Significance
The process of malting in the making of beer is under fine biological regulation. Here, we report the dissection of a malting quality quantitative trait locus (QTL) known for its association with variation in the yield of β-glucan and malt extract. A key gene, HvTLP8, has been identified in the QTL2 region that is expressed differentially in barley malt and feed varieties, based on analyses of both mRNA and protein. The present finding that the affinity of recombinant HvTLP8 for insoluble barley β-glucan is redox-dependent suggests a key role for redox in the malting process. This discovery represents a significant step forward in understanding the biochemistry of malting.
Keywords: Hordeum vulgare, malting, β-glucan, quantitative trait loci, thaumatin-like protein
Abstract
Barley is the cornerstone of the malting and brewing industry. It is known that 250 quantitative trait loci (QTLs) of the grain are associated with 19 malting-quality phenotypes. However, only a few of the contributing genetic components have been identified. One of these, on chromosome 4H, contains a major malting QTL, QTL2, located near the telomeric region that accounts, respectively, for 28.9% and 37.6% of the variation in the β-glucan and extract fractions of malt. In the current study, we dissected the QTL2 region using an expression- and microsynteny-based approach. From a set of 22 expressed sequence tags expressed in seeds at the malting stage, we identified a candidate gene, TLP8 (thaumatin-like protein 8), which was differentially expressed and influenced malting quality. Transcript abundance and protein profiles of TLP8 were studied in different malt and feed varieties using quantitative PCR, immunoblotting, and enzyme-linked immunosorbent assay (ELISA). The experiments demonstrated that TLP8 binds to insoluble (1, 3, 1, 4)-β-D glucan in grain extracts, thereby facilitating the removal of this undesirable polysaccharide during malting. Further, the binding of TLP8 to β-glucan was dependent on redox. These findings represent a stride forward in our understanding of the malting process and provide a foundation for future improvements in the final beer-making process.
Thaumatin-like proteins (TLPs), also designated “pathogenesis-related” (PR) proteins, represent a complex gene family involved in a broad range of defense and developmental processes in plants, fungi, and animals (1). TLPs share sequence similarity with thaumatin, an intensely sweet-tasting disulfide protein originally identified in the fruit of the shrub, Thaumatococcus daniellii (2). In plants, TLPs are members of an inducible protein cascade that includes 17 families of pathogenesis-related proteins, PR-1 to PR-17. Their synthesis in plants is triggered mainly in response to biotic and abiotic stresses but is also developmentally regulated, particularly during fruit ripening (3). Additionally, TLPs are present constitutively in different plant organs, including grain (4).
TLPs have a molecular mass ranging from 21 to 26 kDa and are highly conserved with antiparallel β-sheets and five to eight disulfide bridges formed from conserved cysteines (5). The disulfide bridges are required for correct folding and stability under high temperature or low pH conditions (6). During evolution, TLPs have assumed wide functional diversification, possibly because of their interaction with numerous ligands, such as actin, ice crystals, fungal proteins, and antifreeze proteins. Their functions in plants include antifungal activity (7), freezing tolerance, and protection against osmotic stress (8). Other features include binding to proteins, such as viral CMV-1 protein, yeast glycoproteins, and G protein-coupled receptor (GPCR), to hormones, such as cytokinins (5), and to glucanase (9) and xylanase (10). TLPs with diverse properties have been identified in moss (Physcomitrella), algae (Chlamydomonas), and several land plants (rice, maize, poplar, Arabidopsis) in addition to barley (11).
TLPs are known to be expressed in germinating grain (12, 13) but are not known to play a role in malting—an area of general interest in efforts to understand the 5,000-y-old brewing process. The malting process in beer production involves controlled germination of barley during which hydrolytic enzymes are synthesized and the cell walls, proteins, and starch of the endosperm are largely digested, making the grain more friable for obtaining high-quality malt.
Improved knowledge of the genetics and biochemistry of barley grain has been a major goal of plant breeders in their quest to identify superior traits for malting. Generally, varieties possessing characteristics, such as high levels of starch-degrading enzymes and low protein content, have been used for malting, and varieties lacking these traits have been relegated to feed production (14).
Glucan (1,3;1,4-β-D) (hereafter “β-glucan”) and arabinoxylans are the major components of the nonstarch polysaccharides of barley grain endosperm (15). A low β-glucan content has been considered desirable for malting (16). Excessive amounts lead to the formation of highly viscous wort (the sugar-rich liquid formed before fermentation), a diminished rate of wort filtration, and the formation of undesirable haze (17). Manipulation of malting quality traits, such as extent of malt extraction, diastatic power, and yield of β-glucan and β-glucanase, is challenging for geneticists and breeders alike because of complexities in inheritance and difficulties in evaluation. Measuring malting quality is also laborious and costly, thus limiting its use in cultivar development and attempts to improve this trait (18, 19).
To date, genetic investigations of malting traits have involved studying quantitative trait loci (QTLs) with the result that more than 250 of these gene sets have been located on the seven barley chromosomes (20). Among these, QTL2 is a major component, contributing ca. 29–38% to the variation of β-glucan content and extracted malt, respectively (21). QTL2 is accommodated on the telomeric region of the short arm of chromosome 4H and is located at 15.8 cM (18, 19).
A major focus of our interest lies in identifying QTL2 genes in an effort to understand and potentially improve the malting process. We now report that one such gene, TLP8, harbors a carbohydrate-binding domain that binds β-glucan tightly in a reaction dependent on redox. In this way, TLP8 lowers the content of β-glucan, thereby enhancing malt quality.
Results
Barley QTL2 Has Synteny with Rice Chromosome 3L.
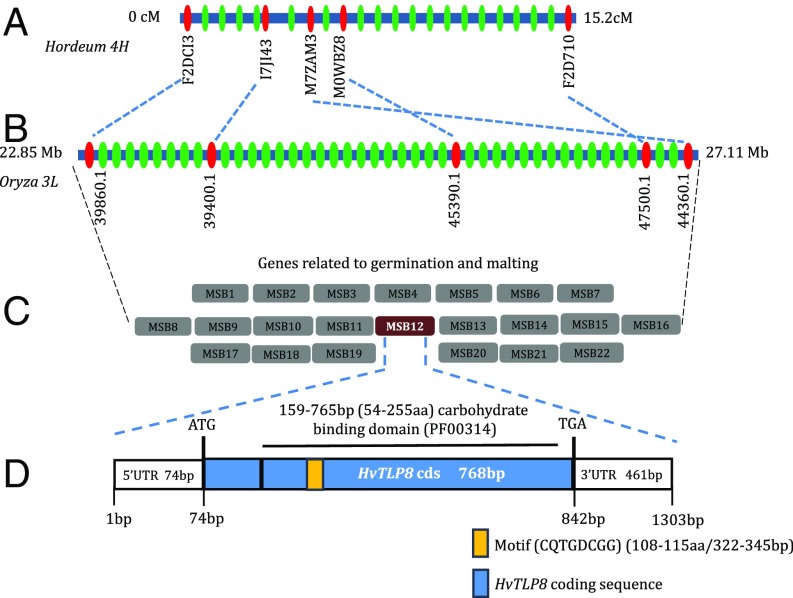
To gain insight into the barley counterpart, we investigated the microsynteny of the corresponding chromosome region of rice (3L). QTL2 spans a region of 15.2 cM in the telomeric region of chromosome 4H; the syntenic region of rice has a corresponding 1.795-Mbp (base pairs 25328757–27123813) coverage on chromosome 3L (Fig. 1 A and B). We found that nearly 80% of markers mapped in QTL2 maintain colinearity with 3L of rice. More than 100 predicted genes were detected in the syntenic region of rice. Each was analyzed using BLAST tools from various expressed sequence tag (EST) databases [HarvEST, National Center for Biotechnical Information (NCBI), and Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Crop EST Database]. Based on this analysis, 22 candidate rice genes were identified that might be involved in malting-related traits, thereby allowing retrieval of the corresponding barley genes (Fig. 1C). Putative functions of the candidate genes were assigned using BLASTx analysis, and their map location and presumed functions were investigated (Table S1). Based on its functional relationship to germination and malting and expression analysis, a TLP was identified as having a possible role during the malting process.
