Significance
Fibrous sheath interacting protein 1 (FSIP1) has recently been identified as a prognostic marker for human epidermal growth factor receptor 2 (HER2)-positive breast cancers. Its increased expression is importantly associated with poor outcomes. Here, we document that FSIP1 is a signaling partner to HER2, and that FSIP inhibition reduces cell growth and invasiveness in HER2-positive breast cancer cells. Considering its limited expression in normal tissues, FSIP1 may become a potentially druggable target for the therapy of HER2-positive breast cancers, and provide a new mechanism for improved therapeutic responses to current HER2 blockers, such as trastuzumab.
Keywords: GO analysis, KEGG pathway analysis, gene expression signature, breast cancer therapeutics
Abstract
Fibrous sheath interacting protein 1 (FSIP1), a spermatogenesis-related testicular antigen, is expressed in abundance in breast cancers, particularly in those overexpressing human epidermal growth factor receptor 2 (HER2); however, little is known about its role in regulating the growth and metastasis of breast cancer cells. We and others have shown previously that FSIP1 expression in breast cancer correlates positively with HER2-positivity, recurrence, and metastases and negatively with survival. Here, using coimmunoprecipitation and microscale thermophoresis, we find that FSIP1 binds to the intracellular domain of HER2 directly. We further show that shRNA-induced FSIP1 knockdown in SKBR3 and MCF-7 breast cancer cells inhibits proliferation, stimulates apoptosis, attenuates epithelial–mesenchymal transition, and impairs migration and invasiveness. Consistent with reduced proliferation and enhanced apoptosis, xenotransplantation of SKBR3 cells stably transfected with sh-FSIP1 into nu/nu mice results in reduced tumor volumes compared with sh-NC transplants. Furthermore, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) mapping using sh-FSIP1 gene signature yielded associations with extracellular matrix protein pathways, and a reduction in SNAI2 protein expression was confirmed on Western blot analysis. Complementarily, interrogation of the Connectivity Map using the same gene signature yielded, as top hits, chemicals known to inhibit epithelial–mesenchymal transition, including rapamycin, 17-N-allylamino-17-demethoxygeldanamycin, and LY294002. These compounds phenocopy the effects of sh-FSIP1 on SKBR3 cell viability. Thus, FSIP1 suppression limits oncogenesis and invasiveness in breast cancer cells and, considering its absence in most other tissues, including normal breast, may become a potential target for breast cancer therapy.
Therapy for breast cancer has evolved considerably over the past decade through the use of chemotherapy, radiation and endocrine therapy, and targeted therapies, such as trastusumab (Herceptin) for human epidermal growth factor receptor 2 (HER2)-positive cancers (1). Nonetheless, some patients fail targeted therapy and/or experience recurrence, prompting the continuing search for new prognostic markers (2). Confirming previous data (3), our group recently documented that the testicular antigen fibrous sheath interacting protein 1 (FSIP1) was expressed abundantly in breast cancer tissues (4), and that its abundance is correlated positively with HER2 expression and tumor invasiveness and negatively with disease-specific survival (5).
A component of the fibrous sheath of sperm flagellum, FSIP1 is known to assemble AKAP4, an A-kinase protein that binds the regulatory subunit of protein kinase A (PKA) (6, 7). Whereas the function of FSIP1 in tumorigenesis is poorly understood, AKAP4 is known to be expressed in several cancers, including multiple myeloma and prostate and breast cancers (6, 8). PKA activation also has been shown to confer resistance to trastuzumab (9). More recent studies have implicated FSIP1, together with some other spermatogenesis-associated genes, in the regulation of chromosome segregation (4). The depletion of FSIP1 thus results in abnormal mitosis, which includes the enhancement of multipolar spindles and mitotic transit time (4). FSIP1 is also a known target for steroid receptor coactivator-3 (SRC-3), an oncogene associated with breast cancer and a coactivator for nuclear receptors, such as estrogen receptor (ER) α (10).
Several key findings implicate FSIP1 not only as a marker of breast cancer severity, but also as a potential therapeutic target. First, FSIP1 expression correlates with the abundance of HER2 in breast cancers. HER2-overexpressing SKBR3 cells thus display higher FSIP1 levels than MCF-7 cells (5). Second, FSIP1 expression is notably higher in breast cancer tissue with high Ki67 expression, indicating a correlation with cell proliferation (5). Third, FSIP1 is detectable in the serum of breast cancer patients, and high serum FSIP1 is associated with worse postoperative disease-specific survival (5). Finally, with the exception of the testes and pituitary, FSIP1 displays limited expression in noncancerous tissues, including normal breast (https://gtexportal.org/home/gene/FSIP1).
Consequently, we were prompted to investigate whether this spermatogenesis-associated protein could serve as a potential target for breast cancer therapy. In the present study, we found that FSIP1 binds directly to HER2. We also explored the effect of inhibiting FSIP1 expression in two breast cancer cell lines, SKBR3 and MCF-7. Short hairpin (sh) RNA-induced FSIP1 knockdown was found to inhibit tumor cell proliferation and migration, wound healing, and epithelial–mesenchymal transition, and to induce cell cycle arrest and apoptosis. It also reduced tumor volume when SKBR3 cells were transplanted into nu/nu mice. Finally, the FSIP1 inhibition gene signature mapped to extracellular matrix component genes in both Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) mapping, and on Connectivity Mapping (CMAP) computation yielded chemicals that phenocopied the survival inhibitory effect of sh-FSIP1.
