Significance
A core question in evolutionary biology is how mutation and selection adapt and constrain species to specialized habitats. We sequenced the genome of the sand rat, a desert rodent susceptible to nutritionally induced diabetes, and discovered an unusual chromosome region skewed toward G and C nucleotides. This region includes the Pdx1 homeobox gene, a transcriptional activator of insulin, which has undergone massive sequence change, likely contributing to diabetes and adaptation to low caloric intake. Our results imply that mutation rate varies within a genome and that hotspots of high mutation rate may influence ecological adaptation and constraint. In addition, we caution that divergent regions can be omitted by conventional short-read sequencing approaches, a consideration for existing and future genome sequencing projects.
Keywords: desert rodent, type 2 diabetes, homeobox, Pdx1, gene conversion
Abstract
The sand rat Psammomys obesus is a gerbil species native to deserts of North Africa and the Middle East, and is constrained in its ecology because high carbohydrate diets induce obesity and type II diabetes that, in extreme cases, can lead to pancreatic failure and death. We report the sequencing of the sand rat genome and discovery of an unusual, extensive, and mutationally biased GC-rich genomic domain. This highly divergent genomic region encompasses several functionally essential genes, and spans the ParaHox cluster which includes the insulin-regulating homeobox gene Pdx1. The sequence of sand rat Pdx1 has been grossly affected by GC-biased mutation, leading to the highest divergence observed for this gene across the Bilateria. In addition to genomic insights into restricted caloric intake in a desert species, the discovery of a localized chromosomal region subject to elevated mutation suggests that mutational heterogeneity within genomes could influence the course of evolution.
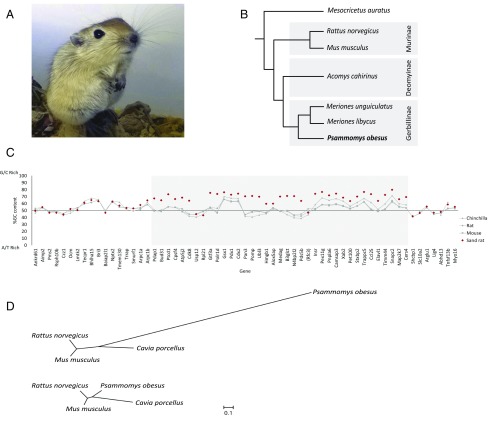
Arid environments impose extreme physiological demands on animals because of low food and water availability. The sand rat Psammomys obesus (Fig. 1A) is a member of the subfamily Gerbillinae, most species of which live in deserts and arid environments (Fig. 1B). P. obesus has emerged as a model for research into diet-induced type II diabetes because, if provided with high carbohydrate diets, the majority of individuals become obese and develop classic diabetes symptoms, in the most extreme cases leading to pancreatic failure and death (1–4).
Fig. 1.
The sand rat and its genomic hotspot of mutation. (A) Juvenile sand rat P. obesus. (B) Cladogram of representative murid rodents indicating the phylogenetic position of sand rat. (C) GC content of genes around the ParaHox cluster of sand rat and other rodents (Mus musculus, Rattus norvegicus, Chinchilla lanigera) revealing a chromosomal hotspot of GC skew in sand rat (shaded in gray). Genes shown in inferred ancestral gene order; parentheses around Rfc3 indicate this gene has been transposed to a different genomic location in sand rat. Sand rat GC values based on transcriptome and genome sequences; when partial only alignable sequence is compared. (D) Unrooted phylogenetic trees inferred from synonymous changes (dS) only from concatenated alignments of 26 genes in the mutational hotspot (Upper) and 100 random genes (Lower).
