Significance
Precise cell division is crucial for all cellular life. In the pneumococcus, which kills more than a million people annually, the protein MapZ was recently identified as a marker for the division site. By developing a suite of tools, including a chromosome labeling system and CRISPR/Cas9-based perturbation of chromosome segregation, we mapped the pneumococcal cell cycle in detail. We show that MapZ is not involved in division site selection but is important for setting the correct angle of the division plane. Importantly, we demonstrate that the pneumococcus has a unique chromosomal organization and that timely replication and segregation of the chromosomal origin to the future cell division site is a key determinant for correct positioning of cell division proteins.
Keywords: cell division, Streptococcus pneumoniae, SMC, chromosome organization, FtsZ
Abstract
Accurate spatial and temporal positioning of the tubulin-like protein FtsZ is key for proper bacterial cell division. Streptococcus pneumoniae (pneumococcus) is an oval-shaped, symmetrically dividing opportunistic human pathogen lacking the canonical systems for division site control (nucleoid occlusion and the Min-system). Recently, the early division protein MapZ was identified and implicated in pneumococcal division site selection. We show that MapZ is important for proper division plane selection; thus, the question remains as to what drives pneumococcal division site selection. By mapping the cell cycle in detail, we show that directly after replication both chromosomal origin regions localize to the future cell division sites, before FtsZ. Interestingly, Z-ring formation occurs coincidently with initiation of DNA replication. Perturbing the longitudinal chromosomal organization by mutating the condensin SMC, by CRISPR/Cas9-mediated chromosome cutting, or by poisoning DNA decatenation resulted in mistiming of MapZ and FtsZ positioning and subsequent cell elongation. Together, we demonstrate an intimate relationship between DNA replication, chromosome segregation, and division site selection in the pneumococcus, providing a simple way to ensure equally sized daughter cells.
In eukaryotic cells, DNA replication, chromosome segregation, and cell division are tightly coordinated and separated in time (1–3). In most bacteria, this is less obvious as these processes occur simultaneously. However, in the last decade, it has become evident that the bacterial cell cycle is a highly regulated process in which both cell-cycle proteins as well as the chromosome have defined spatial and temporal localization patterns (4, 5). The tubulin-like protein FtsZ (forming the Z-ring) is key for initiating divisome assembly in virtually all bacteria (6). Accurate cell division is mostly exerted through regulation of FtsZ positioning in the cell. However, the mechanisms that control FtsZ positioning can be highly diverse among bacterial species. In well-studied rod-shaped model organisms, such as Bacillus subtilis and Escherichia coli, precise formation of the Z-ring at midcell is regulated by the so-called Min-system and nucleoid occlusion (7, 8). These are both negative regulators of FtsZ polymerization, which prevent premature Z-ring formation and cell division near cell poles and across unsegregated chromosomes, respectively (9, 10). These two regulatory mechanisms have been found in many bacteria. However, in some species, other dedicated proteins are used for this purpose, including MipZ in Caulobacter crescentus (11), SsgB in Streptomyces coelicolor (12), and PomZ in Myxococcus xanthus (13). It is important to note that none of these FtsZ regulation mechanisms are essential for bacterial growth, and other mechanisms of cell-cycle control must therefore also exist (14–16). In this context, it has been suggested that there are important links between different cell-cycle processes, such as DNA replication and Z-ring assembly (15–19).
As for the opportunistic pathogen Streptococcus pneumoniae, the orchestration of replication and chromosome dynamics remains largely unknown. Ovococcal S. pneumoniae lacks a nucleoid occlusion system and has no Min-system (20, 21). Recently, MapZ (or LocZ) was proposed to be a division site selector in S. pneumoniae (22, 23). This protein localizes early at new cell division sites and positions FtsZ by a direct protein–protein interaction (22). MapZ is binding peptidoglycan (PG) via an extracellular domain and is also a protein substrate of the master regulator of pneumococcal cell shape, the Ser/Thr kinase StkP (22–24). Together, this suggests that for division site selection in S. pneumoniae, FtsZ is controlled via the MapZ beacon at midcell (15, 25, 26). Furthermore, the mechanisms of chromosome segregation in pneumococci also seem to be different from rod-shaped model bacteria; S. pneumoniae harbors a single circular chromosome with a partial partitioning system that only contains the DNA-binding protein ParB with parS binding sites but lacks the ATPase ParA. Furthermore, the ubiquitous condensin protein SMC is not essential (27). Although both ParB and SMC are involved in chromosome segregation in pneumococci, parB and smc mutants have minor growth defects and a low percentage of anucleate cells (1–4%) (27, 28). In contrast, in B. subtilis, deletion of smc is lethal at normal growth conditions (29). To gain more understanding of the progression of the pneumococcal cell cycle, we therefore investigated the relationship between DNA replication, chromosome segregation, and division site selection in this pathogen. We show that MapZ is not involved in division site selection as suggested before but is crucial for correctly placing the Z-ring perpendicularly to the length axis of the cell. By establishing tools to visualize the replisome and different genetic loci, we show that there is an intimate relationship between DNA replication, chromosome segregation, and division. Importantly, we demonstrate that correct chromosomal organization acts as a roadmap for accurate division site selection in pneumococcus and possibly other bacteria.
Results
MapZ Identifies the Division Plane but Does Not Select the Division Site.
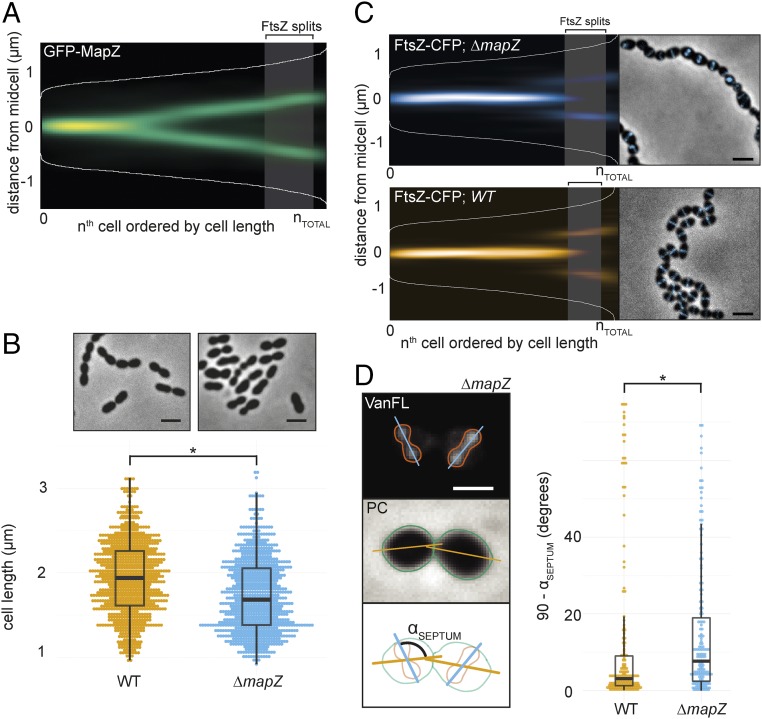
MapZ has been proposed as the division site selector in S. pneumoniae, and Z-ring positioning and angle were shown to be perturbed in mapZ-null mutants (22, 23). In contrast to what can be expected for a protein involved in division site selection, ΔmapZ mutants are not elongated but are on average shorter than wild-type cells with relatively minor distortions in cell morphology (22, 23), raising questions on what the actual biological function of MapZ is (30). To reassess the ΔmapZ phenotype, we fused MapZ at its N terminus to a monomeric superfolder green fluorescent protein (GFP). Using the cell-segmentation software Oufti (31), to detect cell outlines and fluorescent signals, in combination with the newly developed R-package SpotprocessR to analyze the microscopy data (see Materials and Methods), GFP-MapZ localization was determined in exponentially growing cells (balanced growth). Note that by balancing growth by rediluting exponentially growing cells several times, pneumococcal cell length becomes an accurate proxy for the cell-cycle state (21, 24). Cells were ordered by length, and this order was plotted against the position of GFP-MapZ on the long axis of the cells as a density histogram (Fig. 1A). In line with previous reports (22, 23), GFP-MapZ localized to the division site (Fig. 1A). As a new cell wall is synthesized at midcell (20), MapZ seems to move from the current division site, probably via attachment to PG, and ends up at the interface between the new and old cell halves. This position will eventually become the future division site where the Z-ring assembles. It should be noted that we do not observe three MapZ-rings, which is in line with previous results from S. pneumoniae D39 (23) but different from what has been found for S. pneumoniae R800 (22). Deleting mapZ in the encapsulated D39 genetic background led to irregularly shaped and shorter, sometimes branched or clustered cells (Fig. 1B). Similar observations were made in serotype 4 strain TIGR4 and in the unencapsulated R6 laboratory strain (SI Appendix, Fig. S1 A and B).