Results
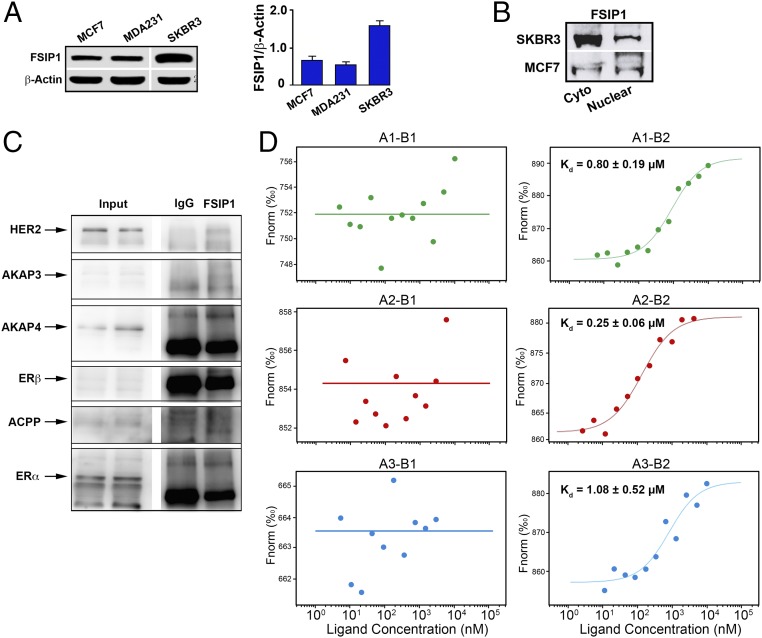
Clinical data show that 45% of HER2+ and 24% of HER2− breast tissues immunolabel for FSIP1. We confirmed that ER−; PR−; HER2high SKBR3 cells express higher levels of FSIP1 protein compared with ER+; PR+; HER2low MCF-7 and ER−; PR−; HER2low MDA-MB-231 cells (Fig. 1A). Furthermore, FSIP1 was present in both the cytosol and nuclear compartments (Fig. 1B). Considering that high FSIP1 was also found to correlate with a poor prognosis in HER2+ breast cancer patients (5), we tested whether FSIP1 directly interacts with HER2. Coimmunoprecipitation using an antibody to FSIP1 and immunoblotting with an anti-HER2 antibody revealed a bound product (Fig. 1C). In contrast, no bound products with FSIP1 were seen on immunolabeling for AKAP3, acid phosphatase, prostate (ACPP), ERα, ERβ, and surprisingly, AKAP4 (Fig. 1A) (6, 7).
Fig. 1.
The intracellular domain of HER2 binds FSIP1. (A) Western blots showing greater expression of FSIP1 protein in extracts of SKBR3 cells, compared with MCF-7 and MDA-MB-231 cells. Densitometry of blots is shown (mean ± SEM, n = 3 biological replicates). (B) Western blots using cytosolic and nuclear fractions extracted from SKBR3 or MCF-7 cells, showing FSIP1 localization to both cellular compartments. (C) Immunoprecipitation using resin coated with FSIP1 antibody or IgG, following immunoblotting of the complex by antibodies against HER2, AKAP3, AKAP4, ERβ, ACPP, and Erα. (D) Microscale thermophoresis assay (NanoTemper) in which biomolecular interactions are quantitated by examining the motion of the molecules along a microscopic temperature gradient induced by an infrared laser. Changes in the molecular hydration shell, charge, or size are measured using a fluorescent probe (NT-647) bound covalently to the extracellular (B1) or intracellular (B2) domain of HER2. Binding of three recombinant FSIP1 fragments—FSIP11-350 (A1), FSIP1351-581 (A2), and FSIP1191-581 (A3)—were studied. There was a concentration-dependent binding between the intracellular B2 (but not B1) domain of HER2 and all three FSIP1 fragments, with the best binding noted with the C-terminal A2 fragment (FSIP1351-581). Also shown are dissociation constants (Kd).
To study FSIP1-HER2 binding further, we used a microscale thermophoresis assay (NanoTemper) in which biomolecular interactions are quantitated by examining the motion of the molecules along a microscopic temperature gradient induced by an infrared laser. Changes in the molecular hydration shell, charge, or size are measured using a fluorescent probe (NT-647) bound covalently to one of the binding partners, in our case the extracellular (B1) or intracellular (B2) domain of HER2. We studied the binding of three recombinant FSIP1 fragments—FSIP11-350 (A1), FSIP1351-581 (A2) and FSIP1191-581 (A3)—and found that none bound to the extracellular B1 domain of HER2. However, we did note concentration-dependent binding between the intracellular B2 domain of HER2 and all three FSIP1 fragments, with the best binding noted with the C-terminal A2 fragment (FSIP1351–581) (Kd = 0.25 ± 0.06 µM) (Fig. 1D). Although the submicromolar affinity between HER2 and FSIP1 may be the result of our testing of recombinant proteins in a cell-free system, these Kd values may indeed reflect relatively weak (albeit functionally relevant) interaction between the two molecules in vivo.
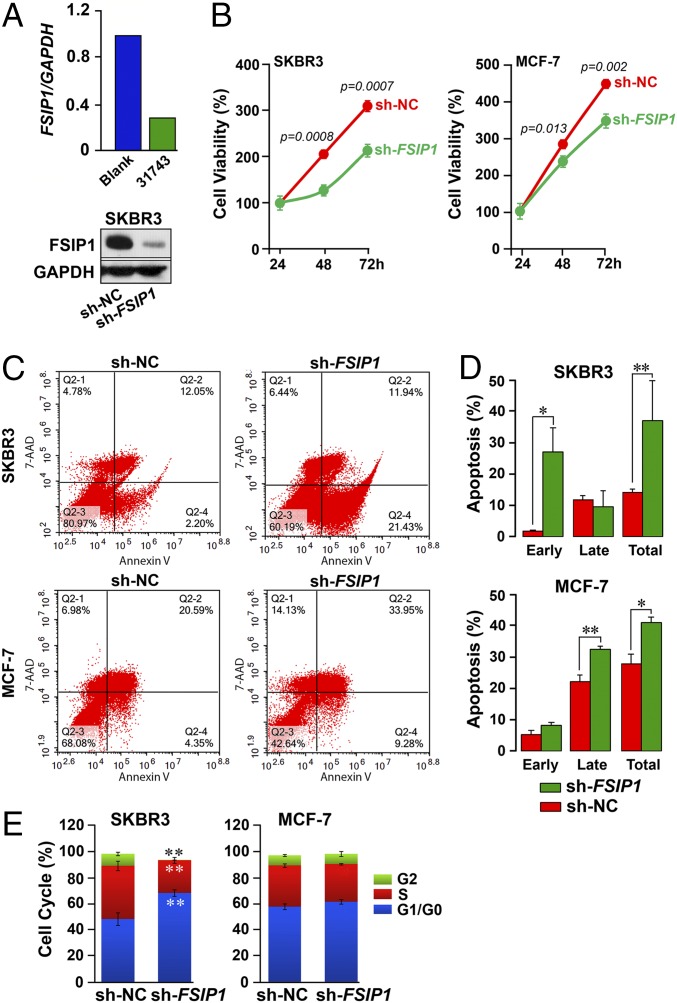
We tested the effect of inhibiting FSIP1 expression, for which we designed three shRNA constructs: 31740, 31741, and 31743 (Genechem). Of these, 31743 (hereafter referred as sh-FSIP1) elicited a highly significant decrease in FSIP1 mRNA (quantitative PCR) and protein (Western immunoblot) in SKBR3 cells (Fig. 2A), and was used for inhibition in further experiments, together with sh-NC (scrambled shRNA control). SKBR3 and MCF-7 cells, seeded on six-well culture plates, were transfected at 40–50% confluence with sh-FSIP1 or sh-NC (TurboFect; Fermentas), and proliferation was measured using the CCK-8 Cell Counting assay (Sigma-Aldrich). FSIP1 depletion significantly decreased proliferation at 48 h in both cell lines, with the greatest effect an ∼45% reduction by 72 h in SKBR3 cells (P values shown; Fig. 2B).
