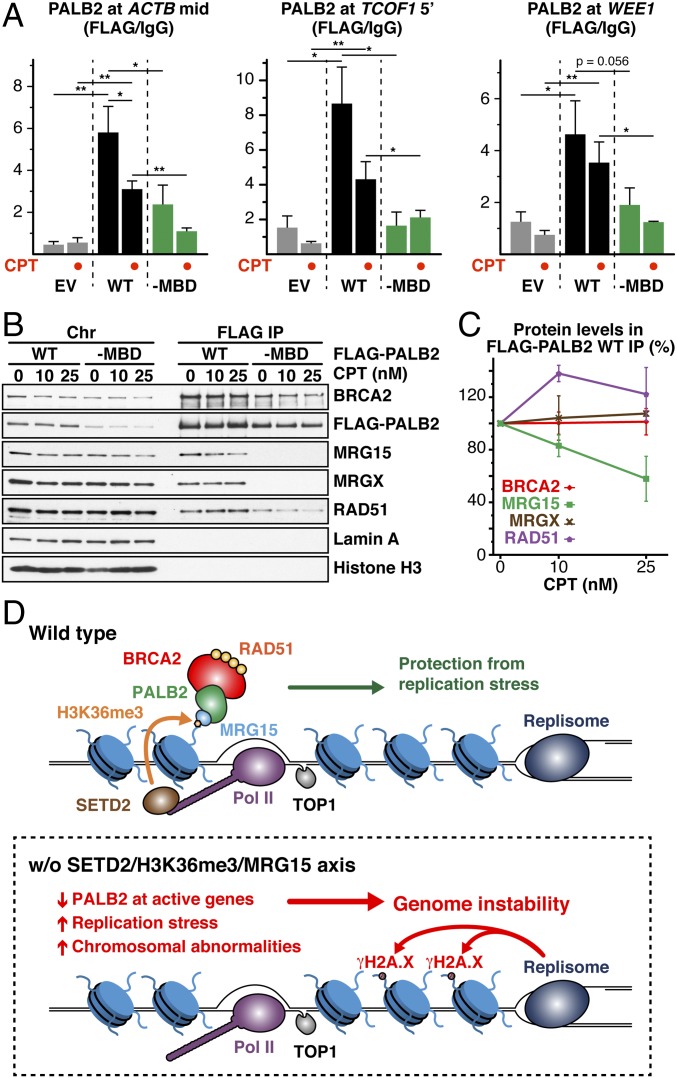
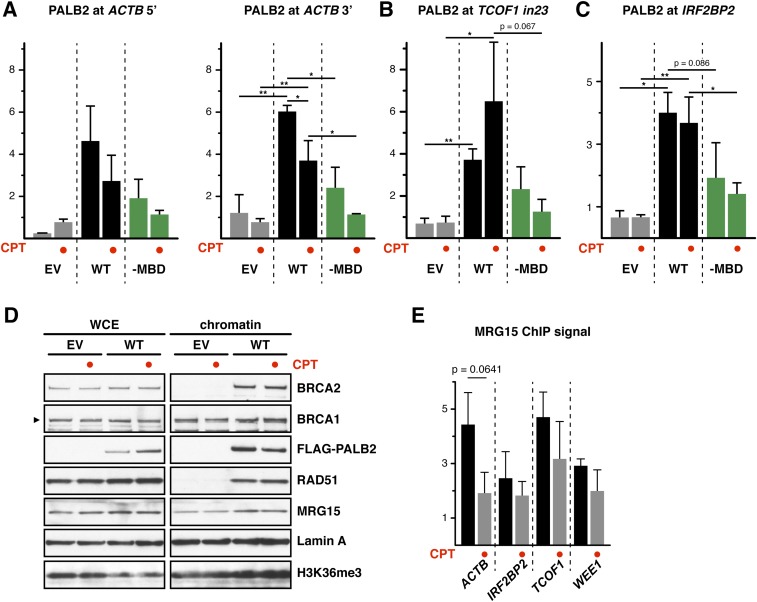
Lastly, we investigated whether CPT treatment elicits additional PALB2 chromatin association at genic regions. Unexpectedly, our ChIP-qPCR analyses revealed a decrease, rather than an increase, of PALB2WT occupancy at the majority of tested loci in CPT-treated cells (Fig. 5A and Fig. S6 A–C). In line with this observation, we found an overall reduction of chromatin-associated PALB2 under the same conditions, but no change of H3K36me3 was visible (Fig. S6D). Also, a reduced interaction between chromatin-associated PALB2 and MRG15 (Fig. 5 B and C) and lower MRG15 association with active genes (Fig. S6E) were found in CPT-treated cells, indicating that MRG15 plays an important role in controlling the dynamic association of PALB2 with active genes. Interestingly, PALB2–MBD also displayed a consistent reduction in its chromatin association upon CPT treatment (Fig. 5A and Fig. S6 A–C), suggesting that PALB2 displacement further involves MRG15-independent mechanisms. These observations demonstrate that the dynamic regulation of PALB2 chromatin association is highly complex.
Fig. 5.
Chromatin-associated PALB2 is mobilized in response to CPT treatment. (A) ChIP-qPCR quantification of FLAG-PALB2 variants. Mean values ± SD (n = 3, with triplicate qPCR reactions). Where indicated, EUFA1341 cells expressing FLAG-PALB2 variants were treated with CPT. Asterisks indicate two-tailed paired Student's t test; *P < 0.05, **P < 0.01. (B) Pull-down experiment showing the interaction between chromatin-associated PALB2 and MRG15 upon CPT treatment. (C) Quantification of BRCA2, MRG15, MRGX, and RAD51 levels in FLAG-PALB2 WT IP. Mean values ± SD (n = 2). (D) Proposed function of constitutive PALB2 chromatin association.