In searching for the molecular basis of this unusual phenotype, attention has been paid to the Pdx1 homeobox gene, also called Ipf1, Idx1, Stf1, or Xlox (5–9), the central and most highly conserved member of the ParaHox gene cluster (10). Pdx1 is the only member of the Pdx gene family in tetrapods and encodes a homeodomain that has been invariant across their evolution. Mammalian Pdx1 is expressed in pancreatic beta cells and encodes a homeodomain transcription factor that acts as a transcriptional activator of insulin and other pancreatic hormone genes (11, 12). A pivotal role in insulin regulation is also reflected in the association of heterozygous Pdx1 mutations with maturity-onset diabetes of the young (MODY4) and type II diabetes mellitus in humans (13). Contrary to the usual conservation, several studies have reported inability to detect Pdx1 in multiple gerbil species, including P. obesus, by immunocytochemistry, Western blotting, or PCR. However, Pdx1 is readily detectable in the closely related spiny mouse, Acomys cahirinus (Fig. 1B), leading to the hypothesis that the gene has been lost within the Gerbillinae subfamily, contributing to the compromised ability to regulate insulin in the sand rat (14–16). Such a conclusion would raise further questions, because in addition to its adult functions, Pdx1 is also essential for pancreatic development in the embryo. For example, targeted deletion in mice causes loss of pancreas and anterior duodenum and is lethal (9, 17). In humans, pancreatic agenesis has been reported in a patient with a homozygous frameshift mutation before the Pdx1 homeobox and in a compound heterozygous patient with substitution mutations in helices 1 and 2 of the homeodomain (18–20).
Results
To resolve the conundrum of a putatively absent “essential” gene, we sequenced the P. obesus genome by using a standard shotgun strategy (Illumina), using a combination of short and long insert libraries, initially at 85.5× coverage (SI Appendix, SI Materials and Methods, section 1). This assembly lacked a Pdx1 gene, supporting the prevailing hypothesis of a loss of the Pdx1 gene in gerbils. However, a synteny comparison between P. obesus and other mammals delineated a contiguous block of 88 genes (SI Appendix, Fig. S2) missing from the assembly including several genes essential to basic cellular functions, such as Brca2 and Cdk8, in addition to Pdx1. This finding led us to suspect that standard short read sequencing may have given an incomplete genome assembly, even at high coverage. To resolve whether the gene absence reflected a large-scale deletion or an unusual genomic region, we sequenced the transcriptomes of P. obesus liver, pancreatic islets, and duodenum, which contained transcripts for many of the missed genes (SI Appendix, Tables S4–S6). Furthermore, these transcripts show unusually high GC content in most cases, indicating that a large contiguous stretch of elevated GC had either been underrepresented in initial sequencing data or had failed to assemble correctly, most likely due to nucleotide compositional bias. We term such cryptic or hidden sequence “dark DNA.” We therefore isolated GC-rich P. obesus genomic DNA by cesium chloride gradient centrifugation, sequenced this fraction after limited amplification by using Illumina MiSeq overlapping paired-end reads, and reassembled the genome incorporating this longer-read sequence data (SI Appendix, SI Materials and Methods, section 1.5). This approach gave a refined assembly with a total size of 2.38 Gb and a scaffold N50 of 10.4 Mb (Table 1 and SI Appendix, SI Materials and Methods, sections 1, 3, 4, and 6), including much of the dark DNA region in several scaffolds, and containing genes syntenic to a region of chromosome 12 in rat and a region of chromosome 5 and the subtelomeric region of chromosome 8 in mouse. Analysis indicates that the region was initially omitted by standard genome assembly methods because of lower read coverage of GC regions coupled with short sequence read lengths. Comparison of GC content between species demonstrates that sand rat genes are elevated in GC content across this chromosomal region, syntenic to 12 Mb of the rat genome (Fig. 1C and SI Appendix, SI Materials and Methods, section 9). This large region encompasses a 250-kb repeat-rich scaffold containing the sand rat ParaHox cluster and its well-characterized genomic neighbors. We inferred a high W (weak, A/T) to S (strong, G/C) allelic mutation rate in this region of the P. obesus genome compared with randomly selected genomic regions or homologous regions in other species of rodent (Fig. 1D and SI Appendix, SI Materials and Methods, section 12 and Tables S11 and S12). The existence of a localized GC-biased stretch of the P. obesus genome is striking and of far-reaching importance, and implies the existence of elevated and biased mutational pressure, acting in one region of a mammalian genome. Gene conversion, caused by the nonreciprocal exchange of information during meiosis, is the best characterized process known to cause GC-biased mutation (21).
Table 1.