Fig. 1.
MapZ sets the pneumococcal division plane but is not involved in division site selection. (A) Fluorescence microscopy of 2,976 cells and 7,020 spot localizations were quantified and analyzed using Oufti and SpotprocessR (see Materials and Methods). The distance of GFP-MapZ (strain RR101) from midcell was plotted in a heatmap where all localizations are ordered by cell length, and the color saturation represents the density of the localizations. The shaded area represents the point in the cell cycle where 50% of the FtsZ proteins move to the 1/4 positions of the cell (124 ± 9.04% of the mean cell length; see Materials and Methods). (B) Cell size distribution of wild-type D39 and ΔmapZ cells (strain RR93), representing measurements of 1,692 and 705 cells, respectively. (Top) Phase contrast microscopy images. (Scale bar, 2 µm.) (C) The localization of FtsZ-CFP in wild-type (strain RR23) and ΔmapZ (strain RR105) cells as shown by histograms and micrographs from overlays of phase contrast images with CFP signal. (Scale bar, 2 µm.) The plots are based on data from 617 cells/957 localizations for ΔmapZ and 1,717 cells/2,328 localizations for wild type. The shaded area represents the point where 50% of FtsZ moves to the 1/4 positions in wild-type cells. (D) The angle of the septum relative to the length axis of the cells is larger and more variable in ΔmapZ cells. (Left) Wild-type D39 and ∆mapZ cells (strain RR93) were stained with Van-FL. Fluorescence image (Top), phase contrast image (Middle), and a schematic drawing of the analysis (Bottom) are shown. The angle, αSEPTUM, was measured automatically by measuring the angle between the long axis of the bounding boxes of the cell outlines and the long axis of the bounding box of the Van-FL signal. The angles of both wild-type and ΔmapZ cells are significantly different from 90° (sign test, P ≤ 2.2 × 10−16), however the septum angle is significantly more skewed in ΔmapZ cells (P < 0.05, Kolmogorov–Smirnov test). (Scale bar, 1 µm.) (Right) The αSEPTUM plotted in wild-type cells and ∆mapZ cells. A total of 177 and 181 cells were measured for wild type and ΔmapZ, respectively. The asterisks denote statistically significant differences.
To examine FtsZ localization, we constructed a C-terminal monomeric red fluorescent protein (mCherry) fusion to FtsZ expressed from its own locus as the only copy (32). Although the FtsZ-mCherry strain showed a normal cell size distribution in a wild-type background, when combined with the ΔmapZ mutant, a clear synthetic phenotype arose and cells were misformed (SI Appendix, Fig. S2 A and B), suggesting that the previously described mapZ phenotype in the presence of FtsZ fusions should be interpreted with caution (30). Therefore, we replaced FtsZ-mCherry by a more functional FtsZ-CFP or FtsZ-mKate2 (FtsZ-RFP) fusion (SI Appendix, Fig. S2 C and D) and reassessed FtsZ localization in ΔmapZ cells. As reported before (22, 23), localization of FtsZ to future division sites occurs when MapZ is already localized at this position (Fig. 1 A and C). Note, however, that in stark contrast to MapZ, which gradually moves as the new cell wall is synthesized, FtsZ is highly dynamic and remodels quickly from the previous to the future division site. Thus, there is only a short period of the cell cycle where MapZ and FtsZ colocalize (compare Fig. 1 A and C) (22, 23). To compare the localization of FtsZ in the wild-type and mutant cells, we first determined the relative cell size where FtsZ localization is at the 1/4 positions in 50% of the cells in the wild-type background using eight datasets of both FtsZ-CFP and FtsZ-RFP localizations (totaling 26,986 cells). The cell size where 50% of FtsZ splits was strikingly consistent between the datasets (1.97 ± 0.137 µm). Because the cell sizes of mutant cells differ from wild-type cell sizes, we converted the splitting point to a percentage of the mean cell length (PSPLIT_N = 124 ± 9.04%) and visualized this as a shading area in both the wild-type and ΔmapZ density histograms (Fig. 1 A and C). Importantly, FtsZ localization over the length axis of the cell was not affected in ∆mapZ cells, suggesting that MapZ is not essential for accurate timing of Z-ring assembly. To gain more insights into the role of MapZ during septum formation, we stained cells with fluorescently labeled vancomycin (Van-FL) to image sites of cell-wall synthesis (33) and measured the angle of the areas in the cell enriched with Van-FL relative to the long axis. Interestingly, the angle of the septum was not perfectly perpendicular to the cell length in wild-type cells (median difference from 90° αSEPTUM = 3.08 ± 1.47°), and this difference from 90° was found to be significant (one-sided sign test, P ≤ 2.2 × 10−16). Notably, however, in ΔmapZ cells, this angle was significantly more skewed than wild type (Fig. 1D, median difference from 90° αSEPTUM = 7.65 ± 1.27°, P = 0.014, Kolmogorov–Smirnov test). Measuring the angle of FtsZ-CFP in the same manner as with Van-FL confirmed that the angle of the Z-ring was significantly more skewed in ∆mapZ cells (SI Appendix, Fig. S2E).
These results are in line with previous observations (22, 23) and could explain the variability in cell shapes observed in ΔmapZ mutants. The observed cell-shape alterations are reminiscent of E. coli mutants lacking certain low-molecular weight penicillin-binding proteins (LMW-PBPs) such as PBP5 that have defected division plane selection and mislocalized Z-rings (34). LMW-PBPs modify PG by trimming amino acid linkages from the glycan side chains (35). Because MapZ has a large extracellular PG binding domain and is controlled by the Ser/Thr kinase StkP (22, 36), which is proposed to be a key player in tuning peripheral and septal PG synthesis (24, 37), it is tempting to speculate that MapZ has a function in cell wall remodeling and subsequently maintaining the perpendicular Z-ring plane. Although this remains to be verified experimentally, it has been shown that mutations in PG hydrolases as well as depletion of Pbp2b involved in cell elongation can lead to nonparallel division and cell-shape alterations in S. pneumoniae (30, 38, 39), similar to the phenotype of ΔmapZ cells.
The Replisome of S. pneumoniae Is Dynamic Around Midcell.
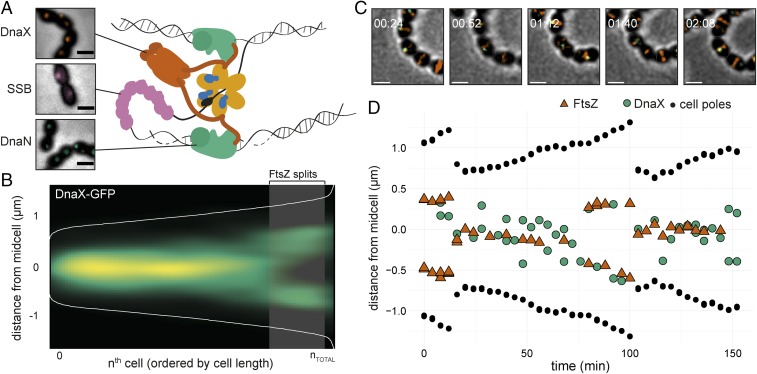
Because the ΔmapZ mutant has moderate effects on division site selection under our experimental conditions, another system must be in place. Because S. pneumoniae lacks the canonical systems, we hypothesized that ovococci might coordinate division via chromosome replication and segregation (17). To test this, we first aimed at imaging the DNA replication machine (replisome) and constructed inducible, ectopic fusions of the single-strand binding protein (SSB), the β sliding clamp (DnaN), and the clamp loader (DnaX) with GFP or RFP (mKate2). Fluorescence microscopy showed enriched signals as bright diffraction limited spots for all fusions, mainly localized in the middle of the cells, similar to what has been observed for B. subtilis and E. coli (40, 41) (Fig. 2A). Notably, the background signal of SSB-RFP was higher than the background of the other fusions, as also reported for E. coli (41). Chromosomal replacements of the fusion constructs with the original gene could only be obtained for dnaX, but not for ssb and dnaN, suggesting that the fusion tags of these two latter genes are not fully functional. To validate that the localizations of the fusions represent biologically active replisomes, we examined their colocalization patterns. As expected, the ectopically expressed fluorescent fusions of DnaX, DnaN, and SSB to RFP colocalize with the functional DnaX-GFP fusion in live cells (91% colocalization or more; SI Appendix, Fig. S3A).