Fig. S6.
(Related to Fig. 5) (A–C) Occupancy of FLAG-PALB2 at ACTB (A), TCOF1 (B), and IRF2BP2 (C) loci was analyzed by FLAG ChIP-qPCR. FLAG-PALB2 signal is shown as the FC over the IgG control. Where indicated, EUFA1341 cells stably expressing FLAG-PALB2 WT or -MBD mutant were treated for 17 h with 10 nM CPT. Mean values ± SD (n = 3, with triplicate qPCR reactions). Statistical significance was determined using the two-tailed paired Student’s t test. *P < 0.05, **P < 0.01. (D) The chromatin association of the indicated proteins and the levels of H3K36me3 were examined following fractionation of CPT-treated EUFA1341 cells complemented with WT FLAG-PALB2. (E) MRG15 occupancy at the ACTB, IRF2BP2, TCOF1, and WEE1 loci was determined by ChIP-qPCR. MRG15 signal is shown as the FC over the IgG control. Mean values ± SD (n = 3, with triplicate qPCR reactions). Where indicated, EUFA1341 cells stably expressing WT FLAG-PALB2 were treated for 17 h with 10 nM CPT.
Evidence has emerged in recent years for the importance of histone modifications, which mark chromatin independently of DNA damage, in the regulation of HR. For example, dimethylation of histone H4 at lysine 20 (H4K20me2) contributes to the direct recruitment of HR suppressor 53BP1 (39), whereas nonmethylated H4K20 (H4K20me0), which is only found in newly replicated DNA, plays a critical role in the recruitment of TONSL–MMS22L, promoting HR repair in S phase (40). Also, H3K36me3 supports the constitutive association of LEDGF (p75) with active genes, which upon DNA damage recruits the repair factor CtIP to facilitate HR repair (24, 26). Our study establishes that H3K36me3 tethers the MRG15–PALB2 complex to undamaged chromatin, in a manner that makes PALB2 immediately available at active genes and, in this way, protects these more vulnerable regions of the genome from DNA damage (Fig. 5D). We propose that, in the event of DNA stress arising from DNA replication, such as that induced by irreversible TOP1-DNA adducts at active genes, PALB2 in complex with BRCA2 and RAD51 is mobilized and, by actively interacting with BRCA1, facilitates the protection and/or repair of nearby replication forks experiencing stress (41, 42). Interestingly, we noticed enhanced levels of BRCA2 and RAD51 association with PALB2–MBD but relatively decreased association of these proteins with PALB2–cc (Fig. 2B). These observations could be explained by a model in which full activation of PALB2 through stable interactions with BRCA1, BRCA2, and RAD51 is restricted by its association with MRG15, contributing to the prevention of undesired recombination events in the absence of DNA stress. We envision that PALB2 in complex with MRG15 acts as a local DNA stress sensor, whereas DNA stress swiftly converts it into an effective linker of BRCA1 and BRCA2, providing an elegant mechanism to ensure genome stability. Our study further provides a rationale for the development of small molecules blocking PALB2 binding to MRG15, which may potentiate the effect of CPT-based chemotherapy in the treatment of PALB2-proficient cancers.
Materials and Methods
Cell Lines and Plasmids.
All cell lines were grown at 37 °C in an incubator containing 5% CO2, using DMEM supplemented with 10% (vol/vol) FBS, penicillin (100 U/mL), and streptomycin (0.1 mg/mL). Plasmids for expression of PALB2, MRG15, and ChAM; HEK293 cells with conditional expression of FLAG-EGFP-PALB2; U2OS cells with constitutive expression of FLAG-PALB2 variants; and EUFA1341 cells complemented with FLAG-PALB2 were generated using standard methods. Oligonucleotides used for site-directed mutagenesis are listed in Table S1. Please refer to SI Materials and Methods for detailed procedures.
Table S1.
List of mutagenic DNA oligonucleotides used in this study
| Protein | Amino acid change(s) | Direction | Sequence |
| MRG15 | W53A | Forward | 5‘-CTTCATACATTACAGTGGTTGGAATAAAAATGCGGATGAATGGGTTCCG-3‘ |
| Reverse | 5‘-CGGAACCCATTCATCCGCATTTTTATTCCAACCACTGTAATGTATGAAG-3‘ | ||
| I245E/L246E | Forward | 5‘-TAAATTTGAGAGACCACAGTATGCTGAAGAGGAGGCAGATCATCCCGATGCACCCATGTC-3‘ | |
| Reverse | 5‘-GACATGGGTGCATCGGGATGATCTGCCTCCTCTTCAGCATACTGTGGTCTCTCAAATTTA-3‘ | ||
| PALB2 | L21P | Forward | 5‘-GGAAAAGTTAAAGGAGAAACCCGCATTCTTGAAAAGGG-3‘ |
| Reverse | 5‘-CCCTTTTCAAGAATGCGGGTTTCTCCTTTAACTTTTCC-3‘ | ||