Metrics of sand rat raw genomic sequencing data and final genome assembly
| Genome sequencing and assembly | Value |
| Total no. of paired-end reads | 724,377,486 |
| Total no. of mate-pair reads | 1,780,436,140 |
| Total bases sequenced | 394,396,928,120 |
| Estimated sequencing coverage, x | 87.6 |
| No. of scaffolds >2 kb | 1,737 |
| Total length of assembly, bp | 2,381,209,849 |
| Longest scaffold, bp | 54,616,910 |
| Mean scaffold length, bp | 15,794 |
| Scaffold N50, bp | 10,461,538 |
| Scaffold L50 | 63 |
| Contig N50, bp | 83,904 |
| Percentage of assembly in scaffolds, % | 98.6 |
Coverage was calculated by using an estimated genome size of 2.51 Gb based on a k-mer analysis (SI Appendix, SI Materials and Methods, section 1.3) and is based on paired-end sequencing data only.
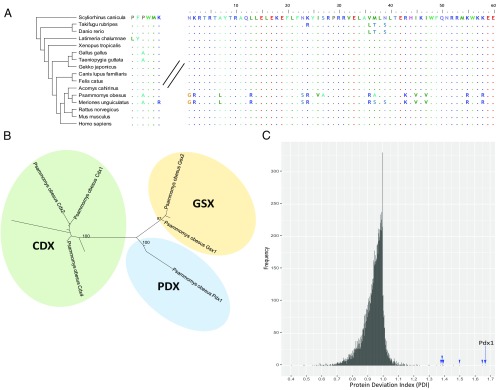
The full coding sequence of the P. obesus Pdx1 gene was deduced from the refined genome and transcriptome assemblies, and the gene was found to be expressed in sand rat pancreatic islets and duodenum (SI Appendix, SI Materials and Methods, section 7). The 60-aa homeodomain of Pdx1 shows 100% conservation across other mammals for which data are available; however, in P. obesus, there are remarkable 15-aa differences in the homeodomain, making it the most divergent Pdx1 gene discovered in the Bilateria (Fig. 2A). All but one of the amino acid changes are caused by A/T to G/C mutation. The N-terminal and C-terminal regions are also divergent with numerous deletions, although the hexapeptide motif used in heterodimer formation with TALE proteins is conserved (Fig. 2B). Additional RNA sequencing of Mongolian jird (Meriones unguiculatus) duodenum reveals that extensive sequence divergence due to GC-biased mutation in Pdx1 is not unique to sand rat (Fig. 2A). Analysis of synonymous and nonsynonymous mutations in Pdx1 across vertebrates reveals a dN/dS ratio of 2.6 (dN = 39; dS = 15) in the lineage leading to P. obesus and M. unguiculatus (SI Appendix, Fig. S10). High dN/dS ratios are often taken as evidence for positive selection, but can be skewed by mutational processes such as GC-biased gene conversion (22). Despite its radical divergence, Pdx1 is the closest homeodomain by BLASTP, and phylogenetic analysis places it as a rodent Pdx1 on a long branch (Fig. 2B); extensive synteny with the ParaHox region of mouse and rat confirms it is the true and single Pdx1 ortholog (SI Appendix, Table S9). Evidence that the locus is functional includes expression in pancreas and duodenum, and the fact that extensive polymorphism is found in the 3′ untranslated region but is limited in the coding sequence (SI Appendix, Fig. S11), indicating that the coding region is under functional constraint despite extensive mutation. Extreme deviation from the expected sequence explains why antibodies and PCR failed to detect Pdx1 in sand rat, Mongolian jird, and, potentially, other gerbil species (14–16).
Fig. 2.
Molecular divergence of sand rat Pdx1. (A) Alignment of Pdx1 hexapeptide domain and homeodomain sequences across vertebrates. (B) Maximum likelihood tree of ParaHox proteins showing divergent P. obesus Pdx1; species included are sand rat, mouse, zebra finch, spotted gar, amphioxus (full tree in SI Appendix, Fig. S6). (C) Histogram of PDI values for 1:1:1 mammalian orthologs of the sand rat predicted proteins: Pdx1 is marked by an arrow, other genes within the GC-rich region are marked by arrowheads.