Fig. 2.
Localization of the pneumococcal replisome. (A) Localization of DnaX-GFP (RR31), GFP-DnaN (DJS02), and SSB-GFP (RR33). (Scale bar, 1 μm.) A cartoon of the bacterial replication fork at initiation shows the role of DnaX (clamp loader), SSB, and DnaN (β sliding clamp). DNA polymerase is replicating the leading strand (Top) and Okazaki fragments at the lagging strand (Bottom). The cartoon is based on what is known about the replication fork in E. coli. (B) Plotting the localization of DnaX-GFP (RR22) shows that the replisome is localized at midcell. Data are from a total of 3,574 cells and 3,214 unique localizations. The shaded area represents the point in the cell cycle where 50% of FtsZ has localized to the 1/4 positions of the cell. (C) Snap shots from a representative time-lapse movie of strain MK396 (dnaX-GFP, ftsZ-mKate2). Overlays of GFP, RFP, and phase contrast are shown. (Scale bar, 1 µm.) (D) Transcript of the cell shown in C. The distance of FtsZ (red), DnaX (green), and the cell poles (black) to midcell is plotted against time. The data are also shown in Movie S1. Transcripts of more single cells are shown in SI Appendix, Fig. S3.
DnaX-GFP is positioned close to midcell with a similar pattern as FtsZ-RFP, although the DnaX-GFP localization is less precise than FtsZ-RFP (Fig. 2B). To analyze the correlation between DnaX appearance at the new division site and FtsZ splitting, a DnaX-GFP/FtsZ-RFP double-labeled strain was made. A cluster analysis on both DnaX and FtsZ localizations was performed to determine the “fork point” of the cell length where 50% of the proteins are localized at the new division sites. This point was at 1.856 µm for DnaX and 1.998 µm for FtsZ, respectively (1,234 cells analyzed), which is a difference within the resolution limit. Furthermore, plotting the localization densities of both the midcell clusters and 1/4-position clusters against each other shows that there is no significant difference in the fork points between DnaX and FtsZ (SI Appendix, Fig. S3B). To validate these results, we tracked single cells during growth using time-lapse fluorescence microscopy (Fig. 2 C and D, SI Appendix, Fig. S3C, and Movies S1 and S2). This showed that although the replisome(s) is dynamic, it stays in near proximity to the Z-ring. Together, these data demonstrate that the replisome moves to the future cell division sites coincidently with FtsZ and the Z-ring does not linger for cell division to finish (Fig. 2D).
To gain more insight into the nature of the movement of the replisome, we imaged DnaX-GFP in short-time interval movies (1 s, Movie S3) using total internal reflection fluorescence (TIRF) microscopy. We tracked DnaX-GFP using the single-molecule tracking software U-track (42) and analyzed mobility using SpotprocessR (SI Appendix, Fig. S3D). As expected, replisome mobility was significantly lower than that of free diffusing GFP (43). However, compared with ParB-GFP, which binds to the origin of replication (oriC) (27), DnaX-GFP showed a nearly twofold higher mobility (MSD = 2.66 × 10−2 µm2, Dapp = 2.44 × 10−3 µm2/s−1 compared with MSD = 8.8 × 10−3 µm2, Dapp = 3.19 × 10−4 µm2/s−1; SI Appendix, Fig. S3 D and E), indicating that DnaX movement is not strictly confined by the chromosome.
The Pneumococcal Chromosome Segregates in a Longitudinal Fashion.
The on-average midcell localization of the replication machineries in S. pneumoniae suggests that DNA replication at midcell might determine an ordered chromosomal organization. To examine this, methods for tagging chromosome positions in this bacterium were established (SI Appendix, Figs. S4 and S5). We first constructed a chromosome marker system based on fluorescent protein fusions to ParB of plasmid pLP712 (44) from Lactococcus lactis (hereafter named ParBp), which was found to require insertion of only a 18-bp parS binding site (hereafter named parSp) in the pneumococcal genome for visualizing genetic loci by microscopy (SI Appendix, Fig. S4 A and C). The parSp sequence is simpler compared with existing ParB/parS systems and does not require additional host factors (45, 46). Importantly, this system does not perturb DNA replication and is completely orthogonal to S. pneumoniae chromosomal ParB (SI Appendix, Figs. S4B and S5 A–D). Secondly, we adapted the TetR/tetO fluorescence repressor-operator system (FROS) (47) for S. pneumoniae and validated that it does not interfere with DNA replication (SI Appendix, Figs. S4D and S5 A–C). To verify the localization patterns and compatibility of both systems, we constructed a strain containing both parSp and tetO near oriC and showed that parBp-gfp and tetR-rfp foci colocalize (SI Appendix, Fig. S5 E and F).
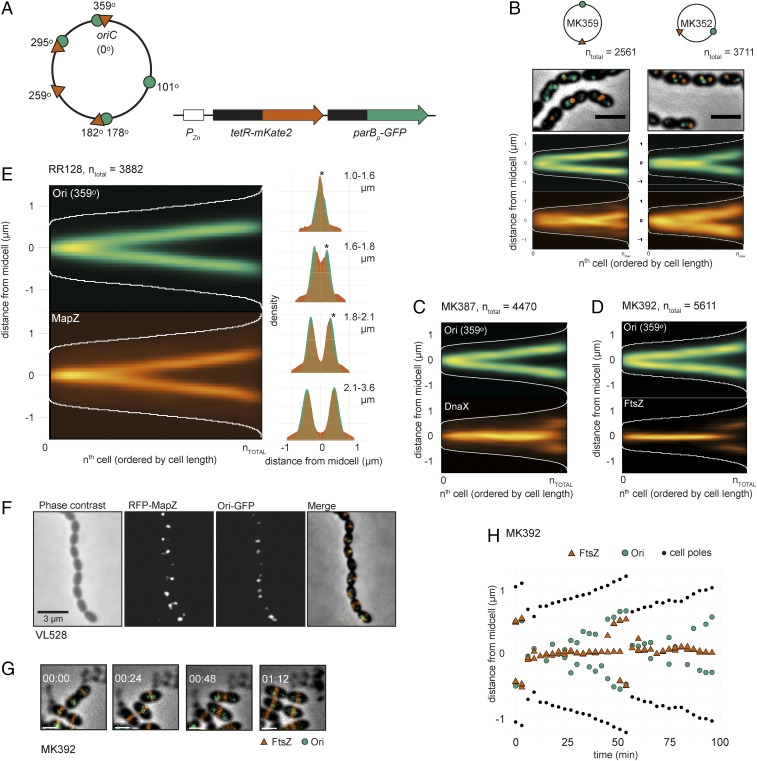
In total, five chromosomal locations were marked using ParBp/parSp and/or TetR/tetO: the origin-region (359°), right arm (101°), ter-region (178° and 182°), and two positions on the left arm (259° and 295°) (Fig. 3A). Using double-labeled strains under balanced growth, the localizations of loci were compared, revealing that the pneumococcal genome is organized in a longitudinal fashion (Fig. 3B and SI Appendix, Fig. S6). The left and right arms move at the same time to the new daughter cells (Fig. 3B). The terminus region seemed less confined in space during the cell cycle (Fig. 3B). Strikingly, the origins never localized near the cell poles, as is common in other bacteria (5, 47, 48), and arrived to future division sites at a very early stage, before DnaX and FtsZ, with a similar timing as MapZ (Fig. 3 C–F). The early segregation of oriC was also observed when single cells were tracked over time (Fig. 3 G and H, SI Appendix, Fig. S7, and Movie S4). Note that this organization is strikingly different from organisms with nucleoid occlusion where FtsZ preferably polymerizes across DNA-empty regions or regions near the terminus that lack Noc protein.
Fig. 3.