| V410P/T413P | Forward | 5‘-GCCTTCTGTTTCCTGCAGAATATTATCCTAGAACACCACGAAGCATGTCC-3‘ | |
| Reverse | 5‘-GGACATGCTTCGTGGTGTTCTAGGATAATATTCTGCAGGAAACAGAAGGC-3‘ | ||
| A427P/I429P | Forward | 5‘-GAGGAAAGTAGCCGTGGAGCCTGTCCCTCAGAGTCATTTGGATGTC-3‘ | |
| Reverse | 5‘-GACATCCAAATGACTCTGAGGGACAGGCTCCACGGCTACTTTCCTC-3‘ | ||
| N418A/C419I | Forward | 5‘-TAGAACAACACGAAGCATGTCCGCTATCCAGAGGAAAGTAGCCGTGGAG-3‘ | |
| Reverse | 5‘-CTCCACGGCTACTTTCCTCTGGATAGCGGACATGCTTCGTGTTGTTCTA-3‘ | ||
| C419P | Forward | 5‘-AGAACAACACGAAGCATGTCCAATCCCCAGAGGAAAGTAGC-3‘ | |
| Reverse | 5‘-GCTACTTTCCTCTGGGGATTGGACATGCTTCGTGTTGTTCT-3‘ | ||
| C419G | Forward | 5‘-AACAACACGAAGCATGTCCAATGGCCAGAGGAAAG-3‘ | |
| Reverse | 5‘-CTTTCCTCTGGCCATTGGACATGCTTCGTGTTGTT-3‘ | ||
| ∆611–629 | Forward | 5‘-TGCTCAGAAAAACCAGTGGAGCCC-3‘ | |
| Reverse | 5‘-TGTGATACTGAGAAAAGACAGTAGTTGC-3‘ | ||
| ∆724–737 | Forward | 5‘-GGCCCTCAAGGCTCCTATGAAAAAG-3‘ | |
| Reverse | 5‘-CATGTCTGTGGTAGGCCTGTCATTATC-3‘ | ||
| ∆611–737 | Forward | 5‘-GGCCCTCAAGGCTCCTATGAAAAAG-3‘ | |
| Reverse | 5‘-TGTGATACTGAGAAAAGACAGTAGTTGC-3‘ | ||
| D611A/F612A | Forward | 5‘-TACTGTCTTTTCTCAGTATCACAGCCGCTCAGTTACCTGATGAAGACTTTGG-3‘ | |
| Reverse | 5‘-CCAAAGTCTTCATCAGGTAACTGAGCGGCTGTGATACTGAGAAAAGACAGTA-3‘ | ||
| L614A/P615A | Forward | 5‘-CTCAGTATCACAGACTTTCAGGCAGCTGATGAAGACTTTGGACCTC-3‘ | |
| Reverse | 5‘-GAGGTCCAAAGTCTTCATCAGCTGCCTGAAAGTCTGTGATACTGAG-3‘ | ||
| F619A/G620A | Forward | 5‘-GACTTTCAGTTACCTGATGAAGACGCTGCACCTCTTAAGCTTGAAAAAGTGAA-3‘ | |
| Reverse | 5‘-TTCACTTTTTCAAGCTTAAGAGGTGCAGCGTCTTCATCAGGTAACTGAAAGTC-3‘ | ||
| L622A/K623A | Forward | 5‘-GTTACCTGATGAAGACTTTGGACCTGCTGCGCTTGAAAAAGTGAAGTCCTGC-3‘ | |
| Reverse | 5‘-GCAGGACTTCACTTTTTCAAGCGCAGCAGGTCCAAAGTCTTCATCAGGTAAC-3‘ | ||
| A1025R | Forward | 5‘-AGGTCCAAGGGATGCAAGAACGTCTGCTTGGTACTAC-3‘ | |
| Reverse | 5‘-GTAGTACCAAGCAGACGTTCTTGCATCCCTTGGACCT-3‘ |
SiRNA-Mediated Knockdown, Chemical Cell Fractionation, Immunoprecipitation, and Immunofluorescence.
ON-TARGETplus SMARTpool siRNAs targeting human BRCA1, MRG15, MRGX, and PALB2 were purchased from Dharmacon and delivered to cells with Dharmafect 1 at a final concentration of 25 nM. The siRNA targeting human SETD2 was previously described (26). Whole-cell extract preparation, chemical cell fractionation, immunoprecipitation, and immunofluorescence analyses were carried out as previously described (14, 43), using antibodies listed in Table S2.
Table S2.
List of antibodies used in this study
| Antibody target | Supplier | Catalog no./name | Application | Dilution |
| BRCA1 | Millipore | OP107 | WB | 1:200 |
| BRCA2 | Millipore | OP95 | WB | 1:1,000 |
| FLAG | Sigma | A8592 | WB | 1:1,000 |
| FLAG | Sigma | F1804 | IF/ChIP | 1:500/2 µg |
| GFP | Sigma | G1544 | WB | 1:2,000 |
| Histone H2A.X | Cell Signaling | 2595 | WB | 1:500 |
| Histone H2A.X | Millipore | 07–627 | ChIP | 2 µg |
| Histone γH2AX [JBW301] | Millipore | 05–636 | WB/IF | 1:1,000/1:500 |
| Histone γH2AX | Millipore | 07–164 | ChIP | 2 µg |
| Histone H2B Ubiquityl Lys120 | Cell Signaling | 5546 | WB | 1:1,000 |
| Histone H3 | Bethyl | A300-823A | WB/ChIP | 1:2,000/2 µg |
| Histone H3 Tri-Methyl Lys36 | Cell Signaling | 4909 | WB/ChIP | 1:1,000/2 µg |
| Histone H3 Phospho Ser10 | Millipore | 06–570 | WB | 1:1,000 |
| Lamin A | Sigma | L1293 | WB | 1:1,000 |
| MRG15/MORF4L1 | Cell Signaling | 12169 | WB | 1:1,000 |
| MRG15 | Kind gift from Olivia Pereira-Smith, UT Health Science Center, San Antonio | ChIP | 2 µg | |
| MRGX/MORF4L2 | Novus Biologicals | NB-300–803 | WB | 1:1,000 |
| PALB2 | Bethyl | A301-246A | WB/ChIP | 1:1,000/3 μg |
| PALB2 | Santa Cruz | sc-160647 | WB | 1:100 |
| PCNA | Sigma | P8825 | WB | 1:2,000 |
| RAD51 [14B4] | Abcam | ab213 | WB | 1:1,000 |
| RAD51 | Yata et al., 2014 (43) | 7946 | IF/WB | 1:1,000/1:5,000 |
| Tubulin | Cell Signaling | 3873 | WB | 1:5,000 |
| Rabbit IgG | Life Technologies | 02–6102 | ChIP | 2–3 µg |
| Anti-mouse IgG HRP conjugated | Dako | P0447 | WB | 1:1,000 |
| Anti-rabbit IgG HRP conjugated | Dako | P0448 | WB | 1:2,000 |
| Anti-mouse IgG Alexa 488 conjugated | Life Technologies | A-11017 | IF | 1:400 |
| Anti-rabbit IgG Alexa 555 conjugated | Life Technologies | A-21430 | IF | 1:400 |
ChIP, chromatin immunoprecipitation; IF, immunofluorescence; WB, Western blotting.