Fig. 1.
Physical map of the barley 4H-QTL2 region and TLP8 gene structure of barley. (A) Physical map of the barley QTL2 region; predicted genes are indicated by green ovals, with solid red ovals indicating some of the loci mapped to depict synteny. (B) Rice chromosome 3L region syntenic to barley QTL2. (C) From more than100 predicted genes in the corresponding QTL2 region of barley, candidates were narrowed to 22 for those functional in germination or malting. (D) Structure of one TLP8 gene, corresponding to MSB12, indicates the presence of a sugar-binding domain (black bar) and a motif (orange box) affecting malting by controlling amount of β-glucan.
Table S1.
Candidate malting synteny barley (MSB) genes MSB1–MSB22 retrieved from syntenic relationship between barley chromosome 4HS (QTL2 region) and corresponding rice chromosome 3L region
| Candidate marker ID | Expression profile | Homology (BLASTX) | E-value | Chromosome location(s) |
| MSB1 | Seed/malting | Amino acid permease family protein | 1.00E-154 | 4HS |
| MSB2 | Seed/malting | Predicted protein: not characterized | 3.00E-180 | 4HS |
| MSB3 | Seed/malting | High-affinity cationic amino acid transporter | 5.00E-88 | 4HS |
| MSB4 | Seed/malting | Glucan endo-1,3-β-glucosidase | 2.00E-134 | 4HS |
| MSB5 | Embryo/malting | Isopropylmalate dehydrogenase | 3.00E-62 | 2H/4HS |
| MSB6 | Seed/malting | High-affinity cationic amino acid transporter 1 | 7.00E-113 | 4HS |
| MSB7 | Seed/malting | β-Tubulin | 0.00E+00 | 5H/4HS |
| MSB8 | Seed/malting | β-Tubulin | 0.00E+00 | 4HS |
| MSB9 | Endosperm/malting | Guanine nucleotide exchange factor in Golgi transport N-terminal | 5.00E-93 | 4HS |
| MSB10 | Embryo/malting | Embryo globulin, storage protein | 6.00E-87 | 4HS |
| MSB11 | Seed/malting | Putative senescence-associated protein | 3.00E-97 | 4HS |
| MSB12 | Endosperm/malting | Thaumatin-like protein | 1.00E-113 | 4HS |
| MSB13 | Embryo, seed/malting | 40S ribosomal protein | 1.00E-47 | 4HL |
| MSB14 | Embryo, scutellum/malting | Predicted: acidic leucine-rich nuclear phosphoprotein | 4.00E-65 | 4HS |
| MSB15 | Endosperm/malting | Low-temperature–responsive RNA-binding protein | 5.00E-54 | 4HS |
| MSB16 | Endosperm/malting | Putative: early responsive to dehydration protein | 5.00E-179 | 4HS |
| MSB17 | Seed/malting | RING finger protein 5 | 5.00E-58 | 4HS |
| MSB18 | Seed/malting | Predicted protein | 5.00E-160 | 4HL |
| MSB19 | Endosperm/malting | Predicted: glucose-induced degradation protein | 8.00E-123 | 4HS |
| MSB20 | Endosperm/malting | Benzothiadiazole-induced homeodomain protein | 0.00E+00 | 4HS |
| MSB21 | Seed/malting | Ubiquitin-conjugating enzyme | 6.00E-101 | 5H |
| MSB22 | Embryo, endosperm/malting | Putative ubiquitin-conjugating enzyme E2 | 3.00E-85 | 4HS |
Information includes expression, homology, and location on barley chromosome 4HS.
Phylogenetic Relationship of Barley TLPs and Gene Structure of QTL2-Associated TLP8.
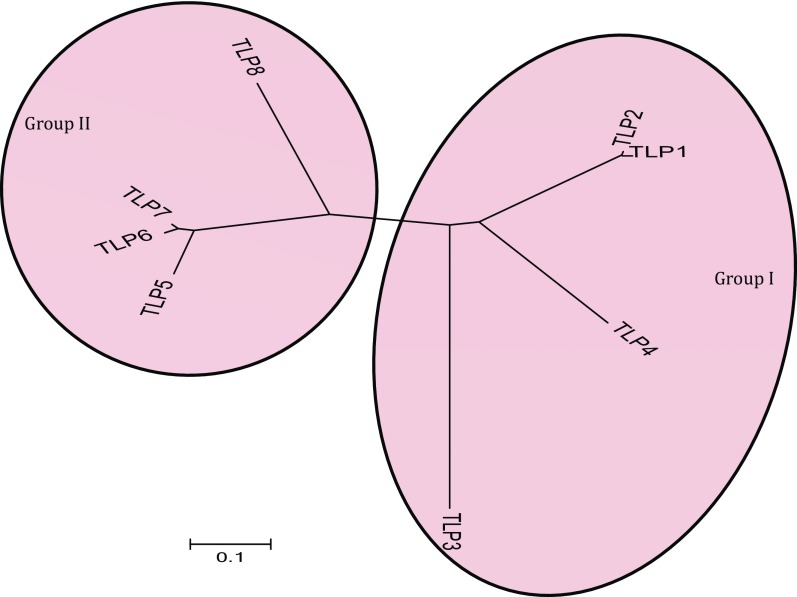
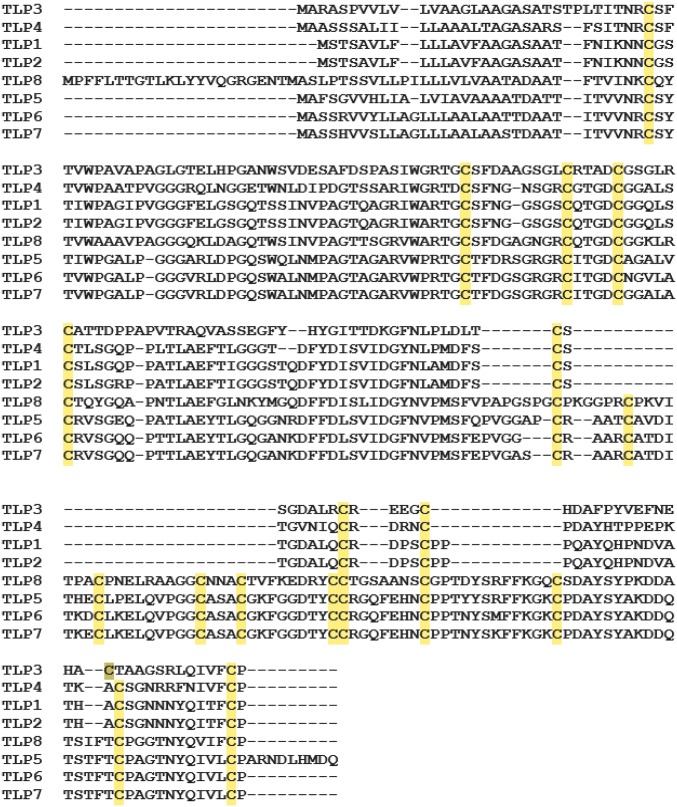
Eight TLP genes (TLP1–TLP8) have been reported to function in disease resistance in barley (22). Phylogenetic analysis of the TLP genes revealed two well-defined groups (I and II). In group I, TLP1 and TLP2 (each with 10 Cys residues) were found to be phylogenetically close to each other, whereas TLP3 and TLP4 (also with 10 Cys residues) were more diverged within the group (Fig. S1). Interestingly, the number of Cys residues was also conserved in group II proteins: TLP5, TLP6, TLP7, and TLP8 (16 Cys residues each). Genetic mapping indicated that barley TLP3, -4, -5, -6, and -7 were clustered on the short arm of chromosome 5H, whereas TLP1, -2, and -8 were found on chromosome 7HL, 7HS, and 4HS, respectively (Table S2).
Fig. S1.