Chromosomal organization in S. pneumoniae. (A) Visualizing specific genetic loci in live cells by fluorescence microscopy was done by developing two independent chromosomal markers systems: TetR-mKate2/tetO (tetO integration sites indicated by red triangles on the chromosome map) and ParBp-GFP/parSp (parSp integration sites indicated by green circles). tetR-mKate2 and parBp-GFP are ectopically expressed from the nonessential bgaA locus under control of the Zn2+-inducible promoter PZn. (B) Localization of the origin and terminus (MK359, Left) and left and right arm (MK352, Right) in exponentially growing cells. Overlays of GFP signal, RFP (mKate2) signals, and phase contrast images are shown. (Scale bar, 2 µm.) The data represent 2,561 cells/3,815 GFP-localizations/2,793 RFP-localizations from MK359 and 3,711 cells/5,288 GFP-localizations/4,372 RFP-localizations from MK352. (C) Localization of the origin (ParBp-GFP/parSp at 359°) and DnaX-RFP (MK387). The data represent 4,470 cells/5,877 GFP-localizations/4,967 RFP-localizations. (D) Localization of the origin (ParBp-GFP/parSp at 359°) and FtsZ-RFP (MK392). The data represent a total of 5,611 cells/6,628 GFP-localizations/26,674 RFP-localizations. (E) Localization of the origin (ParBp-GFP/parSp at 359°) and RFP-MapZ (RR128) on the length axis of the cell shown as heatmaps (Left) and overlay of both localization density plots when the cells are grouped in four quartiles by cell length (Right). Stars indicate a significant difference between GFP and RFP localization (Kolmogorov–Smirnov test, P < 0.05). The data represent 3,882 cells/1,785 GFP-localizations/8,984 RFP localizations. (F) Microscopy images of strain VL528 showing subsequent origin and MapZ splitting. Phase contrast, RFP, GFP, and overlay images are shown. (Scale bar, 3 µm.) (G and H) Time-lapse microscopy shows that the origins move to the next cell halves before FtsZ. Snap shots from a representative time-lapse experiment (G) and plotting of the distances of FtsZ, the origins, and the cell poles relative to midcell in a single cell (H) are shown. (Scale bars in G and H, 1 μm.) More examples of origin and FtsZ localizations in single cells are shown in SI Appendix, Fig. S7.
SMC Is Required for Correct Segregation of oriC and Cell Shape.
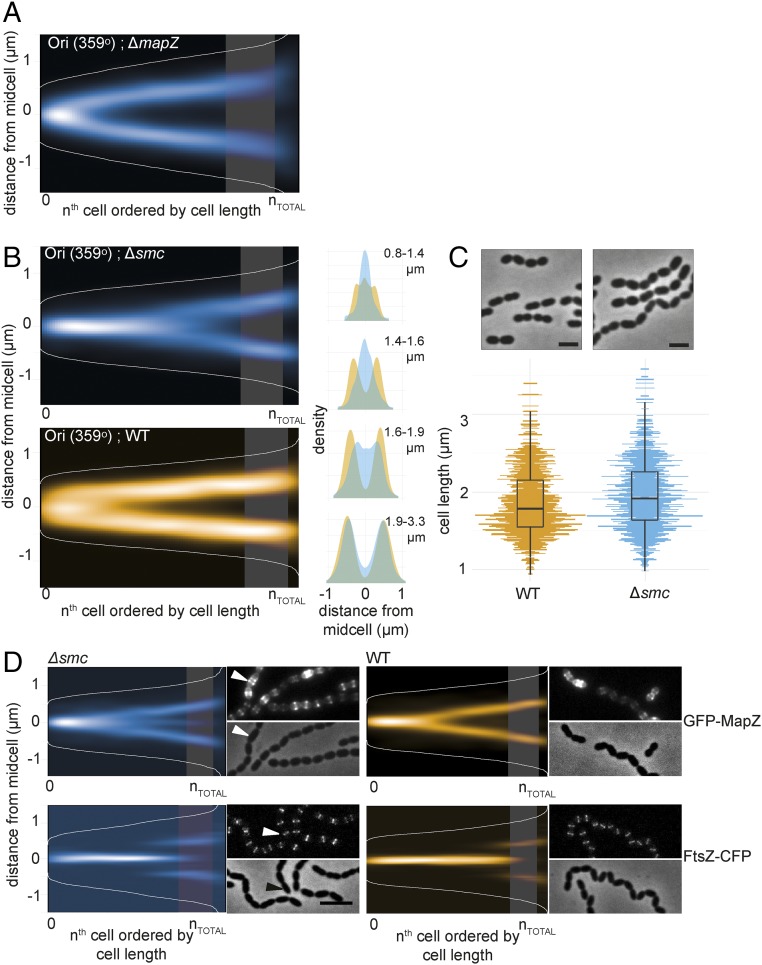
The observation that oriC arrives at future cell division sites before FtsZ opens the question as to whether MapZ has a role in directing the chromosome. However, the origin still localized to the future division site in ΔmapZ (Fig. 4A). The localizations of MapZ and oriC were further analyzed in a wild-type background by sorting the cells into four subgroups according to cell size and plotting the localizations as histograms over the cell lengths (Fig. 3E, Right). This shows that MapZ is localized slightly closer to the old midcell in smaller, newborn cells (Fig. 3E, Right, stars indicate a significant difference in the first three groups of cells, Kolmogorov–Smirnov test, P < 0.05) and indicates that oriC localizes to the new midcell before MapZ. To study this in more detail, we constructed a strain with GFP labeled oriC and two copies of rfp-mapZ (strain VL528) to improve RFP-MapZ signal (Fig. 3E). For this strain, we determined, per single cell, whether the origin or MapZ localizes farthest away from midcell. In 71% of the cells, the origin and MapZ localized together (distance <200 nm). Of the remaining 29%, the origin localized farthest away from midcell than MapZ in 68% of the cells (2,240 cells analyzed). Although the difference is small, this confirms that the origin, on average, localizes to the new midcell slightly before MapZ.
Fig. 4.
SMC is required for origin segregation and accurate division site selection. (A) Localization of the chromosomal origin (ParBp-GFP/parSp at 359°) in a ΔmapZ background shows that MapZ does not affect origin segregation (RR99). The data represent 2,976 cells/7,020 localizations. The shaded areas in all density histograms in this figure represent the point in wild-type cells where 50% of FtsZ localizes to the 1/4 positions of the cell. (B) The origins (ParBp-GFP/parSp at 359°) are segregated at a later stage in the cell cycle in Δsmc compared with wild type. The localizations are shown as heatmaps when cells are sorted according to length (Left) and as overlay of both localization density plots when the cells are grouped in four quartiles by cell length (Right). The data represent 2,012 cells/3,815 localizations for wild-type (MK359) and 3,908 cells/5,192 localizations for the Δsmc mutant (MK368). (C) Phase contrast images of wild-type D39 and Δsmc cells (AM39). (Scale bar, 2 µm.) Comparison of cell lengths between the wild type (1,407 cell analyzed; Top) and Δsmc (1,035 cells analyzed; Bottom). (D) Localization of GFP-MapZ and FtsZ-CFP in wild type versus Δsmc. Fluorescence and phase contrast micrographs are shown along with heatmaps. (Scale bar, 3 μm.) The arrowhead in the micrograph points to a cell with clearly mislocalized MapZ. Data represent 2,560 cells/5,314 localizations (∆smc, GFP-MapZ, RR110), 1,300 cells/2,257 localizations (∆smc, FtsZ-CFP, RR84), 3,908 cells/3,128 localizations (wild type, GFP-MapZ, RR101), and 3,422 cells/29,464 localizations (wild type, FtsZ-CFP, RR70).
Given the early movement of the origin to the future cell division sites, we wondered whether instead the chromosome- or nucleoid-associated proteins could play a role in guiding Z-ring positioning. In many prokaryotes, condensin-like proteins called Structural Maintenance of Chromosomes (SMC) play a role in the organization and compaction of the chromosome (49). In S. pneumoniae, deletion of smc leads to ∼2% anucleate cells and problems in chromosome segregation (27, 28). To specifically investigate how the absence of SMC affected chromosome organization, the origin, terminus, and left/right arm chromosome positions were determined in Δsmc cells. In line with what has been found using temperature-sensitive or degradable alleles in B. subtilis (50, 51), the origin region arrived to the new midcell at a considerable delay when SMC was absent (Fig. 4B). Quantitative analysis of the origin localization showed a significant different localization of oriC in Δsmc vs. wild type (P < 1.5 × 10−3, Kolmogorov–Smirnov test; Fig. 4B). Eventually, however, the origins still segregated to their correct localization in subsequent larger cells. Also, segregation of the left arm, right arm, and terminus did not differ significantly from wild type (SI Appendix, Fig. S8 B–D). Thus, S. pneumoniae SMC is specifically important for the early segregation of oriC. SMC-GFP showed, in line with previous observations (27), a nucleoid-like pattern with foci in a subset of cells with poorly defined localizations (SI Appendix, Fig. S8A).