PALB2 Affinity Purification–MS.
In brief, HEK293 cells were grown for 1 h in the presence of 2 μg/mL doxycycline to induce FLAG-EGFP-PALB2 expression. Whole-cell lysate was prepared and precleared with IgG agarose beads. FLAG-EGFP-PALB2–containing complexes were captured using GFP-Trap_A (Chromotek) from the precleared whole-cell lysate. Proteins were eluted twice, and elution fractions were pooled before in-solution digestion and quantitative liquid chromatography (LC)–MS/MS analysis. The dataset is available from ProteomeXchange (www.ebi.ac.uk/pride/archive/) with identifier PXD006391. Please refer to SI Materials and Methods for detailed procedures.
Cell Survival Assay and IC50 Values.
In 96-well plates, EUFA1341 cells complemented with FLAG-PALB2 variants were exposed to increasing concentrations of APH (0–20 µM), CPT (0–100 nM), HU (0–2 mM), or MMC (0–2 µM). After 4 d, cell proliferation was measured using WST-1 reagent (Roche Applied Science). Dose–response curves were fitted from the data pool of three independent experiments and the IC50 values calculated using Prism 6 (Graphpad Software). Please refer to SI Materials and Methods for detailed procedures.
Metaphase Spread Analysis.
EUFA1341 cells complemented with FLAG-PALB2 variants were grown to ∼70% confluence and treated with CPT (10 nM) for 17 h. Colcemid (0.1 µg/mL; Millipore) was added to the media and cells harvested after another 3.5 h. Cells were swollen with 0.56% KCl (6 min, room temperature), fixed with methanol/acetic acid (3:1), and dropped onto a microscope slide. Air-dried slides were stained with 0.4% Giemsa (Sigma) and mounted with Histomount (National Diagnostic). Images were captured with an Olympus BX60 microscope, using N-MSI-420–20 camera and Nuance software version 2.10.0 (Cambridge Research & Instrumentation).
ChIP.
In brief, EUFA1341 cells complemented with FLAG-PALB2 variants were harvested and fixed with 1% formaldehyde. Nuclei were isolated and genomic DNA was fragmented by partial micrococcal nuclease digestion. The majority of fragments were 150–900 bp in size (1–6 nucleosomes). The digested nuclei were further extracted, lysed by sonication, and cleared by centrifugation. A total of 20 µg of digested chromatin was incubated overnight at 4 °C with 2 µg of antibody. Immunocomplexes were captured with magnetic protein G beads, washed, and ChIP DNA eluted. After cross-link reversal, the samples were extracted twice with Phenol/Chloroform and Ethanol precipitated. The PCR analysis was performed on a Rotorgene Q Real-Time PCR System (Qiagen) using the SensiFAST SYBR No-Rox kit (Bioline). Please refer to SI Materials and Methods for full procedures. Where indicated, cells were treated with 10 nM CPT, 0.5 µM APH, or 4 µg/mL α-Am.
ChIP-Seq and GSEA.
DNA ChIP fraction was sequenced on the Applied Biosystems SOLiD platforms (SOLiD 5500) and analyzed. The dataset is available from the National Center for Biotechnology Information, Sequence Read Archive (NCBI-SRA) (https://www.ncbi.nlm.nih.gov/Traces/sra/) under accession no. SRP105310. ChIP-seq data of HeLa-S3 H3K36me3 and the corresponding control sample were obtained from ENCODE project (SRA; SRX067462 and SRX067410, respectively). GSEA was performed using the GSEA software provided by the Broad Institute (software.broadinstitute.org/gsea/msigdb/annotate.jsp). Please refer to SI Materials and Methods for detailed procedures.
Statistics and Quantitative Analysis.
For experiments reproduced at least three times, statistical significance was determined using the indicated test. Data were analyzed using Excel (Microsoft Software) and Prism 6 (Graphpad Software). Quantitative analyses of Western blots (WBs) were performed using the Fiji distribution of ImageJ. For chromatin-bound protein quantitative analyses, the data presented were normalized to the protein level present in the input and expressed as percentage of the control condition.
SI Materials and Methods
Cell Culture and Cell Lines.