Fig. 2.
Inhibition of FSIP1 expression reduces cell proliferation and induces apoptosis in breast cancer cells. (A) Short hairpin RNA-mediated knockdown of FSIP1 (sh-FSIP1) in SKBR3 cells, showing reduced mRNA (qPCR) and protein (Western blot) expression (sh-NC: scrambled control; 3 technical replicates for qPCR). (B) CCK-8 Assay (Sigma-Aldrich) used to determine viable SKBR3 or MCF-7 cells following transfection with sh-NC or sh-FSIP1 (mean ± SEM, n = 3 biological replicates; P values against sh-NC shown). SKBR3 or MCF-7 cells transfected with sh-NC or sh-FSIP1 double-stained for Annexin V and 7-AAD and analyzed by FACS (FACSAria III, BD Bioscience). Cells in early and late apoptosis were defined as Annexin V+:7-AAD− (Q2-4) or double-positive (Q2-2) cells, respectively. (C and D) Representative FACS plots (C) and mean ± SEM for apoptotic cells in Q2-4 (early), Q2-2 (late) or both quadrants (total) (D) are shown (repeated three times; *P ≤ 0.05; **P ≤ 0.01). (E) Cell cycle FACS analysis shows that sh-FSIP1 transfection results in greater number of cells in the G0/G1 phase, and none in G2, compared with sh-NC transfectants. No differences were noted in MCF-7 cells. Mean ± SEM, n = 3 biological replicates; two-tailed Student's t test, **P ≤ 0.01.
We also assessed the effects of reduced FSIP1 expression on the survival of SKBR3 and MCF-7 cells using the Annexin V/7-AAD apoptosis assay (KeyGen Biotech). Double-stained cells were subject to flow cytometry (FACSAria III; BD Biosciences). Cells in early apoptosis were defined as Annexin V+:7-AAD− (Q2-4), and those in late apoptosis as double-positive (Q2-2) (Fig. 2C). The total percentage of apoptotic cells was significantly higher in sh-FSIP1 transfectants than in sh-NC in both SKBR3 and MCF-7 cells (P = 0.01 and 0.03, respectively), with increases noted in early and late apoptosis, respectively (Fig. 2 C and D). Consistent with the apoptosis, there was a significant increase in G1/G0 of sh-FSIP1 transfected SKBR3 cells compared with sh-NC cells (P = 0.0015) (Fig. 2E). There were also lesser cells in the S and G2/M phases of the SKBR3 transfectants (P = 0.0015 and 0.00015, respectively). No difference in MCF-7 cells was noted (Fig. 2E).
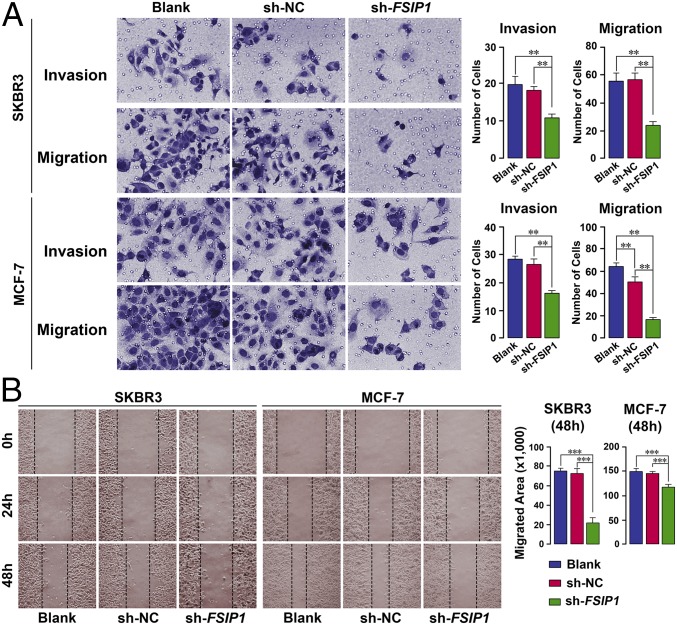
We directly studied in vitro migration and invasiveness in 24-well plates inserted with 8.0-mm-pore transwells (EMD Millipore). FBS-free medium containing ∼25,000 cells was placed in the top chamber of the insert, with complete medium (10% FBS) with or without placental growth factor (PlGF) in the lower chamber. After a 6-h incubation, migrated cells adhering to the underside of the membrane were methanol-fixed and stained with crystal violet. The in vitro invasion assay was performed similarly, except that the membranes were coated with Matrigel and prehydrated in serum-free medium, and cells were incubated for 36 h before fixation and staining. Highly significant decreases in both migration and invasiveness were noted in sh-FSIP1–transfected cells compared with untreated or sh-NC cell groups (Fig. 3A).