In the current data, we also found that Δsmc cells are longer, more irregularly shaped and form long chains (Fig. 4C). The same observation was also made upon deletion of smc in strains TIGR4 and R6 (SI Appendix, Fig. S1 A and B). This suggests that smc mutants are somehow defective, not only in chromosome segregation but also in cell division. We therefore compared the localization of MapZ and FtsZ in wild-type and Δsmc cells (Fig. 4D). MapZ showed an obvious mislocalization; part of the MapZ-rings arrived at the new septa at a later stage, whereas a large fraction stayed at midcell in larger cells (11% of the MapZ localizations in the largest quartile of the cell population stayed at midcell in ∆smc cells vs. 4% in wild-type cells; Fig. 4D). For FtsZ, the effect is less pronounced, but a small difference in localization accuracy is observed at the time when new Z-rings were formed (Fig. 4D). This subtler difference in FtsZ localization between wild-type and Δsmc cells is not surprising, though, given that in the Δsmc cells, the origins have started segregating at the time of FtsZ assembly (Fig. 3B). Note that the angle of the division ring is not affected in Δsmc cells (SI Appendix, Fig. S1C). Together, these results suggest that SMC and/or origin localization is important for timely and precise positioning of the cell division machinery in S. pneumoniae.
Knowing that both origins and MapZ localize very early in the pneumococcal cell cycle to the future division sites and that individual perturbation of origin localization or MapZ cause division problems, we deleted both smc and mapZ to understand more about the link between them. The double-mutant strain was readily obtained, although the strain had severe defects in growth and cell shape (SI Appendix, Fig. S1 C and D). Notably, the phenotype of the double mutant looked like a combination of the single mutants; like ΔmapZ, the cells were on average smaller with large cell shape variation due to nonperpendicular division ring formation (SI Appendix, Fig. S1C), and like Δsmc, they displayed a chaining phenotype probably reflecting problems with timing of division ring formation leading to consecutive problems in timing of division and cell wall splitting (SI Appendix, Fig. S1D). These observations suggest that MapZ and SMC have independent roles in pneumococcal cell division, where SMC is important for timely localization of the division site, whereas MapZ is involved in placement of the division ring at a correct angle.
Proper Localization of oriC Is Crucial for Division Site Selection in S. pneumoniae.
We show above that deletion of smc caused a cell division defect in S. pneumoniae distinct from the ΔmapZ phenotype. To untangle whether this was a direct effect of SMC or whether it was caused by the resulting chromosome organization defect, we exposed S. pneumoniae to sublethal concentrations of ciprofloxacin to disturb chromosome organization while keeping smc intact. Ciprofloxacin is a broad-spectrum antibiotic which blocks the activities of type II topoisomerases and thereby affects DNA supercoiling and chromosome decatenation (52). Strikingly, when exponentially growing cells are transferred to a nonlethal concentration of ciprofloxacin (0.4 µg/mL), cells rapidly increase in cell length and form longer chains compared with untreated cells (Fig. 5 A and B and SI Appendix, Fig. S9). Origin splitting was clearly delayed in ciprofloxacin-treated cells, and the timing and accuracy of Z-ring formation was severely affected (Fig. 5C). Moreover, localization of the replisome was less confined to the center of the cells, as was observed for Δsmc cells (SI Appendix, Fig. S10). Note that, at the ciprofloxacin concentration used in this experiment, replication elongation is reduced, but new rounds of replication are still initiated (53).
Fig. 5.
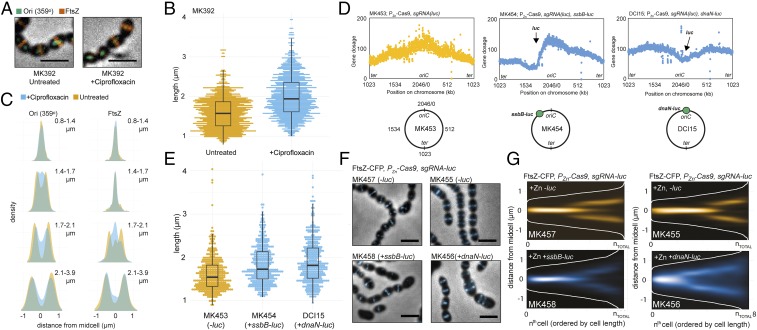
Perturbed chromosome segregation delays cell division. (A–C) Comparison of S. pneumoniae D39 wild-type cells treated or untreated with sublethal concentrations (0.4 µg/mL) of ciprofloxacin for 60 min. (A) Images of strain MK392 (ParBp-GFP/parSp at 359°, FtsZ-RFP) with overlay of phase contrast, with GFP signals representing the origin and RFP signal representing FtsZ-RFP. (Scale bar, 2 µm.) A micrograph with more cells of the same strain can be found in SI Appendix, Fig. S9. (B) Cell length comparison of ciprofloxacin-treated (total 1,138) and nontreated (total 1,402) cells (MK392). (C) Subcellular localization of the origin (Left) and FtsZ (Right) in MK392 cells treated (blue) or untreated (yellow) with ciprofloxacin. Localization density plots when the cells are grouped in four quartiles by cell lengths are shown. Data represent 1,138 cells/2,518 RFP-localizations/2,762 GFP-localizations for cells treated with ciprofloxacin and 1,402 cells/2,940 RFP-localizations/2,540 GFP-localizations for untreated cells. (D–G) Comparisons of cells with or without Cas9-nuclease cut chromosome. The expression of Cas9 (together with a constitutively expressed single-guide RNA directed to the luc-gene) was induced with Zn2+ for 2.5 h in cells with or without the luc gene located on the chromosome. The luc gene was inserted either in the origin region (0°) or at the left arm (301°). In cells without the luc gene, no Cas9-mediated cutting will occur. (D) Whole genome marker frequency analysis of strains without luc (MK453), luc at 301° (ssbB-luc, MK454), or luc at the origin (dnaN-luc, DCI15). The number of mapped reads (gene dosage) is plotted as a function of the position on the circular chromosome. The chromosomal position of the inserted luc gene is indicated in the plot and on the schematic chromosome maps. (E) Cell size comparison of cells with and without cut chromosomes. The number of cells measured were 643 for the noncut strain (MK453), 393 for the strain cut at 301° (MK454), and 383 for the strain cut near the origin (DCI15). (F) Overlay of FtsZ-CFP signals with phase contrast images shows that cell morphologies are affected in cells with cut chromosomes. (Scale bar, 2 µm.) (G) Localization of FtsZ-CFP in cells with a cut chromosome (MK 458 and MK456) and intact counterparts (MK457 and MK455, respectively) shown as heat maps where cells are ordered according to cell length. The data represent 1,323 cells/3,117 localizations for MK455, 1,128 cells/3,509 localizations for MK456, 721 cells/1,133 localizations for MK457, and 683 cells/1,209 localizations for MK458.
Finally, we also perturbed the DNA biology by cutting the chromosome at two different locations—close to oriC (at 0°) and on the left arm (at 301°)—using an inducible CRISPR/Cas9 system (see Materials and Methods). Whole-genome marker frequency analysis of these strains after induction of Cas9 for 2.5 h showed the expected cleavage of the chromosomal DNA at these two positions in the respective strains, and major alterations in the replication patterns were observed (Fig. 5D). Cutting of the chromosome had drastic effects on cell morphology, and we observed frequent mistimed FtsZ localization (Fig. 5 F and G) and increased cell sizes (Fig. 5E). The effects on DNA replication were more pronounced when Cas9 was targeted to oriC compared with the left arm location, and subsequently, proper control of Z-ring formation was completely lost in the former case. It should be noted that generation of double-strand breaks will impose severe stress on the cells and probably elicit numerous stress responses. Nevertheless, these results further confirm that normal chromosome segregation, and origin segregation in particular, is key for well-timed Z-ring assembly and cell division progression in S. pneumoniae.