HEK293, HeLa Kyoto, HT-1080, U2OS, and EUFA1341 cell lines were grown in DMEM supplemented with 10% (vol/vol) FBS, penicillin (100 U/mL), and streptomycin (0.1 mg/mL). All cells were grown at 37 °C in an incubator containing 5% CO2. HEK293 Flp-In T-REx cells were cotransfected with pOG44 and pcDNA5/FRT/TO/FLAG-EGFP-PALB2 or pcDNA5/FRT/TO/FLAG-EGFP vectors, followed by selection with blasticidin (15 µg/mL) and hygromycin B (150 μg/mL). To induce protein expression, cells were grown for 1 h in the presence of 2 μg/mL doxycycline. U2OS Flp-In T-REx cells were transfected with pSUPERIOR.puro/P2shRNA plasmid (14), and a cell line conditionally expressing the P2shRNA was cloned following puromycin selection (1 μg/mL). Established U2OS Flp-In T-REx P2shRNA cells were then used to generate stable isogenic cell lines by cotransfection with pOG44 and pcDNA5/FRT/GW/FLAG-PALB2 (WT or -MBD) vectors, followed by selection with blasticidin (15 µg/mL) and hygromycin B (150 μg/mL). EUFA1341 cells were transfected with pCEP4-GW/FLAG-PALB2 vectors and stable cell lines expressing FLAG-PALB2 variants generated following hygromycin selection (300 μg/mL); cells were later maintained using 150 µg/mL hygromycin. U2OS cells with CRISPR knockout of SETD2, conditional overexpression of KDM4A, and constitutive overexpression of WT or mutant (K36M) histone variant H3.3 were maintained as previously described (26, 44).
Plasmids.
For GFP-fusion expression in HEK293T cells, MRG15 in pDONR221, ChAM in pENTR1A, or PALB2 in pENTR3C was transferred to pcDNA-DEST53 (Life Technologies). For FLAG-PALB2 fusion expression in U2OS Flp-In T-REx P2shRNA cells, PALB2 in pENTR3C was transferred to pcDNA5/FRT-GW/N3×FLAG, a modified pcDNA5/FRT vector (Life Technologies) with an N3×FLAG-Gateway cassette at HindIII/XhoI sites. For FLAG-PALB2 fusion expression in EUFA1341 cells, PALB2 in pENTR3C was transferred to pCEP4-GW/N3×FLAG, a modified pCEP4 vector (Life Technologies) with an N3×FLAG-Gateway cassette at HindIII/XhoI sites. MRG15, ChAM, and PALB2 point mutations were introduced in Gateway entry vectors using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol and were confirmed by DNA sequencing. Mutagenic oligonucleotide sequences are listed in Table S1. pcDNA5/FRT/TO/FLAG-EGFP-PALB2 was previously described (14).
FLAG-EGFP-PALB2 Affinity Purification–MS.
HEK293 Flp-In T-REx cells expressing either FLAG-EGFP or FLAG-EGFP-PALB2 (∼15 × 106 cells) were collected by scraping on ice and were washed first with cold PBS and with cold PBS supplemented with 1× protease inhibitor mixture (Sigma, P2714). Cell pellets were collected by centrifugation for 5 min at 500 × g, 4 °C, and lysed into NET150 buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, and 2 mM EDTA) supplemented with 0.5% n-Dodecyl β-d-maltoside detergent (Millipore, 324255), 1 mM DTT, 1× protease inhibitor mixture (Sigma, P2714), phosphatase inhibitors (20 mM NaF, 20 mM β-glycerophosphate, 1 mM Na3VO4), lysine deacetylase inhibitor (5 mM sodium butyrate, NaB), 1 mM MgCl2, and 125 U/mL of benzonase (Novagen, 71206–3). After 30 min of incubation on ice, cell debris were removed by 30 min of centrifugation at 16,100 × g, 4 °C. The supernatant containing whole-cell protein extract was collected and precleared onto IgG agarose beads (Sigma, A0919) for 1 h, rotating at 4 °C. After centrifugation for 5 min at 500 × g, 4 °C, precleared whole-cell protein lysate was collected and incubated with GFP-Trap_A (Chromotek) to perform GFP pull-down. After 1 h of protein binding on beads by rotating at 4 °C, GFP-Trap beads were collected by 5 min of centrifugation at 500 × g, 4 °C, and washed three times in NET150 buffer supplemented with 0.1% n-Dodecyl β-d-maltoside detergent, 1 mM DTT, 1× protease inhibitor mixture, phosphatase inhibitors (20 mM NaF, 20 mM β-Glycerophosphate, 1 mM Na3VO4), lysine deacetylase inhibitor (5 mM NaB), and 1 mM MgCl2. Proteins were eluted off beads using 0.2 M glycine, pH 2.3, for 5 min rotating at 4 °C. After centrifugation at 500 × g for 5 min, at 4 °C, the protein eluate was collected and neutralized with 1 M Tris·HCl, pH 8.8. The elution procedure was repeated once, and the eluted fractions were pooled before in-solution digestion and LC–MS/MS analysis.
Proteomic Analysis.
Protein digestion.
GFP pulled-down proteins were denatured in 4 M urea dissolved into 50 mM triethylammonium bicarbonate (TEAB), reduced with 10 mM Tris-(2-carboxyethyl)phosphine (TCEP) for 30 min at room temperature, alkylated with 50 mM chloroacetamide for 30 min at room temperature in the dark, digested with endoproteinase Lys-C (Roche) for 2 h at 37 °C, followed by trypsin digestion for 16 h at 37 °C. Before the trypsin digestion, the urea concentration was diluted down to 1 M into 50 mM TEAB, and calcium chloride was added at 1 mM final. The digestion steps were performed in a Thermomixer compact (Eppendorf) shaking at 650 rpm. Trypsin digestion was stopped by addition of trifluoroacetic acid (TFA) to a final concentration of 1%. Digested samples were centrifuged for 30 min at 16,100 × g at 4 °C to remove aggregates. Peptide mixtures were further desalted using handmade C18 tips, as follows: C18 tips were washed twice with 100% acetonitrile after centrifugation for 5 min at 376 × g at room temperature. Peptides were then loaded onto C18 tips by centrifugation at 4,000 × g at room temperature. After two washing steps with 0.1% TFA solution, desalted peptides were eluted into 50% acetonitrile/0.1% TFA solution and dried using a SpeedVac.