Fig. 3.
FSIP1 inhibition attenuates breast cancer cell migration and invasiveness. (A) For the migration assay, top and bottom chambers of 24-well plates inserted with 8.0-mm-pore transwells (Millipore) contained, respectively, FBS-free or complete medium (10% FBS). For the invasion assay, the above membranes were coated with Matrigel and prehydrated in serum-free medium (Materials and Methods). Crystal violet-stained cells adhering to the underside of the membranes are shown. Highly significant decreases in both migration and invasiveness were noted upon sh-FSIP1 transfection compared with untreated (blank) or sh-NC cells. (B) Migration was also assessed using the scratch assay, wherein SKBR3 or MCF-7 cells plated at 90–100% confluence were subjected to the scratch test. Image J was used to measure the closure rate over 24 and 48 h following sh-NC or sh-FSIP1 transfection (Materials and Methods). Mean ± SEM; n = 10 samples per treatment for the migration assay and three replicates for each time point for the invasion assay; two-tailed Student's t test, **P ≤ 0.01, ***P ≤ 0.001.
Complementarily, migration was assessed using the in vitro scratch assay, which involves creating a “scratch” in a cell monolayer, followed by image capture at regular intervals to evaluate cell migration to close the scratch (11). MCF-7 and SKBR3 cells at 90–100% confluence were plated uniformly in 18 wells and subjected to the scratch test. Measurement of rate using Image J revealed that although the scratch closed progressively over 24 and 48 h with untreated and sh-NC cells, this closure was markedly attenuated at both time points with sh-FSIP1 cells (Fig. 3B).
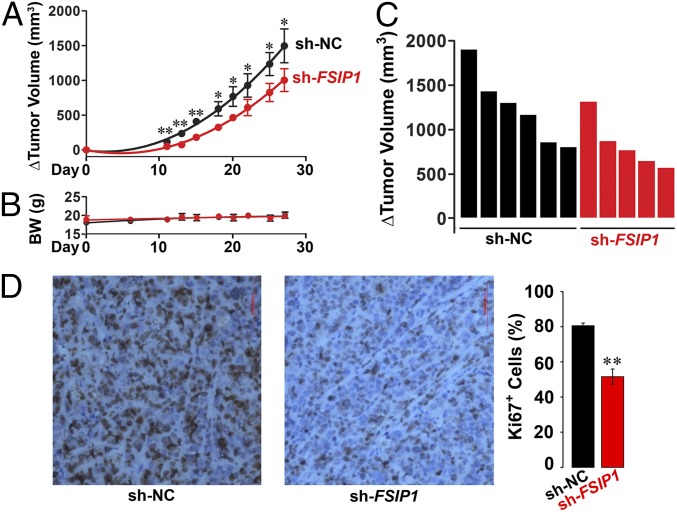
For proof-of-concept that reduced FSIP1 expression inhibits tumor growth in vivo, we xenotransplanted HER2high SKBR3 cells stably transfected with either sh-FSIP1 or sh-NC into BALB/c nu/nu mice (12). Sequential measurements of tumor volume by calipers over a 27-d period showed reduced growth of sh-FSIP1–transfected tumors compared with sh-NC-transfected tumors (Fig. 4A). No effects on body weight were noted (Fig. 4B). These data are also shown as Waterfall plots at 27 d (Fig. 4C). This effect was associated with reduced Ki67 immunostaining, indicative of reduced cell proliferation in vivo (Fig. 4D). These actions of FSIP1 on tumor growth are likely coupled with reduced tumor invasiveness. Although we have not measured invasiveness (or metastasis) directly in vivo, our data showing the effect of FSIP1 inhibition on tumor migration and invasiveness in three in vitro assays would suggest a strong effect on tumor metastasis in vivo. Thus, collectively our data show that shRNA-mediated reduction of FSIP1 expression attenuates breast cell proliferation, migration, and invasiveness and increases apoptosis, with a prominent net reduction in tumor growth in vivo noted in HER2high SKBR3 cells.
Fig. 4.
Breast cancer cells stably transfected with sh-FSIP1 display reduced growth when xenotransplanted in nude mice. SKBR3 cells stably transfected with sh-FSIP1 or sh-NC were injected into the flanks of BALB/c nu/nu mice and tumor volume measured by calipers for 27 d, after which the mice were killed. (A–C) Change (Δ) in tumor volume (A) and biweekly body weight (B) measurements, are shown together with Waterfall plot comparing Δ tumor volume in individual mice (C). (D) Ki67 staining of tissues. Mean ± SEM; n = 5 and 6 mice injected with sh-FSIP1 and sh-NC transfectants, respectively. Two-tailed Student's t test, *P ≤ 0.05, **P ≤ 0.01.