Discussion
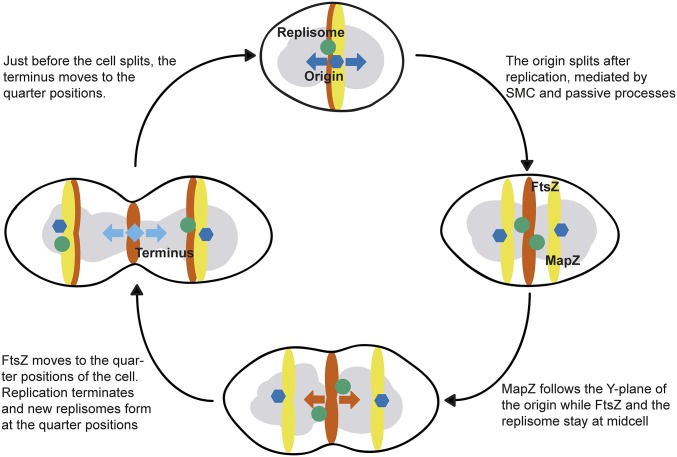
By detailed mapping of DNA replication and chromosome segregation in live S. pneumoniae cells, we found that DNA replication initiation coincides with Z-ring formation and that proper segregation of the chromosomal origin is crucial for division site selection. We show that the pneumococcal chromosome is organized in a longitudinal fashion (ori-left/right-ter-ter-left/right-ori; Fig. 3 and SI Appendix, Fig. S6) with specific subcellular addresses for each locus. In contrast to, for example, B. subtilis and C. crescentus (5, 47, 48), the origins never localize near the cell poles in S. pneumoniae, and the organization is in this aspect more similar to the situation in slow-growing E. coli (54). Importantly, the newly replicated origins immediately mark the future cell division sites while the terminus remains at midcell. This organization is somewhat reminiscent of the chromosomal organization in B. subtilis and E. coli but is slightly simpler, as every replicated locus eventually segregates to midcell before a new round of replication initiates (Fig. 6). Segregation of the chromosomal origin was dependent on SMC, and deletion of smc caused a marked delay in origin segregation, which in turn led to alterations in the timing of localization of important cell division proteins such as MapZ and FtsZ. Mistimed MapZ and FtsZ ultimately resulted in larger cells, which form chains. Importantly, the observed cell division defects are not caused by the deletion of smc per se; treatment of the cells with sublethal concentrations of ciprofloxacin or a CRISPR/Cas9-induced segregation block also caused similar cell division defects (i.e., larger cells and chaining). Together, this indicates that timely segregation and positioning of the chromosomal origin at the quarter position in cells is important for orchestrating pneumococcal cell division.
Fig. 6.
A schematic model for division site selection in the pneumococcus. The bulk chromosome is shown in gray, whereas the chromosomal origin, left/right arm, and terminus are indicted as a dark blue hexagon, green circle, and light blue diamond, respectively. MapZ is shown in yellow and FtsZ in orange. Four key stages of the pneumococcal cell cycle, with a newborn cell on the top, are included.
Recently, it was found that MapZ localizes to future division sites before FtsZ and positions the Z-ring correctly via protein–protein interactions (22, 23). We found that MapZ gradually moves with a similar timing as the chromosomal origins, but MapZ is not important for correct oriC positioning. On the other hand, perturbation of oriC segregation clearly resulted in altered MapZ localization, thus indicating the pivotal role of chromosomal origin positioning for proper cell division coordination. Notably, the PG-binding protein MapZ plays an important role in establishing the correct division plane, as the angle of the Z-ring to the cell length axis was frequently skewed when mapZ was deleted (Fig. 1D). Taken together, this suggests that although timely oriC positioning determines the timing of assembly and position of the cell division machinery, MapZ is a ruler for the correct angle of the division ring across the cell (Fig. 6). This explains the variable size and shape of ΔmapZ cells as well as the cell division defect resulting from the mislocalized origins in Δsmc and ciprofloxacin-treated cells. Note, however, that although oriC segregation is clearly delayed in both the Δsmc strain as well as the ciprofloxacin-treated cells, it eventually segregates to the future division sites in time before Z-ring assembly. This means that there are additional cues, and not solely SMC or topoisomerases, which are involved in segregation and localization of oriC, explaining why the cell division defects resulting from smc deletion or ciprofloxacin treatment are not too severe.
How the origin finds the future division site and how it can modulate the localization of division proteins such as MapZ and FtsZ are unclear. We cannot rule out an as-of-yet unknown protein factor playing a role in keeping the newly replicated origins near the future division site and MapZ or by bridging FtsZ to the DNA replication initiation complex. A link between replication and Z-ring assembly has previously been suggested for B. subtilis (17), however no mechanism is yet known. In E. coli, it has been shown that the chromosomal terminus region (Ter macrodomain) is linked to the cell division apparatus by proteins MatP, ZapA, and ZapB (55, 56). Similar links between the chromosome and division apparatus may also be present in other bacteria, although, in our model, we observe that segregation of the terminus occurs after FtsZ repositioning. Another hypothesis is that coupled transcription–translation–transertion of membrane proteins encoded near oriC aid in transitory attachment of the chromosome to the membrane (57, 58). Alternatively, physical, entropy-driven processes might be at play. In this respect, it is tempting to speculate that the origin region, which was recently shown to be highly structured and globular in shape (59), is pushed outside the region of active DNA replication and remains rather stationary in the crowded cytoplasm (60). The large globular structure of the origin can then act as a landmark for FtsZ polymerization and Z-ring formation. This hypothesis is in line with previous cytological observations demonstrating the absence of nucleoid occlusion in S. pneumoniae and efficient Z-ring formation over the nucleoid (28, 61). The here-described division site selection mechanism by chromosomal organization is a simple way to coordinate DNA replication, chromosome segregation, and division without the need for specialized regulators of FtsZ. Future research should reveal if this mechanism is also in place in other bacteria and whether the intimate relation between chromosome segregation and cell division can be used to treat bacterial infections using combination therapy targeting both processes.
Materials and Methods
Bacterial Strains, Growth Conditions, and Transformation.
Bacterial strains used in this study are listed in SI Appendix, Table S1. S. pneumoniae was grown in C+Y medium (62) at 37 °C without shaking. When appropriate, 0.1 mM ZnCl2 and 0.01 mM MnCl2 was added for induction. To reduce oxygen stress, S. pneumoniae was plated inside Columbia agar supplemented with 2% defibrinated sheep blood (Johnny Rottier). When appropriate, antibiotics were added for selection: 1 µg/mL tetracycline, 100 µg/mL spectinomycin, or 250 µg/mL kanamycin. E. coli was grown in LB medium with shaking. When appropriate, 100 µg/mL ampicillin or 100 µg/mL spectinomycin was used for selection.
Plasmids and ligation products were transformed into E. coli using a standard heat shock transformation of chemically competent cells. Plasmids from E. coli were routinely checked by PCR and sequencing. Plasmids, ligation products, or assembled DNA were transformed into S. pneumoniae by addition of the competence stimulation peptide (CSP) as described previously (53). Constructs introduced directly into S. pneumoniae were checked by PCR and sequencing.
All oligonucleotides used for cloning are listed in SI Appendix, Table S2. Construction of plasmids and strains are described in SI Appendix, SI Materials and Methods.
Growth assays were performed in 96-well plates using a Tecan Infinite 200 PRO instrument. OD595nm was measured every 10th minute.
Preparation of Cells for Microscopy.
S. pneumoniae cells were stored as exponential phase frozen cultures. Frozen stock was inoculated 1:100 in C+Y medium and pregrown to OD600 ∼ 0.1. For time-lapse microscopy, cells of OD600 ∼ 0.1 were washed and diluted 1:100 in fresh C+Y and allowed to grow for 45 min. After this, the cells were washed once with fresh C+Y. For snap shots, cells were diluted once again 1:100 in fresh C+Y (with inducing agent, if applicable) and grown to exponential phase to achieve balanced growth. For expression of the chromosome marker systems and CRISPR/Cas9 system, cells were induced for 2.5 h using 0.1 mM ZnCl2 and 0.01 mM MnCl2 in the growth medium.
Fluorescence Microscopy.
For snap shots, cells were grown as described above to achieve balanced growth and subsequently concentrated and brought onto a multitest slide carrying a thin layer of 1.2% agarose in C+Y. Imaging was performed on a DV Elite microscope (GE Healthcare) with a sCMOS camera using SSI Solid State Illumination through a 100× oil immersion objective (phase contrast). The following filtersets were used: mCherry/mKate2 (Chroma, excitation at 562–588 nm, emission at 602–648 nm, Quad/mCherry polychroic), GFP (Chroma, excitation at 461–489 nm and emission at 501–559 nm, Quad polychroic), and CFP (Chroma, excitation at 400–454 nm and emission at 463–487 nm, CFP/YFP/mCherry polychroic). Several images, capturing >500 cells (exact number per experiment indicated in the figure legends), were acquired using Softworx (Applied Precision) and further processed to TIFF files using FIJI (fiji.sc) (63, 64).