LC-MS/MS analysis.
Peptides were resuspended in 10% formic acid. They were separated on an Ultimate 3000 UHPLC system (Thermo Fischer Scientific) and electrosprayed directly into a QExactive mass spectrometer (Thermo Fischer Scientific) through an EASY-Spray nano-electrospray ion source (Thermo Fischer Scientific). The peptides were trapped on a C18 PepMap100 precolumn (300 µm i.d. × 5 mm, 100 Å, Thermo Fisher Scientific) using solvent A (0.1% Formic Acid in water) at a pressure of 500 bar. The peptides were separated on an in-house packed analytical column (75 µm i.d. packed with ReproSil-Pur 120 C18-AQ, 1.9 µm, 120 Å, Dr. Maisch GmbH) using a linear gradient [length, 120 min; 7–28% solvent B (0.1% formic acid in acetonitrile); flow rate, 200 nL/min]. The raw data were acquired on the mass spectrometer in a data-dependent mode (DDA). Full-scan MS spectra were acquired in the Orbitrap (scan range, 350–2,000 m/z; resolution, 70000; AGC target, 3e6; maximum injection time, 50 ms). After the MS scans, the 20 most intense peaks were selected for Higher-energy collisional dissociation (HCD) fragmentation at 30% of normalized collision energy. HCD spectra were also acquired in the Orbitrap (resolution, 17,500; AGC target, 5e4; maximum injection time, 120 ms) with first fixed mass at 180 m/z. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006391.
Database searching and data filtering.
Tandem mass spectra were searched using SEQUEST within Proteome Discoverer 1.4 (ThermoFischer Scientific, version 1.4.0.288) against a nonredundant protein sequence database for Homo sapiens containing 20,160 protein sequence entries (UniProt, release 2015–09-25), in which the PALB2 protein sequence (UniProt accession no. Q86YC2) was replaced by the PALB2 protein sequence fused to FLAG-EGFP tag in its terminal region (named as P0000_DEF USER). Additionally, contaminant protein sequences such as endoproteinase Lys-C (UniProt accession no. Q7M135, lysyl endopeptidase, Lysobacter enzymogenes) and trypsin (UniProt accession no. P00761, trypsin, Sus scrofa) were inserted in the database. During database searches, cysteine residues were considered to be fully carbamidomethylated (+57 Da statically added), methionine considered to be oxidized (+16 Da dynamically added), and two missed cleavages were permitted. Peptide mass tolerance was set at 50 ppm and 0.02 Da on the precursor and fragment ions, respectively. The protein identification list was filtered at a false discovery rate below 1%.
Data analysis.
PALB2 interacting protein partners were identified by quantitative proteomic and statistical analyses, after sorting out proteins significantly enriched in the FLAG-EGFP-PALB2 protein dataset versus FLAG-EGFP (negative control). The quantitative analysis was based on a label-free quantitation method using Normalized Spectral Abundance Factor (NSAF) as a measure of relative protein abundance within the protein mixture. spectral abundance factor (SAF) and NSAF values were calculated as previously described (45); as the number of spectral counts (PSM) that identify a protein, divided by the protein length (L), the PSM/L value represents SAF, which is then divided by the sum of PSM/L for all proteins in the experiment. NSAF values were calculated after contaminant proteins such as keratins, endoproteinase lys-C, trypsin, and immunoglobulins were removed. For better visualization of the data, NSAF values were multiplied by 100 (NSAF*100). The statistical analysis was performed on NSAF values from three biological replicate experiments, using a Power Law Global Error Model (PLGEM) (46). The Signal-to-Noise (STN) ratio calculated from PLGEM analysis indicates the level of enrichment for each protein identified in FLAG-EGFP-PALB2 versus FLAG-EGFP pull-down. The P value indicates the significance of the enrichment. Before log transformation of the data, zero values were replaced by a minimum value in the dataset. Scatter and bubble plots were generated in Excel.
Nucleosome Pull-Down Assays.
GFP-tagged ChAM and MRG15 variants were affinity-purified from HEK293T cells. To avoid contamination with endogenous histones, the proteins were purified from the cytoplasmic fraction as follows: Cells were lysed in ice-cold sucrose buffer (10 mM Tris·HCl, pH 7.5; 20 mM KCl; 250 mM Sucrose; 2.5 mM MgCl2; 10 mM benzamidine hydrochloride; 10 mM NaF; 1 mM Na3VO4; 10 mM Na-β-glycerophosphate; Sigma protease inhibitor mixture) and the intact nuclei pelleted by centrifugation for 5 min at 500 × g, 4 °C. The cytoplasmic fraction was collected and, following addition of 150 mM NaCl, was cleared by centrifugation (30 min, 16,100 × g, 4 °C). The proteins were purified using GFP-Trap_A (gta-20, Chromotek) according to the manufacturer recommendations, except beads were washed four times with NETN150 buffer (50 mM Tris·HCl, pH 8.0; 150 mM NaCl; 2 mM EDTA; 0.5% Nonidet P-40; 10 mM benzamidine hydrochloride; 10 mM NaF; 1 mM Na3VO4; 10 mM Na-β-glycerophosphate; Sigma protease inhibitor mixture). Human nucleosomes were partially purified from HEK293T cells. Briefly, chromatin was isolated as described for chemical cellular fractionation and digested with 50 gel units of micrococcal nuclease (M0247S, NEB) per milligram of DNA (12 min, 37 °C). The reaction was stopped with 5 mM EGTA and the nucleosomes suspension cleared by centrifugation (30 min, 16,100 × g, 4 °C). Recombinant nucleosomes were a kind gift of Fabrizio Martino, Centro de Investigaciones Biológicas (CIB), Madrid, and Daniela Rhodes. For human nucleosome pull-downs, 250 µg of partially purified nucleosomes, in 500 µL NETN150 buffer, were mixed with ∼400 ng of immobilized GFP-tagged fusion protein and incubated at 4 °C for 2 h on a rotating wheel. The beads were washed four times with NETN150 buffer and samples analyzed by SDS/PAGE and WB. Recombinant nucleosome pull-downs were carried out as above, except 100 ng of nucleosomes and GFP-fusion protein was used.