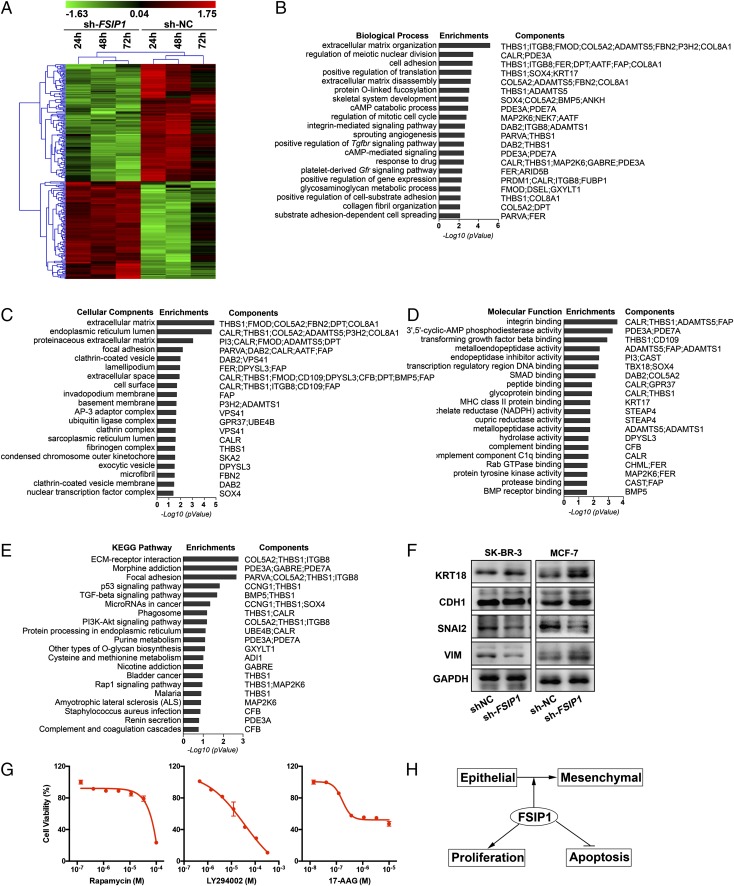
We further explored the mechanism underlying the antitumor actions of FSIP1 inhibition. Affymetrix microarray profiling of gene expression in MCF-7 cells transfected with sh-FSIP1 or sh-NC showed up-regulation of 106 probe sets and down-regulation of 138 probe sets (Fig. 5A and Dataset S1). The gene set was used to probe GO, using biological processes, cellular components, and molecular function as GO terms. This yielded top hits of extracellular matrix organization, extracellular matrix, and integrin binding with P values of 6.86 × 10−6, 1.43 × 10−5, and 2.66 × 10−4, respectively (Fig. 5 B–D). To further validate this bioinformatic output, we probed KEGG with the same gene set for pathway analysis. Once again, the top hit for FSIP1 inhibition was ECM–receptor interactions (P = 1.74 × 10−3) (Fig. 5E).
Fig. 5.
FSIP1 inhibition tracks with extracellular matrix components. (A) Microarray analysis (Affymetrix) of mRNA extracted from MCF-7 cells transfected with sh-NC or sh-FSIP1 for 24, 48 and 72 h, shown as a heat map of regulated genes (gene clusters noted; Materials and Methods). (B–E) A gene signature was derived from this dataset (Dataset S1), which was used to interrogate the GO (B–D) or KEGG (E) databases. The GO terms included biological processes, cellular components, and molecular functions. Note that the top hit in both GO and KEGG related to extracellular matrix proteins. (F) Western immunoblot confirmation of the expression of extracellular matrix interacting gene products involved in epithelial–mesenchymal transition, namely KRT18, CDH1, SNAI2, and vimentin (VIM) (GAPDH as loading control), in SKBR3 and MCF-7 cells transfected with sh-NC or sh-FSIP1 . (G) Effect of the top three hits obtained on CMAP interrogation (Table S1)—rapamycin, 17-AAG, and LY294002—on cell viability after treatment for 3 d (MTT assay). (H) Schematic showing that FSIP1 induces epithelial–mesenchymal transition, accelerates proliferation, and inhibits apoptosis, accounting for the increased aggressiveness of FSIP1-positive breast cancers noted in humans (5).
In view of the potential role of extracellular matrix components, and effects on migration and invasiveness, we chose to focus on proteins involved in epithelial–mesenchymal transition. Western blot analysis showed an increase in keratin 18 (KRT18), with no change in E-cadherin (CDH1), in sh-FSIP1 transfectants, but with a reduction in the epithelial marker SNAI2, consistent with reduced epithelial–mesenchymal transition and cell migration (Fig. 5F).
With the sh-FSIP1 gene signature at hand, we used CMAP to discover other chemicals that could mimic the FSIP1 inhibition pathway. CMAP provides a unique strategy for unraveling new actions of drugs currently used for other medical conditions (13–15). The approach uses a gene signature to query the CMAP for gene expression profiles (https://portals.broadinstitute.org/cmap/). Nonparametric, rank-based pattern matching using the Kolmogorov–Smirnov statistic is used to examine shared mechanisms of action to reveal both mimics and antimimics of any biological intervention (15). We have previously used this strategy to unravel an action of the commonly used class of drugs for osteoporosis and skeletal metastasis—the bisphosphonates—on the EGFR family of tyrosine kinase receptors (14).
A global gene expression profile from a transforming growth factor-β (TGFβ)-induced epithelial–mesenchymal transition model was previously used to identify potential modulators, including rapamycin, 17-N-allylamino-17-demethoxygeldanamycin (17-AAG), and LY294002. The same three chemicals appeared within the first five hits with our sh-FSIP1 gene signature, with P values of ≤6 × 10−5, indicating that they likely modulate epithelial–mesenchymal transition (Table S1), although this has not been tested by us directly. Nonetheless, we found that the three compounds reduced the viability of SKBR3 cells (Fig. 5G).
Table S1.