Time-Lapse Microscopy.
A C+Y 10% polyacrylamide slide was preincubated for 2 h at 37 °C in C+Y (0.1 mM ZnCl2 when applicable). We spotted 0.5 µL of the cells (prepared as described above) on the C+Y-polyacrylamide slide inside a Gene Frame (Thermo Fisher Scientific) and sealed it with a cover glass. The temperature at the incubation chamber was set to 30 °C to optimize fluorescence and stable growth. Time-lapse microscopy was performed as described previously (65) using UltimateFocus (Deltavision) every 2 min to ensure stable imaging. The time-intervals used are indicated at each experiment. Image stacks were acquired using Softworx (Applied Precision) and further processed to TIFF files using FIJI (fiji.sc) (63, 64).
TIRF Microscopy.
Slides were prepared for time-lapse microscopy as described above. Imaging was done using a DV Elite microscope (Applied Precision), using the standard GFP filter sets (Chroma) and Quad polychroic. A 60× DIC oil immersion TIRF objective (1.47 N.A.) was used, and imaging was done at 100% TIRF depth with 0.005 s excitation of a 50-mW laser (488 nm).
Cell Outline and Fluorescence Detection.
Cell outlines were detected using Microbetracker (66) (microbetracker.org) and Oufti (31) (oufti.org). Spots were detected using Microbetracker’s spotFinderZ and Oufti’s Spotfinder tool, with the exception of the experiments where interspot distances were measured. Here we used peak fitter, a Gaussian-fit tool that is part of the ImageJ plugin iSBatch (67), to also obtain information about the full-width half-max (fwhm) and SEs of the location. For larger fluorescent bodies, such as MapZ and FtsZ, Oufti’s Object Detection tool was used to obtain information about the localization and shape. For all microscopy experiments, random image frames were used for analysis, and all experiments were repeated at least two times.
Plotting and Statistical Analysis of the Localization Histograms.
The cell outline, object detection, and spot localization data were further processed using the R-package SpotprocessR (https://github.com/vrrenske/spotprocessR), which enables plotting of the cell outlines and spot localizations (x,y coordinates inside the cell), together with further grouping of the cells and statistical comparison of spot localizations. For comparing the localization of spots inside the cell with detected objects, the center of the minimal bounding box of the object was defined as the localization of the object and further processed the same way as spot localizations.
For the snap shots, the cells were standardly ordered by cell size, and localizations on the length axis were plotted as a density plot. When two localizations were compared, the combined datasets were grouped into quartiles based on cell size, and localizations on the length axis were plotted as dual-color density maps for each group. The similarity of the x and y localization distribution shape and median were tested using a two-sided Kolmogorov–Smirnov-test, as were the similarities in cell length distribution.
Determination of the FtsZ Splitting Point.
To compare mutant and wild-type strains, the relative cell size where FtsZ splits in wild-type cells was used as a fiduciary marker. To determine this, eight different datasets with a total of 26,986 cells were analyzed. Of these datasets, two contained the localization of FtsZ-CFP (4,016 cells) and six FtsZ-RFP (2,2970 cells). For all datasets, the cell length was determined wherein 50% of the cells of FtsZ had moved to the 1/4 positions of the cells. To separate the midcell localizations from the 1/4 position localizations, a Gaussian Mixture Model-based cluster analysis was performed on the localizations per cell length for each dataset using the R package Mclust (68, 69). For each cluster, the density per cell length was determined for both the 1/4 position cluster and the midcell cluster. After correction for the number of localizations per cluster, the point in cell length was found where the density functions intercept. This point, PSPLIT, is the point where 50% of the localizations are at midcell and 50% of the localizations are at the 1/4 positions of the cell. PSPLIT was determined for all eight datasets, where the weighted average (corrected for the difference in the number of cells per dataset) turned out to be 1.97 µm ± 0.13 µm. To be able to use PSPLIT as a marker in mutant strains, where the cell size is often very different from wild type, PSPLIT was normalized to a percentage of the mean cell length. For this, each splitting point of each dataset was divided by the average cell size of the dataset and multiplied by 100%. The final normalized splitting point PSPLIT_N was 124% ± 9.04% of the average cell length of a dataset. This range in cell length was shaded in each graph where mutants and wild-type cells are compared as a fiduciary marker.
Determination and Statistical Analysis of the FtsZ/Septum Angle.
To determine the angle of either FtsZ or the septum labeled with fluorescent vancomycin, Oufti’s Object Detection tool was used to outline the fluorescent bodies. The angle of the longest side of the minimal bounding box of the cell outline and the objects were determined. The difference between these two angles was determined as the angle of the fluorescent object (αobject). Because the direction of the angle has no meaning in this measurement (an angle of 85° is the same as an angle of 95° in a cell upside-down), the final object angles were defined as . Here, if αFTSZ/SEPTUM = 0, the angle of the fluorescent object is perpendicular to the length axis of the cell. To determine whether in the resulting nonnormal distributions αFTSZ/SEPTUM was significantly higher than zero, a one-sided sign test was performed where H0: α = 0 and H1: α > 0 and CI = 95%. The sample distribution shapes and medians within experiments were compared with each other using a two-sided Kolmogorov–Smirnov test.
Analysis of Time-Lapse Movies.
For the time-lapse data, Oufti was used to obtain movies of single cells, and SpotprocessR was used to obtain a plot of the corresponding cells, showing the localization of the spots and cell poles over time. Using SpotprocessR, the localizations inside all individual cells were plotted as the distance from midcell for each time point in the time-lapse movie.
Colocalization Analysis.
For colocalization, the distances between spots in each cell were measured using SpotprocessR. Three categories were defined to determine whether two spots colocalized: (i) Colocalization was defined as an overlap of the fwhm of the Gaussian fit with the distance between the centers of the Gaussian fits. When the distance between the centers was smaller than their fwhm, they were defined as being at the same location. (ii) Possible colocalization was defined as spots where the Gaussians fwhms touch but do not overlap with their center distances, making them close but not certainly colocalizing. (iii) Noncolocalization was defined as the spots where the Gaussians fwhms do not overlap at all.
Movement Analysis.
The movement of the single spots in the short-interval time-lapse movies was tracked using u-track (42). The u-track output was combined with the cell outlines defined by Oufti using SpotprocessR, which plotted the tracks inside the individual cells and calculated and plotted the displacement of individual spots. The collective MSD was calculated using the formula:
where i is the nth track, j is the mth time point, and x(τ) is the position of the tracked point at timepoint τj, or by determining the variances of the histograms of all displacements at different time intervals. The apparent diffusion coefficient Dapp was estimated by fitting a Gaussian curve to a histogram of all displacements as reported previously by Parry et al. (60).
Code Availability.
The code used for image analysis is available (https://github.com/vrrenske/spotprocessR).
Marker Frequency Analysis by Whole-Genome Sequencing.
Marker frequency analysis by whole genome sequencing was performed essentially as described before (28, 53). All strains used for marker frequency analysis (MK453, MK454, DCI15, MK350, MK422, and MK423) were grown under the same conditions as used when growing cells for microscopy analysis; cells of OD600 = 0.4 were diluted 100-fold in C+Y medium with 0.1 mM ZnCl2 and incubated for 2.5 h until OD600 = 0.15. Cells were then harvested by centrifugation for 5 min at 6,500 × g at 4 °C. Genomic DNA was isolated using the Wizard Genomic DNA Purification Kit (Promega) as described previously (28). Fragmentation was performed using Covaris S2 instrument, and libraries were prepared using NEBNext Ultra DNA Library Kit for Illumina before sequencing on an Illuminia HiSEq. 2000 Platform 50 bp single-end [all performed at the Genomics Core Facility, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany]. Using Rockhopper (70), raw reads were mapped onto the genome of S. pneumoniae D39. The genome was split in segments of 1,000 bp, and the number of reads mapped onto each 1,000-bp segment was plotted as a function of the position on the chromosome.
Data Availability.
The raw sequencing data can be accessed at https://seek.sysmo-db.org/investigations/93#datafiles.