Cell Survival Assay and IC50 Values.
EUFA1341 cell lines stably expressing FLAG-PALB2 variants were seeded in 96-well plates at a density of ∼2,000 cells per well and cultured 24 h before treatments. For the dose–response curves, cells were treated with 0–20 µM APH (Fisher BioReagents, BP615-1), 0–100 nM CPT (Calbiochem, 208925), 0–2 mM HU (Sigma-Aldrich, H8627), or 0–2 µM MMC (Sigma-Aldrich, M4287) for 4 d. Cell proliferation was measured using WST-1 reagent (Roche Applied Science), as previously described (14). Two technical replicates were performed for each of three experiments. The dose–response curves were fitted to the data pool and the IC50 values calculated using Prism 6 (Graphpad Software).
Foci Quantification.
RAD51 and γH2AX foci were automatically quantified using the robust FoCo algorithm, as previously described (47). Foci pictures from Olympus BX60 of each individual experiment were all acquired at the same exposure time, allowing for significant comparisons. Noteworthy, foci were filtered with respect to a minimal radius and intensity for better exclusion of background signal. Cell nuclei were also discriminated using a minimal radius in pixel for DAPI signal.
ChIP.
For ChIP, EUFA1341 cells expressing FLAG-PALB2 variants were harvested with trypsin and washed twice with ice-cold PBS. Approximately 2 × 107 cells in 2 mL PBS, pH 7.2 (Gibco), were fixed for 8 min at room temperature with 1% formaldehyde (F1635, Sigma) in PBS and quenched for 5 min with 125 mM Glycine (G8898, Sigma). After two washes with ice-cold PBS, cells were incubated for 10 min on ice in 2 mL of lysis buffer (10 mM Pipes, pH 7.5; 85 mM KCl; 0.5% Nonidet P-40; 10 mM benzamidine hydrochloride; P2714 Sigma protease inhibitor mixture). Isolated nuclei were washed once with micrococcal nuclease (MNase) buffer (10 mM Tris·HCl, pH 7.5; 15 mM NaCl; 60 mM KCl; 1.5 mM CaCl2; 3 mM MgCl2; 10 mM benzamidine hydrochloride; Sigma protease inhibitor mixture) and then incubated for 30 min at 37 °C with 200 gel units of micrococcal nuclease (M0247S, NEB) in 400 µL of MNase buffer; the reaction was stopped by addition of 20 µL 0.5 M EDTA and 5 min of incubation on ice. The digested nuclei were pelleted, resuspended in 1.2 mL of ChIP buffer (20 mM Tris·HCl, pH 7.5; 150 mM NaCl; 1 mM EDTA; 0.5 mM EGTA; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% SDS; 10 mM benzamidine hydrochloride; Sigma protease inhibitor mixture), and incubated for 10 min on ice. We sonicated 4 × 300 µL aliquots in 1.7 mL Axygen tubes using a cooled Bioruptor (Diagenode) on high setting for 30 cycles of 30 s on and 30 s off. Samples were cleared by centrifugation for 10 min at 16,100 × g, 4 °C. A total of 30 µL of chromatin was reversed cross-linked, DNA purified with a QIAquick spin column, quantified, and analyzed by agarose gel electrophoresis. An amount of digested chromatin equivalent to 20 µg DNA was mixed with 2 µg of control mouse IgG or mouse anti-FLAG antibody (F1804, Sigma) in 500 µL of ChIP buffer and incubated overnight at 4 °C on a rotating wheel. A total of 30 µL of protein G Dynabeads blocked with 5 mg/mL BSA in ChIP buffer was added to each sample. After 2 h of incubation at 4 °C on a rotating wheel, the beads were washed twice with low-salt wash buffer (20 mM Tris·HCl, pH 8; 150 mM NaCl; 2 mM EDTA; 1% Triton X-100; 0.1% SDS; 10 mM benzamidine hydrochloride; Sigma protease inhibitor mixture), twice with high-salt wash buffer (20 mM Tris·HCl, pH 8; 500 mM NaCl; 2 mM EDTA; 1% Triton X-100; 0.1% SDS; 10 mM benzamidine hydrochloride; Sigma protease inhibitor mixture), once with LiCl wash buffer (20 mM Tris·HCl, pH 8; 250 mM LiCl; 1 mM EDTA; 1% Nonidet P-40; 1% sodium deoxycholate; 10 mM benzamidine hydrochloride; Sigma protease inhibitor mixture), and twice with TE buffer (10 mM Tris·HCl, pH 8.0; 1 mM EDTA). The beads were resuspended in 200 µL elution buffer (TE buffer; 1% SDS) and incubated with shaking for 20 min at 65 °C. After cross-link reversal, the samples were extracted twice with phenol/chloroform and ethanol-precipitated. The PCR analysis was performed on a Rotorgene Q Real-Time PCR System (Qiagen) using the SensiFAST SYBR No-Rox kit (Bioline). Antibodies used for H2A.X, γH2A.X, H3, H3K36me3, and MRG15 ChIP are listed in Table S2.