Phylogenetic relationship of barley TLPs. The branching diagram was generated using MEGA 6 and was based on the deduced full-length amino acid sequence of barley TLPs. (The scale bar shows the frequency of amino acid substitutions between sequences as determined by the Poisson evolutionary distance method and represents an amount genetic change of 0.1.)
Table S2.
Chromosome location, gene annotation, and amino acid sequence information of barley TLPs
| Barley TLP | Gene annotation | Chromosome location and distance, cM | No. of Cys residues | Amino acids | Binding motif | NCBI GenBank Accession No. | Isoelectric point values |
| TLP1 | MLOC_24152.1 | 7HL, 140.86 | 10 | 173 | Yes | AY839292 | 4.33 |
| TLP2 | MLOC_77363.1 | 7HL, 138.83 | 10 | 173 | Yes | AY839293 | 4.51 |
| TLP3 | MLOC_60646.1 | 5HS, 42.87 | 10 | 175 | No | AY839294 | 5.02 |
| TLP4 | MLOC_20378.1 | 5HS,14.23 | 10 | 231 | No | AF355455 | 5.69 |
| TLP5 | MLOC_2781.1 | 5HS,13.26 | 16 | 236 | No | AY839295 | 6.04 |
| TLP6 | MLOC_75397.1 | 5HS, 14.51 | 16 | 226 | No | AF355456 | 7.33 |
| TLP7 | MLOC_18451 | 5HS, 14.79 | 16 | 227 | No | AF355457 | 7.36 |
| TLP8 | MLOC_39318.1 | 4HS, 3.47 | 16 | 255 | Yes | AF355458 | 8.11 |
The mRNA levels of the 22 candidate genes were compared in malting and feed varieties. Semiquantitative RT-PCR was performed in an initial screening of putative malting genes, and RNA samples extracted at different stages of malting were used as templates in subsequent analyses. Only TLP8 from 22 genes was expressed differentially across the malting and feed varieties, with higher levels of transcripts in malting and lower levels in feed varieties. TLP8, located in the QTL2 region of barley chromosome 4H, is 1,303 bp long and has a 74-bp 5′ UTR, a 461-bp 3′ UTR, and a 768-bp long coding sequence (cds) with no intron (Fig. 1D). The TLP8 cds encodes a 255-a.a. polypeptide with a conserved carbohydrate-binding domain (GH64-TLP-SF, Pfam domain number PF00314) from amino acids 54–255 or 159–765 bp. An eight-a.a. (amino acids 108–115 or 322–345 bp), 24-bp-long, carbohydrate-binding, structural motif located within the thaumatin signature (amino acids 102–119 or 304–357 bp) forms a β-hairpin that might play a role in carbohydrate binding. A motif search revealed that the surface-binding motif, PR-5 (CQTGDCGG; amino acids108–115), is exclusive to barley TLP1, -2, and -8 (Fig. 1D and Table S2). Genetic analyses using the IPK barley database revealed a single copy of barley TLP8 in the genome.
Expression Profiles of Barley TLPs in Different Tissues.
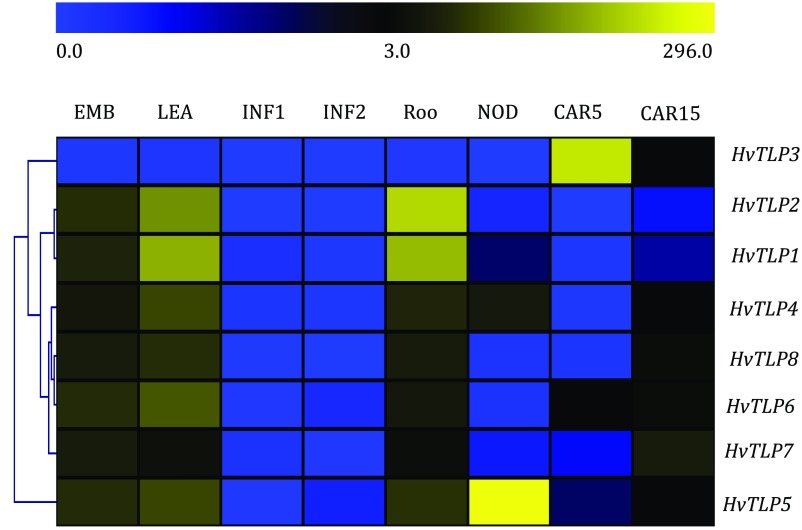
The available RNA-sequencing (RNA-seq) data provided information important for predicting the function of these genes. We conducted a comprehensive expression analysis of barley TLP genes to aid in understanding their role in regulating development. We did so by investigating their spatiotemporal expression pattern in the morexGenes-Barley RNA-seq database (https://ics.hutton.ac.uk/morexGenes) (Fig. S2). Expression of TLP2, -4, -5, -6, -7, and -8 was found to be higher in embryo (EMB), shoot (LEA), and root (ROO), implying their requirement in development of these tissues. These genes also showed higher expression in CAR15 tissue [developing grain, bracts removed; 15 days post anthesis (DPA)], suggesting that they may play a role in grain development. Expression of TLP3 was highest in CAR5 tissue (developing grain, bracts removed; 5 DPA). No expression of barley TLPs was detected in INF1 and INF2 tissue. Expression of TLP1, -4, and -5 was also elevated in NOD tissue (tillers at the six-leaf stage; third internode). Analysis of these results indicates that TLP gene expression during the different stages and in specific tissues might function in barley embryo and grain development.
Fig. S2.
Heat map representation and hierarchical clustering of barley TLPs. The expression of HvTLPs was analyzed in eight Morex tissues from 4-d embryo (EMB); 10-cm-shoot stage (LEA); (5-mm young developing inflorescences (INF1), 1- to 1.5-cm developing inflorescences (INF2), roots of the seedlings at 10-cm shoot stage (ROO); tillers at the six-leaf stage (NOD); 5 DPA, developing grain bract removed (CAR5); and 15 DPA, developing grain bracts removed (CAR15).The FPKM expression values were used to generate heat maps.
TLP8 Is Highly Expressed in Barley Malting Varieties and Is Related to Levels of β-Glucan.
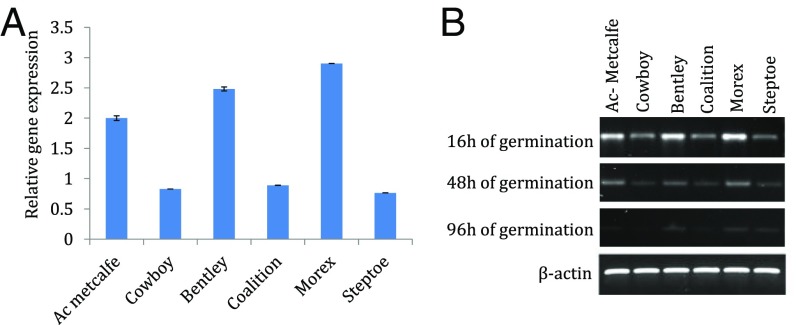
Among the eight barley TLPs, only TLP8 is associated with barley chromosome 4H-QTL2. Analysis of Morex RNA-seq data indicated that the gene is expressed in developing grain and embryo. Further analysis of the expression pattern of TLP8 was conducted with 16-h–imbibed seeds at different stages of germination using semiquantitative RT-PCR and qRT-PCR (Fig. 2). This approach revealed a differential expression of TLP8 at different times of development in varieties used for malt (AC Metcalfe, Bentley, Morex) and those used for feed (Cowboy, Coalition, Steptoe). Its expression was highest in the initial 16 h of germination, decreased at 48 h, and showed almost no expression at 96 h. TLP8 had higher expression in malting varieties and lower expression in feed varieties (Fig. 2A). Overall, malting varieties showed approximately a threefold higher expression of TLP8 than feed varieties.