These findings indicate that GC-biased mutation has driven radical changes in an otherwise highly conserved homeobox gene; these changes could be maladaptive and constrain the physiological capability of the sand rat, or adaptive enhancing ability to live in arid regions. To test whether the extent of sequence divergence is unusual for sand rat proteins, we calculated a “protein deviation index” (PDI) (SI Appendix, SI Materials and Methods, section 5) for all 1:1 mammalian orthologs by dividing mouse-human protein sequence identity by mouse-sand rat sequence identity (Fig. 2C). This analysis is distinct from identifying the fastest evolving proteins and specifically identifies proteins that have undergone uncharacteristic divergence in sand rat. We find the majority of sand rat proteins are highly similar to mouse or human (mode PDI = 1.0); in contrast, Pdx1 is unusually divergent (mouse-sand rat 54.82%, mouse-human 91.37%; PDI = 1.67). To test whether other genes implicated in glucose metabolism or pancreatic function are also divergent, we compiled a list of 45 candidates from human studies including all genes implicated in monogenic diabetes (23) and genes for which coding sequence variants have been strongly associated with type 2 diabetes (24). Of the 33 genes with clear 1:1:1 orthologs between human, mouse, and sand rat, 32 lie between position 225 and 10,195 in our PDI ranking, indicating that they are not unusually divergent in sand rat. Pdx1 is ranked first and is the most unusually divergent protein identified in the sand rat predicted proteome (SI Appendix, Materials and Methods, section 8 and Tables S8 and S10). Strikingly, 7 of the top 10 highest PDI results correspond to genes located within the mutational hotspot (SI Appendix, Table S8), indicating that GC-biased mutation is contributing to coding sequence divergence across this region.
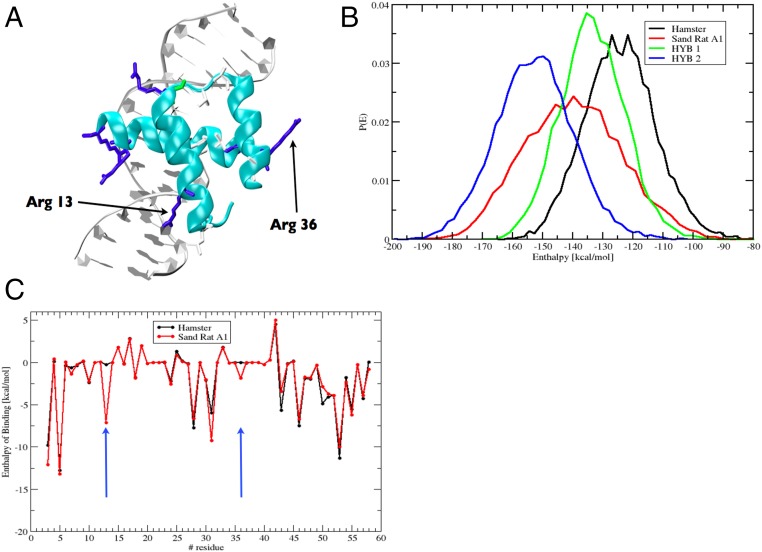
The mutations fixed in sand rat Pdx1 gene do not cause frameshifts or truncations in known domains, and molecular modeling reveals that the sand rat Pdx1 homeodomain has the ability to form all three helices required for DNA binding (Fig. 3A). To examine whether these mutations have resulted in subtle effects on the stability of DNA binding, we deployed molecular dynamics simulations with atomistic representation of Pdx1 homeodomains, DNA target, and solvent. From the postprocessing of the molecular dynamics simulations, we estimated the enthalpy of binding between sand rat and mouse (or other mammal) Pdx1 and monomer DNA binding sites by using the Molecular Mechanics Poisson Boltzmann Surface Area (MM-PBSA) method (SI Appendix, Materials and Methods, section 10). Target DNA sequences used were core Pdx1-binding sites of the mouse insulin A1 promoter and its sand rat ortholog. From 200-ns molecular dynamics simulations, the enthalpy of binding for protein–DNA interaction was calculated to be lower for sand rat than for mouse Pdx1 (mean −140 kcal/mol vs. mean −122 kcal/mol), indicative of sand rat Pdx1 binding DNA more “tightly” than is normal for the mammalian Pdx1 protein (Fig. 3B). One amino acid change was responsible for much of the difference: a Leu-to-Arg substitution in alpha helix 1 (homeodomain position 13), leading to the positive side chain of Arg making a new indirect contact with the phosphate backbone of DNA. A second substitution, Val to Arg in alpha helix 2 (homeodomain position 36), makes a smaller contribution (Fig. 