The 40 most significant hits in sh-FSIP1–transfected MCF-7 cells after CMAP analysis
| Rank | CMAP name | Mean | n | Enrichment | P | Specificity | Percent nonnull | Biological function |
| 1 | 5155877 | 0.689 | 4 | 0.968 | 0.000 | 0.000 | 100 | |
| 2 | Sirolimus (rapamycin) | −0.354 | 44 | −0.574 | 0.000 | 0.055 | 72 | mTor inhibitor |
| 3 | LY-294002 | −0.331 | 61 | −0.46 | 0.000 | 0.086 | 65 | PI3K inhibitor |
| 4 | Withaferin A | 0.634 | 4 | 0.928 | 0.000 | 0.042 | 100 | IKKβ inhibitor |
| 5 | Tanespimycin (17-AAG) | −0.194 | 62 | −0.286 | 0.000 | 0.361 | 51 | HSP90 inhibitor |
| 6 | Fulvestrant | −0.205 | 40 | −0.336 | 0.000 | 0.022 | 55 | Selective estrogen receptor degrader |
| 7 | Baclofen | −0.434 | 5 | −0.782 | 0.001 | 0.000 | 100 | GABAB receptor agonist |
| 8 | Semustine | 0.476 | 4 | 0.841 | 0.001 | 0.068 | 100 | Chemotherapy |
| 9 | Vorinostat | 0.242 | 12 | 0.527 | 0.001 | 0.407 | 58 | Histone deacetylase inhibitor |
| 10 | Equilin | 0.427 | 5 | 0.777 | 0.001 | 0.000 | 100 | Estrogen (for HRT) |
| 11 | Diethylstilbestrol | 0.38 | 6 | 0.712 | 0.002 | 0.047 | 83 | Endocrine disruptor |
| 12 | Berberine | −0.466 | 4 | −0.815 | 0.002 | 0.006 | 100 | Antidiabetic, antibiotic |
| 13 | 3-Nitropropionic acid | −0.45 | 4 | −0.814 | 0.002 | 0.006 | 100 | Mycotoxin |
| 14 | Cefotiam | 0.412 | 4 | 0.81 | 0.002 | 0.006 | 100 | Antibiotic |
| 15 | Naringenin | 0.464 | 4 | 0.809 | 0.002 | 0.011 | 100 | Flavonoid |
| 16 | Mephentermine | −0.173 | 5 | −0.729 | 0.003 | 0.000 | 60 | Cardiac stimulant |
| 17 | Imatinib | 0.59 | 2 | 0.957 | 0.003 | 0.019 | 100 | Tyrosine kinase inhibitor |
| 18 | Phenazopyridine | −0.397 | 4 | −0.796 | 0.003 | 0.013 | 100 | Analgesic |
| 19 | Isotretinoin | 0.415 | 4 | 0.763 | 0.006 | 0.005 | 100 | Retinoid for acne and, rarely, skin cancer |
| 20 | 0179445–0000 | −0.363 | 8 | −0.559 | 0.007 | 0.020 | 75 | |
| 21 | Glafenine | 0.357 | 4 | 0.755 | 0.007 | 0.036 | 100 | Nonsteroidal anti-inflammatory drug |
| 22 | Conessine | −0.252 | 4 | −0.757 | 0.007 | 0.000 | 75 | Histamine H3 antagonist |
| 23 | Thioguanosine | −0.312 | 4 | −0.756 | 0.007 | 0.021 | 75 | Leukemia drug |
| 24 | Rescinnamine | 0.449 | 3 | 0.843 | 0.008 | 0.011 | 100 | Angiotensin-converting enzyme inhibitor |
| 25 | Prasterone | 0.386 | 4 | 0.749 | 0.008 | 0.005 | 100 | Dehydroepiandrosterone |
| 26 | Colecalciferol | −0.372 | 4 | −0.748 | 0.008 | 0.011 | 75 | Vitamin D supplement |
| 27 | Butirosin | −0.302 | 4 | −0.747 | 0.008 | 0.006 | 75 | Antibiotic |
| 28 | Suloctidil | −0.314 | 4 | −0.739 | 0.009 | 0.033 | 75 | Vasodilator |
| 29 | N-Acetylmuramic acid | 0.25 | 4 | 0.736 | 0.009 | 0.006 | 75 | Bacterial cell wall biopolymer |
| 30 | Sulfamonomethoxine | 0.284 | 4 | 0.729 | 0.011 | 0.025 | 75 | Antibiotic and antimicrobial agent |
| 31 | Cinoxacin | −0.274 | 4 | −0.726 | 0.012 | 0.010 | 75 | Quinolone antibiotic |
| 32 | Iloprost | 0.378 | 3 | 0.818 | 0.012 | 0.053 | 100 | Pulmonary arterial hypertension drug |
| 33 | Ouabain | −0.465 | 4 | −0.723 | 0.012 | 0.053 | 75 | Cardiac glycoside |
| 34 | Prestwick-689 | −0.459 | 4 | −0.716 | 0.013 | 0.023 | 75 | Androsterone |
| 35 | Dinoprostone | 0.384 | 4 | 0.71 | 0.014 | 0.031 | 75 | Prostaglandin E2 |
| 36 | Sitosterol | −0.296 | 4 | −0.708 | 0.015 | 0.056 | 75 | Phytosterol |
| 37 | Prestwick-1100 | 0.384 | 4 | 0.703 | 0.016 | 0.007 | 75 | |
| 38 | Galantamine | −0.293 | 4 | −0.698 | 0.017 | 0.000 | 75 | Central acetylcholinesterase inhibitor |
| 39 | Exisulind | 0.476 | 2 | 0.906 | 0.018 | 0.000 | 100 | Antineoplastic agent |
| 40 | CP-944629 | 0.361 | 4 | 0.693 | 0.019 | 0.008 | 75 |
Shown are the mean connectivity score, the number of instances (n), the enrichment score, P values, specificity score, and percent nonnull. The known biological action of each compound is stated.
Discussion
We and others have shown previously that the testicular, spermatogenesis-related antigen FSIP1 is expressed in abundance in various cancers, including multiple myeloma, prostate cancer, and breast cancer (4–6, 8), whereas it is clear from extensive RNA-sequencing studies that only normal testes and pituitary express FSIP1 (https://gtexportal.org/home/gene/FSIP1). In fact, normal breast tissue surrounding the cancer does not express FSIP1 to any appreciable extent (5). In breast cancer tissue, however, FSIP1 expression correlates with HER2-positivity and Ki67 expression (5). Serum FSIP1 levels are also high in ER+ and PR+ breast cancers, and FSIP1-positivity in HER2− and ER+ cancers signals poor postoperative disease-specific survival (5).
Here we report that FSIP1 binds HER2 directly, and that inhibition of FSIP1 expression in HER2-positive breast cancer cells results in reduced cell proliferation, increased apoptosis, and attenuated cell migration and invasiveness. The binding of FSIP1 to HER2, a pleiotropic signaling receptor tyrosine kinase, represents a previously unreported association. Of note is that HER2 activation has diverse effects on signaling pathways involved in breast cancer pathogenesis through as-yet unclear mechanisms. In this context, our identification of an important signaling partner to HER2 that is not directly related to its kinase activity could help explain the pleiotropic signaling capability of HER2. Another implication is that FSIP1 may be not only a prognostic marker for invasive breast cancer, but also a potentially druggable target. We know that the combination of two agents targeting HER2 provides better efficacy compared with a single HER2 modulator; thus, it is possible that FSIP1 inhibition might reflect yet another therapeutic mechanism, possibly through the inhibition of epithelial–mesenchymal transition and cell migration, as noted below.