Fig. 2.
qRT-PCR–based transcript abundance of TLP8 mRNA in barley grains. (A) Bar graph representing qRT-PCR expression profiles of TLP8 in malting and feed varieties of barley in duplicate at 16 h of germination. Ac Metcalfe, Bentley, and Morex are malting varieties in which TLP8 has higher expression; Cowboy, Coalition, and Steptoe are feed varieties in which TLP8 shows lower expression. The x axis represents the malting and feed varieties; the y axis represents the relative gene expression. (B) Gel representation of the expression profiles of TLP8 and the housekeeping gene β-actin at 16 h of germination in malting and feed varieties of barley using semiquantitative RT-PCR.
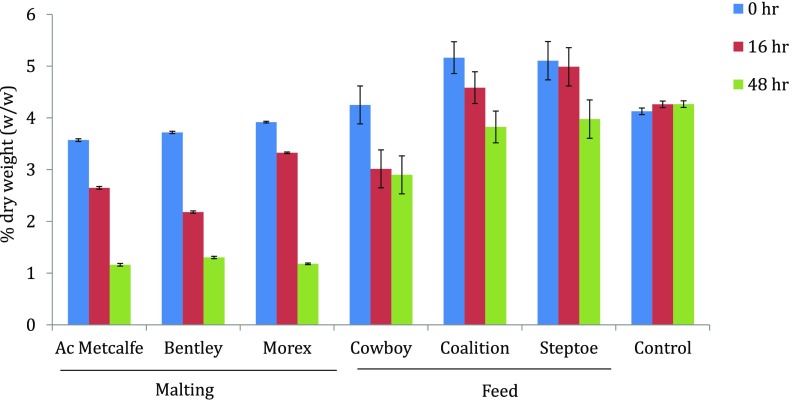
We also found a positive correlation between levels of TLP8 expression and the amount of β-glucan present at specific time points. The amount of β-glucan was determined in malt and feed varieties using well-established McCleary method (Megazyme) at different stages of germination (Fig. S3). Malting varieties had slightly lower levels of β-glucan before germination than feed varieties (Fig. S3). However, after 48 h of germination, the amount of β-glucan was reduced by ∼60% in malt varieties, compared with ∼20% in feed varieties. The corresponding reduction in β-glucan could be attributed to glucanase-type activity of TLP8, because other glucanases were not active in this time frame (16).
Fig. S3.
Amount of β-glucan at different stages of barley seed germination (0, 16, and 48 h). All malting varieties had lower amounts of initial β-glucan than the feed varieties. As germination progresses to 16 and 48 h, the amount of β-glucan drops significantly in all malting varieties; the decrease in feed varieties is significantly less. The x axis represents the malting, feed, and control (standard barley) from the Megazyme kit. The y axis represents the amount of β-glucan as a percentage of total dry weight (wt/wt).
Biochemical Assays for β-Glucan Binding.
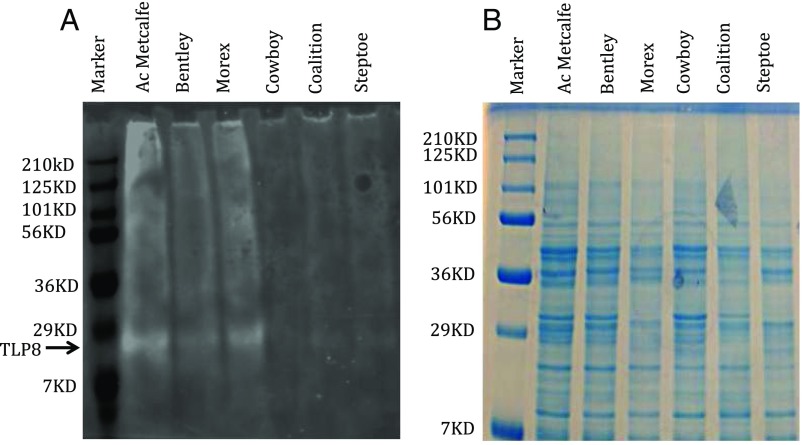
Immunoblots conducted to assess β-glucan binding revealed a slightly larger protein band of ∼300 Da in malt varieties relative to feed counterparts. This alteration in molecular mass was also evident using a glycosylation assay. Thus, under UV illumination, protein bands observed after binding showed high-intensity fluorescence in malting varieties and only weak-intensity fluorescence in feed varieties (Fig. S4). Additionally, a glycosylation prediction program indicated that the carbohydrate-binding motif was potentially glycosylated.
Fig. S4.
Glycosylation of barley TLP proteins and their binding ability with β-glucan. (A) Glycosylation of TLPs in malting and feed varieties was observed by staining with a glycosylation staining kit. Malting varieties (Ac Metcalfe, Bentley, and Morex) show a high degree of fluorescence, indicating higher β-glucan binding than feed varieties (Cowboy, Coalition, and Steptoe). (B) SDS/PAGE gel showing TLP in malting and feed varieties after binding with β-glucan.
In a final approach, we investigated binding of (1, 3) β-glucan (Pachyman polysaccharides) and (1, 3, 1, 4) β-D glucan (barley β-glucan) with enriched barley TLP fractions obtained by ammonium sulfate fractionation. Our analyses indicated that the enriched TLP fraction binds both types of carbohydrate and that binding was generally higher in malt varieties. These results prompted us to overexpress TLP8 in Escherichia coli for further characterization of binding and its specificity for β-glucan.
Role of Redox in Binding Recombinant Barley TLP8 to β-Glucan.
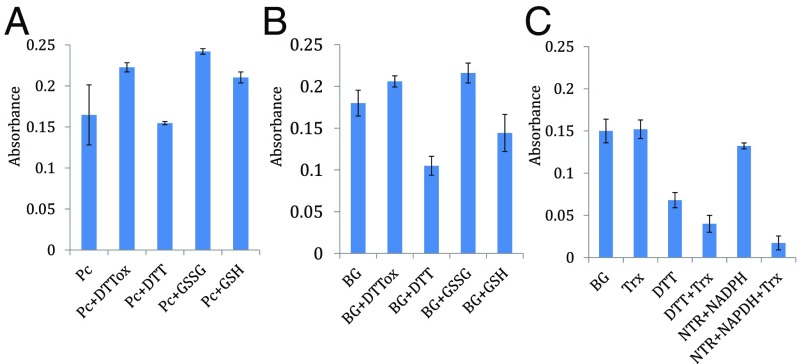
TLP8 is a unique member of group II TLPs that contain 16 Cys residues (Fig. 3 and Table S3). Cys-rich proteins with disulfide bonds, such as TLP8, are often linked to regulation by thiol/disulfide transitions (23). This possibility prompted us to overexpress and investigate the redox properties of recombinant TLP8. We used an enzyme-linked immunosorbent assay (ELISA) to examine the effect of redox agents on the binding of TLP8 to (1, 3) β-glucan (Fig. 4A) and barley β-glucan (Fig. 4B). Analysis of the results showed that binding to both β-glucans was diminished by reducing agents and enhanced with most oxidizing reagents tested. Thus, the binding of untreated TLP8 to β-glucan (38.2%) decreased significantly when exposed to the reductant, DTT (17.3%). A similar reduction in binding was observed when TLP8 was treated with reduced glutathione (GSH) and was maintained in the reduced (sulfhydryl) form with NAPDH and glutathione reductase (GR) (GSH+NADPH+GR) (18.9%) and β-mercaptoethanol (18.0%) (Table 1). Conversely, TLP8 binding was increased with oxidized glutathione (GSSG), dehydroascorbate, oxidized DTT, and hydrogen peroxide (H2O2) but was not affected by the reductant sodium ascorbate or the oxidant sodium tetrathionate (Table 1).
Fig. 3.
Amino acid sequence alignment of group II TLPs of barley. The highlighted sequences indicate 16 conserved Cys residues in TLP5, TLP6, TLP7, and TLP8. Sequence alignment was performed using the muscle alignment tool. GenBank accession numbers for barley TLP proteins are listed in Tables S2 and S3.
Table S3.