ChIP-Seq and GSEA.
For PALB2 ChIP-seq analysis, HeLa Kyoto BRCA2-NFLAP cells (48) were synchronized in S phase using standard double thymidine block procedure. At 3 h after release into thymidine-free medium, cells were fixed for 10 min at room temperature with 1% formaldehyde (F1635, Sigma) and quenched for 10 min with 125 mM Glycine (G8898, Sigma). The DNA–protein complexes were sheared into ∼500-bp fragments by sonication, and endogenous PALB2 was immunoprecipitated using the PALB2 antibody as reported previously (18). DNA ChIP fraction was sequenced on the Applied Biosystems SOLiD platforms (SOLiD 5500). Sequenced single-end 50-bp reads were aligned to the human genome (UCSC hg19) using Bowtie v1.1.0 (49), allowing two mismatches in the first 28 bases per read and outputting only uniquely mapped reads (–n2 –m1 option). Peak calling and visualization were performed using DROMPA version 2.6.4 (50). Peaks were identified by the following criteria: (i) fold enrichment (IP reads/WCE reads) > 2, (ii) ChIP/Input enrichment P value < 0.01 (binomial test), and (iii) ChIP internal enrichment P value < 0.01 (local Poisson test). To eliminate uncertain sites, we ignored regions with low mappability (mappability < 0.3 for a 1-kb window). The dataset is available from the NCBI-SRA (https://www.ncbi.nlm.nih.gov/Traces/sra/) (accession no. SRP105310). GSEA was performed using the GSEA software provided by the Broad Institute (software.broadinstitute.org/gsea/index.jsp). H3K36me3 peaks were called by MACS2 version 2.1.1 (51) with –broad option. The averaged profile (Fig. 1E) was generated by DROMPA3 with total read normalization.
Supplementary Material
Acknowledgments
We thank Dr. H. Joenje (VU University Medical Center, The Netherlands) for sharing the EUFA1341/FA-N fibroblasts; Dr. A. Sartori (Institute of Molecular Cancer Research, Switzerland) for U2OS Flp-In T-REx stable cells; and Prof. C. J. Norbury (Sir William Dunn School of Pathology, United Kingdom) for critical reading of the manuscript. This work was supported by Wellcome Trust Senior Research Fellowship 101009 (to F.E.), Grant-in-Aid for Scientific Research 15K18465 (to R.N.), and Grants-in-Aid for Scientific Research 15H02369, 15K21761, and 15H02369 from MEXT and AMED-CREST (to K.S.). T.C.H. is a recipient of Medical Research Council Grant MC-PC-12001. D.R. is thankful for the support of Singapore Ministry of Education Academic Research Fund Tier 3 Grant MOE2012-T3-1-001 and the NTU Institute of Structural Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Proteomics and ChIP-sequencing datasets are available via ProteomeXchange (identifier PXD006391) and NCBI-SRA (accession no. SRP105310), respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620208114/-/DCSupplemental.
References
- 1.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman N, et al. Breast Cancer Susceptibility Collaboration (UK) PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 5.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 6.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 10.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorslund T, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa T, et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123:1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sy SM, Huen MS, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Gardini A, Baillat D, Cesaroni M, Shiekhattar R. Genome-wide analysis reveals a role for BRCA1 and PALB2 in transcriptional co-activation. EMBO J. 2014;33:890–905. doi: 10.1002/embj.201385567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar GS, et al. Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. J Mol Biol. 2012;422:519–531. doi: 10.1016/j.jmb.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun XJ, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 22.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho S, et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife. 2014;3:e02482. doi: 10.7554/eLife.02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfister SX, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Reports. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 28.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 29.Lu C, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez D, et al. A high-resolution transcriptome map of cell cycle reveals novel connections between periodic genes and cancer. Cell Res. 2016;26:946–962. doi: 10.1038/cr.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J Biol Chem. 2009;284:21127–21131. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie T, Zmyslowski AM, Zhang Y, Radhakrishnan I. Structural basis for multi-specificity of MRG domains. Structure. 2015;23:1049–1057. doi: 10.1016/j.str.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zellweger R, et al. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 36.Dungrawala H, et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeyre C, et al. Nascent DNA proteomics reveals a chromatin remodeler required for topoisomerase I loading at replication forks. Cell Reports. 2016;15:300–309. doi: 10.1016/j.celrep.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saredi G, et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature. 2016;534:714–718. doi: 10.1038/nature18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orthwein A, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yata K, et al. BRCA2 coordinates the activities of cell-cycle kinases to promote genome stability. Cell Reports. 2014;7:1547–1559. doi: 10.1016/j.celrep.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfister SX, et al. Inhibiting WEE1 selectively kills histone H3K36me3-deficient cancers by dNTP starvation. Cancer Cell. 2015;28:557–568. doi: 10.1016/j.ccell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Florens L, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pavelka N, et al. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol Cell Proteomics. 2008;7:631–644. doi: 10.1074/mcp.M700240-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Lapytsko A, Kollarovic G, Ivanova L, Studencka M, Schaber J. FoCo: A simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics. 2015;16:392. doi: 10.1186/s12859-015-0816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lekomtsev S, Guizetti J, Pozniakovsky A, Gerlich DW, Petronczki M. Evidence that the tumor-suppressor protein BRCA2 does not regulate cytokinesis in human cells. J Cell Sci. 2010;123:1395–1400. doi: 10.1242/jcs.068015. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakato R, Itoh T, Shirahige K. DROMPA: Easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells. 2013;18:589–601. doi: 10.1111/gtc.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.