Indeed, inhibition of FSIP1 expression reduced cell migration and invasiveness, an effect mediated through the suppression of extracellular matrix proteins evident on GO and KEGG mapping and confirmed on Western blot analysis. Notably, SNAI2 was down-regulated, KRT18 was up-regulated, and CDH1 showed no change. The reduction in SNAI2 is consistent with reduced epithelial–mesenchymal transition. However, whereas SNAI2 is known to repress CHD1 by binding to the E-box on its promoter (16, 17), sh-FSIP1 transfectants did not display increases in CDH1. In contrast, KRT18, a member of the filament gene family, was elevated on FSIP1 inhibition. In its role in regulating translocation of proteins to cell membranes, KRT18 is normally repressed during epithelial–mesenchymal transition (18). We remain uncertain as to why vimentin, a regulator of motor proteins (19), is regulated reciprocally in SKBR3 and MCF-7 cells. With that said, it is clear from our studies that FSIP1 inhibition not only induces apoptosis and inhibits proliferation to reduce overall cancer cell burden, as noted in vivo, but also attenuates cell migration through an action on proteins responsible for epithelial–mesenchymal transition (Fig. 5H). Studies using in vivo assays for migration and invasiveness should shed further light on the possibility that FSIP1 inhibition also reduces metastasis.
Finally, we used our sh-FSIP1 gene signature to interrogate CMAP, a database of interacting genes, diseases, and drugs, to identify candidate compounds that might trigger similar intracellular mechanisms. CMAP permits the repurposing of existing, often Food and Drug Administration-approved drugs and chemicals, for other uses. For example, CMAP has been used to identify topiramate, an antipsychotic, for potential use in inflammatory bowel disease (20). Likewise, we have found that bisphosphonates, a drug class extensively used for the therapy of osteoporosis and skeletal metastasis, has the potential for adjunctive use in EGFR-driven lung, breast, and other cancers (12, 14). Two antihelminthic agents and a β-blocker similarly track on CMAP with a Gaucher disease gene signature (13).
We were intrigued to find that the CMAP output of FSIP1 inhibition was remarkably similar to that seen with a TGFβ1 cellular model of epithelial–mesenchymal transition (21). Three drugs—rapamycin, 17-AAG, and LY294002, which inhibit mechanistic target of rapamycin (mTOR), heat shock protein 90 (HSP90), and phosphatidylinositide 3-kinase (PI3K), respectively—displayed P values of ≤10−5 with our sh-FSP1 gene signature. All three compounds have been shown to inhibit epithelial–mesenchymal transition, as well as cellular migration and invasion in non–small-cell lung cancer (A549) cells (21), and, in our hands, to reduce the viability of SKBR3 cells. The three drugs have also been shown to inhibit TGFβ1-induced up-regulation of N-cadherin (CDH2) in H358 lung cancer cells, and to induce cell migration and invasion (21). This is not unexpected given the cross-talk between PI3K and mTOR pathways, as well as the effect of HSP90 on stabilizing the TGFβ receptor.
Despite these associations, the mechanism through which FSIP1 inhibition mimics the anti–epithelial–mesenchymal transition triggered by TGFβ1 remains unclear. It is unlikely that this occurs through the inhibition of PKA activation and the subsequent reduction of PI3K activity and mTOR phosphorylation (22), considering that FSIP1 does not bind AKAP4 in our hands. Notwithstanding the absence of a putative mechanism, what is clear from our studies is the potential for targeting FSIP1 with small molecules, particularly for cancers that overexpress FSIP1 and HER2.
Materials and Methods
Human breast carcinoma MCF-7 (luminal, ER+; PR+; HER2−), SK-BR3 (ER−; PR−; HER2+), and MDA-MB-231 (basal-like, ER−; PR−; HER2−) cell lines were obtained from American Type Culture Collection. Anti-FSIP1 (ab87570), anti-HER2 (ab16826), anti-ERα (ab75635), anti-ERβ (ab92306), and anti-ACPP (ab108984) antibodies were purchased from Abcam. Anti-AKAP3 (13907–1-AP) and anti-AKAP4 (24986–1-AP) antibodies were purchased from Proteintech. For coimmunoprecipitation, SKBR3 cell lysates were incubated with resins (Pierce 26149; Thermo Fisher Scientific) immobilized with either anti-FSIP1 antibody or IgG. After washing, eluates were immunoblotted using the indicated antibodies.
For thermophoresis assays, recombinant proteins of the extracellular (B1) and intracellular (B2) domains were N-hydroxysuccinimide (NHS) ester-modified and fluorescently labeled with NT-647. B1 and B2 were incubated with 10 µM–5 nM recombinant FSIP1 fragments (A1, A2, or A3) in PBS. Microscale thermophoresis measurements were performed with a NanoTemper Monolith NT.115. FSIP1 shRNA (31743; sh-FSIP1) and scrambled control shRNA (sh-NC) were designed, synthesized, and cloned into pGPU6/GFP/Neo plasmid (GenePharma) to generate sh-FSIP1 and sh-NC, respectively. These shRNAs were then stably transfected into the respective cancer cells.
Proliferation studies were performed using a Sigma-Aldrich CCK-8 Cell Counting Kit. For apoptosis measurements, cells were cultured at 80% confluence and stained with Annexin V-APC and 7-AAD (Nanjing KeyGen Biotech). Apoptotic cells were identified by flow cytometry (FACSAria III; BD Biosciences). The impact of FSIP1 knockdown on cell migration and invasion was assessed by Transwell migration and invasion assays, respectively. In brief, for the migration assay, stably transfected MCF-7 and SKBR3 cells (25,000 cells/well) were cultured in triplicate in FBS-free medium in the top chambers of 24-well 8.0-mm-pore Transwell plates (EMD Millipore). After 6 h of culture, cells on the top surface of the membrane were removed with a cotton swab. The migrated cells on the bottom surface of the membrane were fixed with methanol, stained with crystal violet solution, and counted in 10 random fields (blinded). Likewise, for the invasion assay, cells were cultured in triplicate in the top chambers, which had been coated with Matrigel for 36 h, to determine the numbers of invading cells.