Amino acid sequences of barley TLP1–8
| HvTLP1: |
| MSTSAVLFLLLAVFAAGASAATFNIKNNCGSTIWPAGIPVGGGFELGSGQTSSINVPAGTQAGRIWARTGCSFNGGSGSCQTGDCGGQLSCSLSGQPPATLAEFTIGGGSTQDFYDISVIDGFNLAMDFSCSTGDALQCRDPSCPPPQAYQHPNDVATHACSGNNNYQITFCP |
| HvTLP2: |
| MSTSAVLFLLLAVFAAGASAATFNIKNNCGSTIWPAGIPVGGGFELGSGQTSSINVPAGTQAGRIWARTGCSFNGGSGSCQTGDCGGQLSCSLSGRPPATLAEFTIGGGSTQDFYDISVIDGFNLAMDFSCSTGDALQCRDPSCPPPQAYQHPNDVATHACSGNNNYQITFCP |
| HvTLP3: |
| MARASPVVLVLVAAGLAAGASATSTPLTITNRCSFTVWPAVAPAGLGTELHPGANWSVDESAFDSPASIWGRTGCSFDAAGSGLCRTADCGSGLRCATTDPPAPVTRAQVASSEGFYHYGITTDKGFNLPLDLTCSSGDALRCREEGCHDAFPYVEFNEHACTAAGSRLQIVFCP |
| HvTLP4: |
| MAASSSALIILLAAALTAGASARSFSITNRCSFTVWPAATPVGGGRQLNGGETWNLDIPDGTSSARIWGRTDCSFNGNSGRCGTGDCGGALSCTLSGQPPLTLAEFTLGGGTDFYDISVIDGYNLPMDFSCSTGVNIQCRDRNCPDAYHTPPEPKTKACSGNRRFNIVFCP |
| HvTLP5: |
| MAFSGVVHLIALVIAVAAAATDATTITVVNRCSYTIWPGALPGGGARLDPGQSWQLNMPAGTAGARVWPRTGCTFDRSGRGRCITGDCAGALVCRVSGEQPATLAEYTLGQGGNRDFFDLSVIDGFNVPMSFQPVGGAPCRAATCAVDITHECLPELQVPGGCASACGKFGGDTYCCRGQFEHNCPPTYYSRFFKGKCPDAYSYAKDDQTSTFTCPAGTNYQIVLCPARNDLHMDQ |
| HvTLP6: |
| MASSRVVYLLAGLLLAALAATTDAATITVVNRCSYTVWPGALPGGGVRLDPGQSWALNMPAGTAGARVWPRTGCTFDGSGRGRCITGDCNGVLACRVSGQQPTTLAEYTLGQGANKDFFDLSVIDGFNVPMSFEPVGGCRAARCATDITKDCLKELQVPGGCASACGKFGGDTYCCRGQFEHNCPPTNYSMFFKGKCPDAYSYAKDDQTSTFTCPAGTNYQIVLCP |
| HvTLP7: |
| MASSHVVSLLAGLLLAALAASTDAATITVVNRCSYTVWPGALPGGGVRLDPGQSWALNMPAGTAGARVWPRTGCTFDGSGRGRCITGDCGGALACRVSGQQPTTLAEYTLGQGANKDFFDLSVIDGFNVPMSFEPVGASCRAARCATDITKECLKELQVPGGCASACGKFGGDTYCCRGQFEHNCPPTNYSKFFKGKCPDAYSYAKDDQTSTFTCPAGTNYQIVLCP |
| HvTLP8: |
| MPFFLTTGTLKLYYVQGRGENTMASLPTSSVLLPILLLVLVAATADAATFTVINKCQYTVWAAAVPAGGGQKLDAGQTWSINVPAGTTSGRVWARTGCSFDGAGNGRCQTGDCGGKLRCTQYGQAPNTLAEFGLNKYMGQDFFDISLIDGYNVPMSFVPAPGSPGCPKGGPRCPKVITPACPNELRAAGGCNNACTVFKEDRYCCTGSAANSCGPTDYSRFFKGQCSDAYSYPKDDATSIFTCPGGTNYQVIFCP |
Fig. 4.
Binding of purified TLP8 with (1, 3, 1, 4) β-D glucan (BG) and (1, 3) β-D glucan (Pc) and the effect of various redox reagents. (A) Binding of recombinant TLP8 protein (RP) with (1, 3) β-D glucan (Pc) in the presence or absence of redox reagents. (B) Binding of recombinant TLP8 protein (RP) with barley (1, 3, 1, 4) β-D glucan (BG) in the presence or absence of redox reagents. (C) Binding of recombinant TLP8 protein (RP) with purified TLP8 reduced with Trx-NTR-NADPH. Treatment details are shown in the footnote to Table 1. Binding percentage was calculated by dividing bound protein by total protein and multiplying by 100.
Table 1.
Effect of redox agents on binding of barley recombinant TLP8 to (1, 3, 1, 4)-β-glucan
| Treatments | % Binding |
| 1. Control | 38.26 |
| Reducing system | |
| 2. Trx | 38.70 |
| 3. DTT | 17.34* |
| 4. DTT + Trx | 10.20** |
| 5. NTR + NADPH | 33.67 |
| 6. NTR + NADPH + TRX | 4.33** |
| 7. GSH | 33.40 |
| 8. GSH + NADPH + GR | 18.97 |
| 9. β-Mercaptoethanol | 18.03 |
| 10. DTT, 5-X | 15.94** |
| 11. Sodium ascorbate | 38.64 |
| Oxidizing system | |
| 12. Dehydroascorbate | 47.25 |
| 13. H2O2 | 44.50 |
| 14. DTToxidized | 43.56 |
| 15. GSSG | 43.27 |
| 16. Sodium tetrathionate | 37.13 |
The control system contained 25 µg recombinant protein (TLP8) and 2.5 mg 1, 3;1, 4- β-D glucan. The other components were added as indicated: 5 µg Trx; 1 mM DTT; 5 mM DTToxidized; 7.5 ug NTR; 0.25 mM NADP; 10 mM GSH; 20 mM H2O2; 25 mM GSSG; 2.25 µg glutathione reductase. *P ≤ 0.05; **P ≤ 0.01 (Student’s t test).
Binding of purified TLP8 to β-glucan was also examined in the presence of thioredoxin (Trx), reduced either with NADPH and NADP-thioredoxin reductase (NTR) (23) or DTT Binding of TLP8 to β-glucan was dramatically decreased with both forms of reduced Trx when TLP8 was treated with a combination of Trx, NTR, and NADPH (4.3%) or with DTT (Fig. 4C and Table 1). Further, no decrease in binding was observed when TLP8 was treated with Trx alone.
Discussion
In the current investigation, we have identified a PR-5 (TLP) gene, TLP8, in the QTL2 region of barley chromosome 4H, a region known to be associated with malting traits (Fig. 1 A–D). Barley contains eight TLPs (Table S2), TLP1–TLP8, with apparent molecular masses ranging roughly from 15–26 kDa (22) and isoelectric points covering a broad pH range, from acidic (TLP1–TLP3) to weakly acidic (TLP4–5) to basic (TLP6–TLP8). The isoforms of barley PR-5 proteins have previously been shown to have antimicrobial and antifungal activity (24).
Phylogenetic analyses revealed that barley TLPs clustered into two groups. TLP1, TLP2, TLP3, and TLP4 form group I, and TLP5, TLP6, TLP7, and TLP8 form group II (Fig. S1). TLP8 is a unique TLP member of group II with 16 Cys residues and a binding motif (Fig. 3 and Table S2). The eight TLPs of barley have been found to be dissimilar in their genomic conformation, resulting in variations in their expression profiles, but all suggestive of a fundamental role in embryo development (Fig. S2). Moreover, greater TLP8 gene expression was observed during germination at 16, 48, and 96 h in the malting varieties than in feed counterparts (Fig. 2B). This pattern indicates a role for TLP8 during the active germination phase when starch and β-glucan are degraded. The higher transcript abundance of TLP8 in malting varieties would lead to enhanced accumulation of the protein, compared with feed varieties. A possible explanation for the differential regulation of TLP8 could lie in the presence of a W-box promoter sequence upstream of the gene. Regulatory consequences of these sequences have been reported for TLPs of rice (25).