Supplementary Material
Acknowledgments
We thank Jeroen Siebring for initial work on parBp; Lieke van Gijtenbeek for providing m(sf)gfp; Oliver Gericke and Katrin Beilharz for technical assistance; GeneCore, EMBL, Heidelberg for sequencing; Jelle Slager and Rieza Aprianto for help with analysis; Dirk-Jan Scheffers for stimulating discussions; and Sophie Martin and Stephan Gruber for constructive feedback on our manuscript. Work in the J.-W.V. laboratory is supported by the European Molecular Biology Organization Young Investigator Program, a VIDI fellowship from the Netherlands Organization for Scientific Research (864.12.001), and European Research Council Starting Grant 337399-PneumoCell. M.K. is supported by The Research Council of Norway Grant 250976/F20.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing data can be accessed at https://seek.sysmo-db.org/investigations/93#datafiles.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620608114/-/DCSupplemental.
References
- 1.Alberts B, et al. The cell cycle. In: Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P, editors. Molecular Biology of the Cell. 6th Ed. Garland Science, Taylor & Francis Group; New York: 2014. pp. 963–1020. [Google Scholar]
- 2.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–447. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 3.Willet AH, McDonald NA, Gould KL. Regulation of contractile ring formation and septation in Schizosaccharomyces pombe. Curr Opin Microbiol. 2015;28:46–52. doi: 10.1016/j.mib.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouet J-Y, Stouf M, Lebailly E, Cornet F. Mechanisms for chromosome segregation. Curr Opin Microbiol. 2014;22:60–65. doi: 10.1016/j.mib.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Rudner DZ. Spatial organization of bacterial chromosomes. Curr Opin Microbiol. 2014;22:66–72. doi: 10.1016/j.mib.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowlett VW, Margolin W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front Microbiol. 2015;6:478. doi: 10.3389/fmicb.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams DW, Wu LJ, Errington J. Cell cycle regulation by the bacterial nucleoid. Curr Opin Microbiol. 2014;22:94–101. doi: 10.1016/j.mib.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 10.Wu LJ, Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 12.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treuner-Lange A, et al. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol Microbiol. 2013;87:235–253. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- 14.Männik J, Bailey MW. Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front Microbiol. 2015;6:306. doi: 10.3389/fmicb.2015.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monahan LG, Liew ATF, Bottomley AL, Harry EJ. Division site positioning in bacteria: One size does not fit all. Front Microbiol. 2014;5:19. doi: 10.3389/fmicb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaritsky A, Woldringh CL. Chromosome replication, cell growth, division and shape: A personal perspective. Front Microbiol. 2015;6:756. doi: 10.3389/fmicb.2015.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajduk IV, Rodrigues CDA, Harry EJ. Connecting the dots of the bacterial cell cycle: Coordinating chromosome replication and segregation with cell division. Semin Cell Dev Biol. 2016;53:2–9. doi: 10.1016/j.semcdb.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Wallden M, Fange D, Lundius EG, Baltekin Ö, Elf J. The synchronization of replication and division cycles in individual E. coli cells. Cell. 2016;166:729–739. doi: 10.1016/j.cell.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 19.Donovan C, Schauss A, Krämer R, Bramkamp M. Chromosome segregation impacts on cell growth and division site selection in Corynebacterium glutamicum. PLoS One. 2013;8:e55078. doi: 10.1371/journal.pone.0055078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinho MG, Kjos M, Veening J-W. How to get (a)round: Mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol. 2013;11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 21.Fadda D, et al. Streptococcus pneumoniae DivIVA: Localization and interactions in a MinCD-free context. J Bacteriol. 2007;189:1288–1298. doi: 10.1128/JB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleurie A, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holečková N, et al. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. MBio. 2014;6:e01700–e01714. doi: 10.1128/mBio.01700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beilharz K, et al. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci USA. 2012;109:E905–E913. doi: 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bramkamp M. Following the equator: Division site selection in Streptococcus pneumoniae. Trends Microbiol. 2015;23:121–122. doi: 10.1016/j.tim.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Grangeasse C. Rewiring the pneumococcal cell cycle with serine/threonine- and tyrosine-kinases. Trends Microbiol. 2016;24:713–724. doi: 10.1016/j.tim.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Minnen A, Attaiech L, Thon M, Gruber S, Veening J-W. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- 28.Kjos M, Veening JW. Tracking of chromosome dynamics in live Streptococcus pneumoniae reveals that transcription promotes chromosome segregation. Mol Microbiol. 2014;91:1088–1105. doi: 10.1111/mmi.12517. [DOI] [PubMed] [Google Scholar]
- 29.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boersma MJ, et al. Minimal peptidoglycan (PG) turnover in wild-type and PG hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host-relevant biofilms. J Bacteriol. 2015;197:3472–3485. doi: 10.1128/JB.00541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paintdakhi A, et al. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol. 2016;99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beilharz K, van Raaphorst R, Kjos M, Veening JW. Red fluorescent proteins for gene expression and protein localization studies in Streptococcus pneumoniae and efficient transformation with DNA assembled via the Gibson assembly method. Appl Environ Microbiol. 2015;81:7244–7252. doi: 10.1128/AEM.02033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 34.Potluri L-P, de Pedro MA, Young KD. Escherichia coli low-molecular-weight penicillin-binding proteins help orient septal FtsZ, and their absence leads to asymmetric cell division and branching. Mol Microbiol. 2012;84:203–224. doi: 10.1111/j.1365-2958.2012.08023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popham DL, Young KD. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr Opin Microbiol. 2003;6:594–599. doi: 10.1016/j.mib.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Manuse S, et al. Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ. Nat Commun. 2016;7:12071. doi: 10.1038/ncomms12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fleurie A, et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 2014;10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsui H-CT, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berg KH, Stamsås GA, Straume D, Håvarstein LS. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol. 2013;195:4342–4354. doi: 10.1128/JB.00184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemon KP, Grossman AD. Movement of replicating DNA through a stationary replisome. Mol Cell. 2000;6:1321–1330. doi: 10.1016/s1097-2765(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 41.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wegmann U, Overweg K, Jeanson S, Gasson M, Shearman C. Molecular characterization and structural instability of the industrially important composite metabolic plasmid pLP712. Microbiology. 2012;158:2936–2945. doi: 10.1099/mic.0.062554-0. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Sergueev K, Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol. 2002;46:985–996. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- 46.Funnell BE, Gagnier L. P1 plasmid partition: Binding of P1 ParB protein and Escherichia coli integration host factor to altered parS sites. Biochimie. 1994;76:924–932. doi: 10.1016/0300-9084(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Montero Llopis P, Rudner DZ. Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA. 2014;111:12877–12882. doi: 10.1073/pnas.1407461111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol. 2014;22:102–110. doi: 10.1016/j.mib.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr Biol. 2014;24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gruber S, et al. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr Biol. 2014;24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez-Moreira E, Balas D, Gonzalez I, de la Campa AG. Fluoroquinolones inhibit preferentially Streptococcus pneumoniae DNA topoisomerase IV than DNA gyrase native proteins. Microb Drug Resist. 2000;6:259–267. doi: 10.1089/mdr.2000.6.259. [DOI] [PubMed] [Google Scholar]
- 53.Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet. 2014;10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espéli O, et al. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 2012;31:3198–3211. doi: 10.1038/emboj.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woldringh CL. The role of co-transcriptional translation and protein translocation (transertion) in bacterial chromosome segregation. Mol Microbiol. 2002;45:17–29. doi: 10.1046/j.1365-2958.2002.02993.x. [DOI] [PubMed] [Google Scholar]
- 58.Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc Natl Acad Sci USA. 2012;109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marbouty M, et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 60.Parry BR, et al. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156:183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Land AD, et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol Microbiol. 2013;90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin B, García P, Castanié M-P, Claverys J-P. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol Microbiol. 1995;15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 63.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Jong IG, Beilharz K, Kuipers OP, Veening J-W. Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J Vis Exp. 2011;(53):3145. doi: 10.3791/3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caldas VEA, Punter CM, Ghodke H, Robinson A, van Oijen AM. iSBatch: A batch-processing platform for data analysis and exploration of live-cell single-molecule microscopy images and other hierarchical datasets. Mol Biosyst. 2015;11:2699–2708. doi: 10.1039/c5mb00321k. [DOI] [PubMed] [Google Scholar]
- 68.Fraley C, Rafterty AE, Brendan Murphy T, Scrucca L. mclust Version 4 for R: Normal Mixture Modeling for Model-Based Clustering, Classification, and Density Estimation. University of Washington; Seattle: 2012. [Google Scholar]
- 69.Fraley C, Rafterty AE. Model-based clustering, discriminant analysis and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 70.McClure R, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data can be accessed at https://seek.sysmo-db.org/investigations/93#datafiles.