3C). We also detect modifications to specific base interactions, with sand rat residues Met54 and Arg58 making new contacts to A and T bases within the TAAT core. Hence, stronger DNA binding is most likely driven by increased contacts with the backbone of DNA, coupled with decreased sequence specificity of DNA interaction. These results suggest that sand rat Pdx1 is divergent in DNA-binding affinity and specificity. Conserved Pdx1-binding sites in well-characterized promoters of three downstream target genes encoding pancreatic hormones (insulin, somatostatin, and glucokinase) show negligible divergence in sand rat compared with mouse, rat, and human (SI Appendix, Materials and Methods, section 11), indicating that Pdx1 divergence alone is likely to be responsible for altered DNA binding affinity and specificity.
Fig. 3.
Molecular modeling of sand rat Pdx1 binding. (A) Molecular model of sand rat Pdx1 homeodomain bound to DNA. The two amino acid changes indicated are the largest contributors to altered enthalpy of binding. (B) Probability distributions of the enthalpy of binding of homeodomain protein–DNA interactions among hamster (normal vertebrate) Pdx1/mouse insulin A1 DNA element (black), sand rat Pdx1/sand rat A1 element (red), hamster Pdx1/sand rat A1 (green), and sand rat Pdx1/mouse A1 (blue) inferred by molecular dynamics simulations and MM-PBSA; sand rat Pdx1 homeodomain has the lowest enthalpy of binding (higher affinity) for each DNA target. (C) Per-site enthalpy of binding comparison between hamster and sand rat Pdx1 revealing contribution of amino acid changes at homeodomain positions 13 and 36 to reduced enthalpy of binding (higher affinity).
Discussion
We show that an unusual genomic region of biased mutation arose in the evolutionary lineage of the sand rat. One consequence of this hotspot of mutation was the generation of GC-bias in the Pdx1 gene of P. obesus; this process forced modification of the Pdx1 protein sequence, likely affecting its ability to regulate transcription of insulin and other pancreatic genes. The sand rat Pdx1 hexapeptide, which mediates cofactor interactions (25), is intact, which may explain why pancreatic development proceeds permitting viable sand rat embryogenesis. We suggest mutation-driven changes have played a role in constraining or adapting the sand rat, and possibly other gerbil species, to arid environments and low caloric intake. Biased gene conversion is a known mechanism that causes GC-biased mutation (21, 26); hence, we suggest this mechanism, driven by elevated localized recombination, is generating a hotspot of skewed base composition. The genomic region we describe here was not detected by standard short-read sequencing approaches, known to be sensitive to nucleotide composition (27). These issues may be circumvented through the use of third generation sequencing technologies offering substantially longer read lengths and reduced nucleotide bias. The possibility remains that other such dark DNA regions could be widespread features of animal genomes, thus far largely overlooked in comparative animal genomics. Indeed, GC-rich genes are also missing from the chicken genome assembly (28, 29). Hotspots of mutation could drive rapid evolutionary change at the molecular level, and it will be important to decipher to what extent such hotspots have constrained and influenced evolutionary adaptation across the animal kingdom.
Materials and Methods
Sand Rat Genome Sequencing.