Structural modeling of barley TLP8 using tobacco PR5d and zeamatin as references is in accord with its grouping with globular proteins. The predicted structure indicates that the protein consists of a flattened β-barrel adjacent to a flexible region. Further, it shows a large negatively charged cleft surrounded by an area rich in loops and coils that are stabilized by disulfide bonds. We conducted a systematic search of the TLP models to locate regions that might interact with (1, 3) β-D glucan (13). The search suggested that the glucan could be docked at the primary polysaccharide-binding site (CQTGDCGG) located in the cleft (26). In our study, this motif was observed exclusively in TLP1, -2, and -8 (Table S2). Further, only TLP8, which has a high degree of binding specificity for β-glucan, mapped in the QTL2 region of chromosome 4H. The lower amounts of β-glucan found in malting varieties could result from the interaction of TLP8 with β-glucan (Fig. S2). In this study we observed that, as germination progressed to 16 and 48 h, the amount of β-glucan decreased considerably in malting relative to feed varieties.
Binding of a carbohydrate moiety to proteins is a common phenomenon known as “glycosylation.” We used a fluorescence-based technique to detect glycosylation in the malt and feed varieties analyzed. A strong glycosylation signal was perceived in malt varieties (Fig. S4), reflective of the CQTGDCGG sugar-binding domain. To assess the relative binding capacity of (1, 3) β-glucan versus (1, 3, 1, 4)-β-D glucan, we overexpressed TLP8 in E. coli and assayed the purified protein. We found that purified TLP8 bound both types of β-glucans (Fig. 4 A and B).
Barley TLP8 belongs to the large (l-) type of TLPs that contain 16 conserved Cys residues. It is well documented that Cys-rich proteins typically form disulfide bonds that may undergo redox transitions (23). Moreover, a comprehensive survey identified TLPs as potential targets of the Trx system (27). This finding prompted us to determine the effect of oxidants and reductants on TLP8 binding. As seen in Table 1, binding was decisively altered by redox treatment. Thus, binding was decreased by treatment with strong reductants, whereas it was either unaffected or enhanced by oxidizing agents. Binding decreased most extensively with Thioredoxin-h (Trx h) that was reduced with either NADPH/NTR or DTT on the one hand and was increased most with dehydroascorbate, hydrogen peroxide, oxidized DTT, and GSSG (Fig. 4C and Table 1). Based on these results, it would seem that the removal of β-glucans during malting would be facilitated under oxidizing conditions.
Changes in the biochemical environment within the grain, including subtle modulation of its oxidation-reduction state along with hormonal signaling, are very important for starch degradation and grain germination. The redox state of a biological system is necessary for its performance as many of the active constituent regulatory proteins are responsive to changes in the system’s redox state (28). It was thus important to determine how oxidizing or reducing agents affect the β-glucan binding activity of TLP8. A variety of redox reactions occur throughout the malting and brewing process. Although oxygen is required during germination and steeping, most postfermentation oxidation reactions are considered detrimental to beer quality. In this study, we observed that β-glucan binding to TLP8 is redox-dependent and its subsequent removal can be facilitated by oxidants or oxidizing conditions, suggesting a key role in the malting process. Therefore, maintaining oxidizing conditions during the germination and wort preparation steps by adding a suitable oxidizing agent could further improve beer quality. Contrariwise, reducing agents can be added at stages when oxygen would have a deleterious effect on quality.
The addition of reducing agents, such as cysteine·HCl, DTT, and β-mercaptoethanol, to malt and brewing mashes has been found to increase the amount of soluble protein, free amino nitrogen, and extract values of the resultant worts (29). This effect was abolished by the addition of oxidizing agents, such as diamide and hydrogen peroxide. Taken together, current results suggest that major aspects of the malting and brewing process can be altered by adjusting the TLP8/β-glucan interaction by modifying redox conditions.
Reduced Trx h has been proposed to facilitate postgermination processes of cereals. The protein functions in the deactivation of enzyme inhibitors, activation of hydrolytic enzymes, and enhancement in the solubility of storage proteins (30). In wheat, it has been shown that the reduction of intramolecular disulfide bonds by Trx h promotes the degradation of major storage proteins in the imbibed seed (31). Further, the regulation of the starch-degrading enzyme, pullulanase, by Trx h has been documented in germinating barley seeds (32). Overexpression of Trx h in barley and wheat grains correlates with an accelerated rate of germination (33, 34). It can be seen from our data that TLP8 has the potential to function in concert with Trx h activity in germinating barley.
Germination is key to the malting process. During this phase extensive cell-wall decomposition and enzyme production take place. Among other things, it is important for the movement of the endosperm-degrading enzymes into the starchy endosperm. Barley endosperm cell walls are composed of about 70% mixed linkage (1, 3, 1, 4)-β-D glucan, which, if not properly degraded, will result in low yields of malt extract and the slow separation of wort during malting and brewing (15). A lower amount of β-glucan during germination enables efficient malt extraction and filtration of wort. The rate of this modification is determined principally by the amount of β-glucan and other enzymes/proteins, which either degrade or sediment this nonstarchy carbohydrate of cell walls. Our experiments indicate that TLP8 lowers β-glucan during the breakdown of cell walls.
This finding has consequences for the beer-making process. Prior research has indicated that two uncharacterized barley TLP isoforms, HvPR5b and HvPR5c, tightly bind (1, 3) β-D glucan (13). These TLP isoforms were found to bind only to water-insoluble (1, 3) β-D glucan, such as pachyman, curdlan, paramylon, zymosan, and Pleurotus ostreatus glucan, but not to chitin, pustulan, cellulose, or yeast invertase. It has been suggested that the binding of TLPs to these carbohydrates could be related to their antifungal activity (13). However, in our study, binding of (1, 3, 1, 4)-β-D glucan and TLP8 was revealed, possibly because of the substantial prevalence of this carbohydrate in barley grains, in which TLP8 may be involved in its regulation.
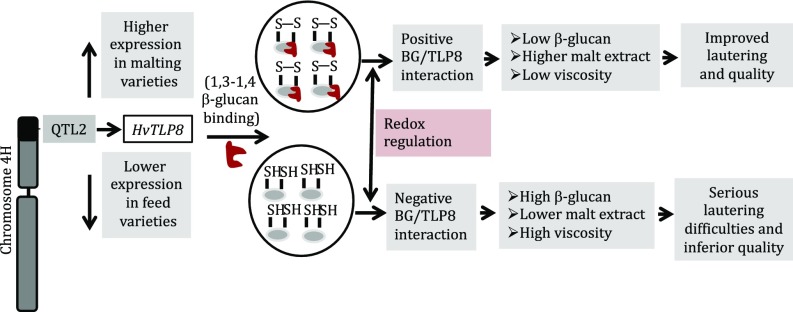
The current study has revealed that TLP8 resides in the QTL2 region of chromosome 4HS, a region known to be associated with malting quality traits such as content of malt extract and β-glucan (19). Our proposed model centers on the function of TLP8 during malting (Fig. 5). We have demonstrated that TLP8 binds (1, 3, 1, 4)-β-D glucan in a redox-dependent manner. Thus, binding was almost completely abolished by reducing agents and was enhanced by oxidants. Based on the present results, TLP8 emerges as a promising candidate gene for cultivar development to enhance the efficiency of malting. Because TLP8 binds to β-glucan, its down-regulation may also present an opportunity to enhance the amount of dietary fiber in cereals. Down-regulation of TLP8 could possibly negate or decrease its binding with (1, 3, 1, 4)-β-D glucan and potentially enhance β-glucan in the grains.
Fig. 5.
Model depicting the interplay of TLP8 and β-glucan during the malting process in barley. The barley chromosome 4H harboring QTL2 contains the TLP8 gene. Differential gene expression modulates the level of β-glucan in the barley genotypes. In vitro experiments indicate that binding of purified recombinant TLP8 and β-glucan is redox regulated. Unequivocal BG/TLP8 binding plays an important role in malt extract yield and the ease of the subsequent lautering and filtration process.