For the wound healing assay, the stable transfectants were cultured in triplicate in 24-well plates. On reaching 90% confluence, the monolayer of each well was scraped in a straight line using a p200 pipet tip. After washing, the cells were cultured in 10% FBS medium for 24 and 48 h. The migrated areas of cells in individual wells were determined using ImageJ.
The protocol for the xenograft studies was approved by the Animal Research and Care Committee of Dalian Medial University. Six-week-old female BALB/c nu/nu mice were obtained from Silaike Laboratory Animal and housed in a specific pathogen-free facility with free access to autoclaved water and food. Individual mice were randomized and injected with 1 × 106 SKBR3 transfectants into their flanks (n = 5–6 per group). The growth of implanted tumors was monitored for 27 d, and the volumes were measured with calipers. Body weight was measured twice per week. Ki67 staining was carried out using standard immunocytochemical protocols.
The impact of FSIP1 silencing on global gene expression was determined using GeneChip Human Transcriptome Array 2.0 (902162; Affymetrix). Data were quantile-normalized and analyzed using GeneSpring 13.1 (Agilent Technologies). Fold changes in single values were transformed to log2. Differentially expressed genes were analyzed by GO analyses using the database for annotation, visualization, and integrated discovery. The potential function and pathways of the differentially expressed genes were also analyzed by KEGG mapping using online tools. The FSIP1 gene signature was subjected to CMAP interrogation, and the top three hits were evaluated for effects on SKBR3 viability using the MTT assay.
Supplementary Material
Acknowledgments
Work at Icahn School of Medicine at Mount Sinai was supported by NIH Grants R01 DK80459 (to M.Z. and L.S.), R01 DK113627 (to M.Z. and L.S.), R01 AG40132 (to M.Z.), R01 AG23176 (to M.Z.), R01 AR65932 (to M.Z.), and R01 AR67066 (to M.Z.).
Footnotes
The authors declare no conflict of interest.
2T.Y. and C.L. are co-senior authors.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621486114/-/DCSupplemental.
References
- 1.Sledge GW, et al. Past, present, and future challenges in breast cancer treatment. J Clin Oncol. 2014;32:1979–1986. doi: 10.1200/JCO.2014.55.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Rorke MA, Murray LJ, Brand JS, Bhoo-Pathy N. The value of adjuvant radiotherapy on survival and recurrence in triple-negative breast cancer: A systematic review and meta-analysis of 5507 patients. Cancer Treat Rev. 2016;47:12–21. doi: 10.1016/j.ctrv.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Chapman KB, et al. Elevated expression of cancer/testis antigen FSIP1 in ER-positive breast tumors. Biomarkers Med. 2013;7:601–611. doi: 10.2217/bmm.13.58. [DOI] [PubMed] [Google Scholar]
- 4.Cappell KM, et al. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol Cell Biol. 2012;32:4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, et al. Expression and clinicopathological significance of FSIP1 in breast cancer. Oncotarget. 2015;6:10658–10666. doi: 10.18632/oncotarget.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiriva-Internati M, et al. AKAP-4: A novel cancer testis antigen for multiple myeloma. Br J Haematol. 2008;140:465–468. doi: 10.1111/j.1365-2141.2007.06940.x. [DOI] [PubMed] [Google Scholar]
- 7.Tortora G, Ciardiello F. Protein kinase A as target for novel integrated strategies of cancer therapy. Ann N Y Acad Sci. 2002;968:139–147. doi: 10.1111/j.1749-6632.2002.tb04332.x. [DOI] [PubMed] [Google Scholar]
- 8.Miller WR. Regulatory subunits of PKA and breast cancer. Ann N Y Acad Sci. 2002;968:37–48. doi: 10.1111/j.1749-6632.2002.tb04325.x. [DOI] [PubMed] [Google Scholar]
- 9.Gu L, Lau SK, Loera S, Somlo G, Kane SE. Protein kinase A activation confers resistance to trastuzumab in human breast cancer cell lines. Clin Cancer Res. 2009;15:7196–7206. doi: 10.1158/1078-0432.CCR-09-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labhart P, et al. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci USA. 2005;102:1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 12.Stachnik A, et al. Repurposing of bisphosphonates for the prevention and therapy of nonsmall cell lung and breast cancer. Proc Natl Acad Sci USA. 2014;111:17995–18000. doi: 10.1073/pnas.1421422111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuen T, et al. Disease-drug pairs revealed by computational genomic connectivity mapping on GBA1-deficient, Gaucher disease mice. Biochem Biophys Res Commun. 2012;422:573–577. doi: 10.1016/j.bbrc.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuen T, et al. Bisphosphonates inactivate human EGFRs to exert antitumor actions. Proc Natl Acad Sci USA. 2014;111:17989–17994. doi: 10.1073/pnas.1421410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 16.Batlle E, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 17.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 18.Toivola DM, Tao GZ, Habtezion A, Liao J, Omary MB. Cellular integrity plus: organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 2005;15:608–617. doi: 10.1016/j.tcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudley JT, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reka AK, et al. Identifying inhibitors of epithelial-mesenchymal transition by connectivity map-based systems approach. J Thorac Oncol. 2011;6:1784–1792. doi: 10.1097/JTO.0b013e31822adfb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YJ, et al. Interplay of PI3K and cAMP/PKA signaling, and rapamycin-hypersensitivity in TGFbeta1 enhancement of FSH-stimulated steroidogenesis in rat ovarian granulosa cells. J Endocrinol. 2007;192:405–419. doi: 10.1677/JOE-06-0076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.