Materials and Methods
Candidate Gene Search and Bioinformatics.
A comparative approach using flanking markers from the QTL2 region of barley chromosome 4H (HarvEST, harvest.ucr.edu/) and the corresponding regions on rice chromosome 3L (rice genome database, www.jcvi.org/cms/research/past-projects/rice-database/overview/) was used. All genes and gene models predicted in the rice syntenic region were analyzed for candidates involving malting traits. Primers designed from candidate genes were used to narrow down candidates to the genes differentially expressed in malting and feed varieties. Additionally, candidate gene sequences were retrieved using localization of Morex WGS contigs in the integrated physical and genetic map of barley [International Barley Genome Sequencing Consortium (IBGSC), 2012; ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/]. To identify TLP genes in barley, we retrieved Arabidopsis TLP sequences (5) and used them as query to BLAST search in the PlantEnsembl (plants.ensembl.org/index.html), IPK (webblast.ipk-gatersleben.de/barley_ibsc/), and NCBI (www.ncbi.nlm.nih.gov/) databases. Conserved domains (glycoside hydrolase family 64) among different species were obtained as described in ref. 35 using the NCBI conserved domain architecture retrieval tool and probable motifs using Motif Scan (myhits.isb-sib.ch/cgi-bin/motif_scan).
RNA Isolation, cDNA Synthesis, and qRT- PCR.
Six commonly used barley malt (AC Metcalfe, Bentley, and Morex) and feed (Cowboy, Coalition, and Steptoe) varieties were obtained from Plant Gene Resources of Canada. All barley varieties were grown in the greenhouse for seed multiplication; RNA isolation was performed using mature barley grains. Seeds were surface sterilized with 70% ethanol and 4% NaOCl and were germinated in the dark on wet filter paper at 21 °C for 16, 48, and 96 h. RNA extraction from seeds was carried out as described elsewhere (36). For cDNA synthesis, a 500-ng sample of DNase-treated RNA was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad).
qRT-PCR was performed as described by in Singh et al. (36) with two biological and three technical replicates for each sample. Relative expression was conducted following the manufacturer’s recommendations with two reference genes (β-actin and GAPDH) and Brilliant III SYBR Green QPCR master mix (Agilent Technologies). Amplification was performed in a 20-µL reaction mixture containing 160 nmol for each primer, 1× Brilliant III SYBR Green QPCR master mix, 15 µM ROX reference dye, and 0.3 µL of cDNA template. The amplification conditions were 95 °C for 10 min (hot start), followed by 30 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Fluorescence reading was taken at 72 °C at the end of the elongation cycle. Primer sequences for RT-PCR were forward: AGCCGGAAATGGATATACTCG and reverse: AGCTCCTAAACTAGCGGTG and for qPCR were forward: CACATTGCCCAATTGTAGATAGC and reverse: AGCTCCTAAACTAGCGGTG. Analysis was performed using web-based, real-time PCR Miner software (ewindup.info/miner/data_submit.htm).
Barley TLP Gene Expression Analysis in Different Morex Tissues.
We used Morex RNA-seq transcriptome data (International Barley Genome Sequencing 2012) with the related log2-transformed fragments per kilobase per million fragments measured (FPKM) values to study TLP gene-expression patterns in the eight different developmental stages, tissues, and inflorescence treatments. EMB was collected from 4-d embryos dissected from germinating grains; ROO was collected from the roots of the seedlings (10-cm shoot stage) and LEA was taken from shoots of the seedlings (10-cm shoot stage). INF1 was taken from young developing inflorescences (5 mm), INF2 was collected from developing inflorescences (1–1.5 cm), NOD was collected from developing tillers at the six-leaf stage (third internode), CAR5 was taken from developing grain (5 DPA), bracts removed, and CAR15 was taken from developing grain (15 DPA), bracts removed (International Barley Genome Sequencing 2012). The heat map of TLP gene expression was developed using the MeV tool (mev.tm4.org) with the average hierarchical clustering method.
Protein Purification, SDS/PAGE, Immunoblotting, and β-Glucan Content.
For protein extraction, barley grains (1 g) from each variety was surface sterilized and germinated in the dark at 21 ± 2 °C under aseptic conditions in a Petri plate with autoclaved Whatman filter paper. Germinated grains were ground with 10 mL of 50 mM sodium phosphate buffer (pH 7.0) and centrifuged at 15,000 × g for 15 min s at 4 °C. The supernatant was subjected to ammonium sulfate fractionation, and the pellet obtained at 30–60% saturation was redissolved in 10 mL of 50 mM sodium acetate buffer (pH 5.0).
For PAGE analysis, proteins were separated using 5% (wt/vol) polyacrylamide stacking and 15% resolving gels, using Mini-Protein gel assembly (Bio-Rad). Gels were stained with Coomassie Brilliant Blue R (Bio-Rad) in 25% (vol/vol) ethanol and 7% (vol/vol) glacial acetic acid and were destained in the same solution without dye. For Western blot analysis, proteins were transferred to PVDF membrane filters (Millipore), blocked with 5% nonfat dry milk for 1 h, and rinsed three times with Tris-buffered saline and Tween 20 (0.05% Tween-20) (TBS-T) buffer. The membrane was incubated with 1:1,000 thaumatin antibody (Abcam) overnight at 4 °C, and further steps were carried out as described in ref. 37. The amount of β-glucan at different stages of germination was determined using the β-glucan kit from Megazyme.
Heterologous Expression of HvTLP8 in E. coli.
To express the putative mature TLP8 protein in E. coli, the ORF of barley TLP8 was codon-optimized and cloned into NdeI- and XhoI-digested pET28a (+) vector by GenScript, resulting in a translational fusion of the protein with six His residues at the N and C termini. This construct was transformed into E. coli strain BL21 competent cells. Expression of the protein and purification were performed as described in ref. 38.
Binding Assay, ELISA, and Glycosylation.
For binding assays, ammonium sulfate fractions of TLP proteins from different barley varieties and E. coli-expressed HvTLP8 were incubated with 5 mg of (1, 3) β-glucan (pachyman; Megazyme) and 2.5 mg of (1, 3, 1, 4) β-D glucan (BG; Sigma-Aldrich) in 50 mM sodium acetate buffer (pH 5.0), to a final volume of 0.5 mL The protein–polysaccharide mixture was incubated at 25 °C for 30 min on an orbital shaker and then was centrifuged at 1,000 × g for 3 min. The supernatant was analyzed for unbound protein using an ELISA-based detection system as described in ref. 37. Proteins bound to β-glucan were estimated by subtraction of residual protein in the supernatant from the values obtained in assays conducted without polysaccharide. The significance of the difference between the binding of normal and redox-treated TLP8 with β-glucan was evaluated by Student's t test. Glycosylation was performed using the Pro-Q Emerald 300 Glycoprotein Gel Stain Kit (Molecular Probes) following the manufacturer’s guidelines.
Redox Treatments of E. coli-Expressed HvTLP8 and Binding with β-Glucan in Vitro.
Pure barley (1, 3, 1, 4) β-D glucan (BG) was obtained from Sigma. Disulfide bond reduction of recombinant TLP8 and binding with BG were determined with (i) oxidants (10 mM dehydroascorbate, 10 mM sodium tetrathiocynate, 5 mM DTT oxidized, 25 mM GSSG, and 20 mM H2O2) and reductants (10 mM GSH, 5 mM reduced DTT, 20 mM β-mercaptoethanol, and 20 mM sodium ascorbate); (ii) the NADP/GSH system composed of 10 mM GSH and 2.25 µg yeast glutathione reductase; (iii) the Trx/DTT system consisting of 5 µg Trx and 1 mM reduced DTT; (iv) the NADP/NTR system consisting 7.5 µg NTR, 0.25 mM NADPH, and 5 µg Trx. Untreated protein without binding (TLP8 RP) was used as control. Redox treatment was carried out for 30 min at room temperature in 150 µL of recombinant TLP8 followed by binding with 2.5 mg BG for 30 min, and RP–BG binding was assessed by ELISA as described above.
Acknowledgments
We thank Kamrul Islam and Prabhjot Nandha for ELISA preparation and Navaldeep Kaur for proofreading. This project was supported by a grant from the Brewing and Malting Barley Research Institute and the Natural Sciences and Engineering Research Council of Canada (to J.S.). P.G.L. was supported by the US Cooperative Extension Service, and B.B.B. was supported by the US Department of Agriculture-Agricultural Experiment Station through the University of California.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701824114/-/DCSupplemental.
References
- 1.Brandazza A, Angeli S, Tegoni M, Cambillau C, Pelosi P. Plant stress proteins of the thaumatin-like family discovered in animals. FEBS Lett. 2004;572:3–7. doi: 10.1016/j.febslet.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.van der Wel H, Loeve K. Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth. Eur J Biochem. 1972;31:221–225. doi: 10.1111/j.1432-1033.1972.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 3.Fils-Lycaon BR, Wiersma PA, Eastwell KC, Sautiere P. A cherry protein and its gene, abundantly expressed in ripening fruit, have been identified as thaumatin-like. Plant Physiol. 1996;111:269–273. doi: 10.1104/pp.111.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edreva A. Pathogenesis-related proteins: Research progress in the last 15 years. Gen Appl Plant Physiol. 2005;31:105–124. [Google Scholar]
- 5.Liu JJ, Sturrock R, Ekramoddoullah AK. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010;29:419–436. doi: 10.1007/s00299-010-0826-8. [DOI] [PubMed] [Google Scholar]
- 6.Petre B, Major I, Rouhier N, Duplessis S. Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol. 2011;11:33. doi: 10.1186/1471-2229-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misra RC, Sandeep, Kamthan M, Kumar S, Ghosh S. A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis. Sci Rep. 2016;6:25340. doi: 10.1038/srep25340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun J, Yu X, Griffith M. Genetic studies of antifreeze proteins and their correlation with winter survival in wheat. Euphytica. 1998;102:219–226. [Google Scholar]
- 9.Grenier J, Potvin C, Trudel J, Asselin A. Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313x.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 10.Fierens E, et al. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem J. 2007;403:583–591. doi: 10.1042/BJ20061291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J, Lv Y, Hou Z, Li X, Ding L. Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants. Plant Growth Regul. 2015;79:299–307. [Google Scholar]
- 12.Hejgaard J, Jacobsen S, Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 13.Osmond RI, Hrmova M, Fontaine F, Imberty A, Fincher GB. Binding interactions between barley thaumatin-like proteins and (1,3)-β-D-glucans. Kinetics, specificity, structural analysis and biological implications. Eur J Biochem. 2001;268:4190–4199. doi: 10.1046/j.1432-1327.2001.02331.x. [DOI] [PubMed] [Google Scholar]
- 14.Molina-Cano JL. The EEC barley and malt committee index for the evaluation of malting quality in barley and its use in breeding. Plant Breed. 1987;98:249–256. [Google Scholar]
- 15.Houston K, et al. A genome wide association scan for (1,3;1,4)-β-glucan content in the grain of contemporary 2-row Spring and Winter barleys. BMC Genomics. 2014;15:907. doi: 10.1186/1471-2164-15-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. The changes of β-glucan content and β-glucanase activity in barley before and after malting and their relationships to malt qualities. Food Chem. 2004;86:223–228. [Google Scholar]
- 17.McClear BV, Glennie‐Holmes M. Enzymic quantification of (1→ 3) (1→ 4) ‐β‐d‐glucan in barley and malt. J Inst Brew. 1985;91:285–295. [Google Scholar]
- 18.Hayes PM, et al. Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor Appl Genet. 1993;87:392–401. doi: 10.1007/BF01184929. [DOI] [PubMed] [Google Scholar]
- 19.Gao W, et al. North American Barley Genome Project Fine mapping of a malting-quality QTL complex near the chromosome 4H S telomere in barley. Theor Appl Genet. 2004;109:750–760. doi: 10.1007/s00122-004-1688-7. [DOI] [PubMed] [Google Scholar]
- 20.Wei K, et al. Genetic mapping of quantitative trait loci associated with beta-amylase and limit dextrinase activities and beta-glucan and protein fraction contents in barley. J Zhejiang Univ Sci B. 2009;10:839–846. doi: 10.1631/jzus.B0920135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han F, et al. Molecular marker-assisted selection for malting quality traits in barley. Mol Breed. 1997;3:427–437. [Google Scholar]
- 22.Reiss E, Horstmann C. Drechslera teres-infected barley (Hordeum vulgare L.) leaves accumulate eight isoforms of thaumatin-like proteins. Physiol Mol Plant Pathol. 2001;58:183–188. [Google Scholar]
- 23.Yano H, Wong JH, Lee YM, Cho M-J, Buchanan BB. A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci USA. 2001;98:4794–4799. doi: 10.1073/pnas.071041998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorjanovic S. Biological and technological functions of barley seed pathogenesis related proteins (PRs) J Inst Brew. 2009;115:334–360. [Google Scholar]
- 25.Hiroyuki K, Terauchi R. Regulation of expression of rice thaumatin-like protein: Inducibility by elicitor requires promoter W-box elements. Plant Cell Rep. 2008;27:1521–1528. doi: 10.1007/s00299-008-0536-7. [DOI] [PubMed] [Google Scholar]
- 26.Doxey AC, Cheng Z, Moffatt BA, McConkey BJ. Structural motif screening reveals a novel, conserved carbohydrate-binding surface in the pathogenesis-related protein PR-5d. BMC Struct Biol. 2010;10:23. doi: 10.1186/1472-6807-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montrichard F, et al. Thioredoxin targets in plants: The first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Marx C, Wong JH, Buchanan BB. Thioredoxin and germinating barley: Targets and protein redox changes. Planta. 2003;216:454–460. doi: 10.1007/s00425-002-0857-7. [DOI] [PubMed] [Google Scholar]
- 29.Jones BL, Budde AD. Effect of reducing and oxidizing agents and pH on malt endoproteolytic activities and brewing mashes. J Agric Food Chem. 2003;51:7504–7512. doi: 10.1021/jf030206u. [DOI] [PubMed] [Google Scholar]
- 30.Hägglund P, et al. The barley grain thioredoxin system - an update. Front Plant Sci. 2013;4:151. doi: 10.3389/fpls.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobrehel K, et al. Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol. 1992;99:919–924. doi: 10.1104/pp.99.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho MJ, et al. Overexpression of thioredoxin h leads to enhanced activity of starch debranching enzyme (pullulanase) in barley grain. Proc Natl Acad Sci USA. 1999;96:14641–14646. doi: 10.1073/pnas.96.25.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong JH, et al. Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA. 2002;99:16325–16330. doi: 10.1073/pnas.212641999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YC, et al. The level of expression of thioredoxin is linked to fundamental properties and applications of wheat seeds. Mol Plant. 2009;2:430–441. doi: 10.1093/mp/ssp025. [DOI] [PubMed] [Google Scholar]
- 35.Kaur S, Dhugga KS, Gill K, Singh J. Novel structural and functional motifs in cellulose synthase (CesA) genes of bread wheat (Triticum aestivum, L.) PLoS One. 2016;11:e0147046. doi: 10.1371/journal.pone.0147046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh M, Singh S, Randhawa H, Singh J. Polymorphic homoeolog of key gene of RdDM pathway, ARGONAUTE4_9 class is associated with pre-harvest sprouting in wheat (Triticum aestivum L.) PLoS One. 2013;8:e77009. doi: 10.1371/journal.pone.0077009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh J, Sharp PJ, Skerritt JH. A new candidate protein for high lysine content in wheat grain. J Sci Food Agric. 2001;81:216–226. [Google Scholar]
- 38.Lamb-Palmer ND, Singh M, Dalton JP, Singh J. Prokaryotic expression and purification of soluble maize Ac transposase. Mol Biotechnol. 2013;54:685–691. doi: 10.1007/s12033-012-9610-z. [DOI] [PubMed] [Google Scholar]