All animal procedures were carried out in accordance with the regulations specified under Protection of Animals Act by the authority in Denmark, European Union, and Novo Nordisk A/S, or the Animals (Scientific Procedures) Act 1986, U.K., and Bangor University Animal Welfare and Ethical Review Board. Sand rat genome sequencing libraries were constructed from a male P. obesus obtained from Hadassah Medical School, Israel. We prepared and sequenced multiple short- and long-insert DNA libraries and sequenced them on an Illumina HiSeq 2000. We also isolated sand rat DNA enriched for GC content through cesium chloride gradient centrifugation, prepared GC-rich DNA libraries, and sequenced using an Illumina MiSeq. In total, we generated ∼398 Gbp of sequencing data, which was assembled by using SOAPdenovo2 (30). Further details are provided in SI Appendix.
Transcriptome Sequencing and Analysis.
Total RNA was extracted and purified by using either Qiagen RNeasy column-based methods (pancreatic islets and liver) or TRIreagent (duodenum). All RNA-seq libraries were prepared by using Illumina chemistry. Pancreatic islet libraries were sequenced individually and as pools on the Illumina GAII. RNA-seq libraries for liver and duodenum were sequenced on the HiSeq 2000 (liver) or the HiSeq 4000 (duodenum). The pancreatic islets transcriptome was assembled by using Trans-ABySS (31) using multiple k-mer sizes (41 up to 79, in increments of 2), and the liver and duodenum transcriptomes were assembled by using Trinity (32) (SI Appendix).
Gene Prediction and Annotation.
We used multiple methods to predict genes in the sand rat genome. Repetitive elements were first masked by using RepeatMasker followed by ab initio gene prediction with AUGUSTUS (33). Homologous proteins from mouse and human were subsequently mapped to the sand rat genome assembly by using TBLASTN, with the aligned sequence being filtered and passed to GeneWise (34) to identify accurate spliced alignments. GLEAN (35) was then used to generate a consensus gene set. These gene models were then further refined by predicting ORFs using genome-guided transcriptome assemblies assembled using TopHat (36) and Cufflinks (37).
Evolutionary Analyses.
Using the gene predictions from our sand rat genome assembly and the assembled tissue transcriptome data, we carried out analyses of coding sequence GC content and GC-biased mutation within coding and intronic regions compared with other rodents. We also conducted an analysis to determine the extent of protein divergence within the sand rat predicted proteome compared with mouse and human. Details of these analyses are described in SI Appendix.
Molecular Modeling.
We used molecular dynamics simulations to calculate the enthalpy of binding of protein–DNA complexes, namely between the sand rat or mouse Pdx1 homeodomain and the sand rat or mouse A1 region of the insulin promoter, using MM-PBSA analyses (SI Appendix).
Supplementary Material
Acknowledgments
We thank Natasha Ng, Gemma Marfany, Thomas Dunwell, Fei Xu, Shan Quah, Anna Gloyn, Christine Hirschberger, Juliane Cohen, Rhys Morgan, Lorna Witty, Monica Martinez Alonso, and Thomas Brekke for assistance and advice, and the Oxford Genomics Centre for GC-rich sequencing. This work was funded principally by the European Research Council under European Union's Seventh Framework Programme (FP7/2007-2013 ERC Grant 268513 to P.W.H.H.), a Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB13000000 (to G.Z.), and Novo Nordisk A/S (coordinated by R.S.H.). E.S. and W.R.T. were supported by the Francis Crick Institute under award FC001179. The Crick receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust.
Footnotes
Conflict of interest statement: J.C., P.G.J., M.T.H., S.V.H.P., S.B., K.S., B.A.F., and R.S.H. are current or former employees of Novo Nordisk.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Short Read Archive (accession nos. SRA502705, SRR5084169, SRR5084170, SRR5092818, SRR5092819, SRR5092820, and SRR5429486) and DDBJ/ENA/GenBank (accession nos. NESX00000000 and NESX01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702930114/-/DCSupplemental.
References
- 1.Schmidt-Nielsen K, Haines HB, Hackel DB. Diabetes mellitus in the sand rat induced by standard laboratory diets. Science. 1964;143:689–690. doi: 10.1126/science.143.3607.689. [DOI] [PubMed] [Google Scholar]
- 2.Bar-On H, Ben-Sasson R, Ziv E, Arar N, Shafrir E. Irreversibility of nutritionally induced NIDDM in Psammomys obesus is related to β-cell apoptosis. Pancreas. 1999;18:259–265. doi: 10.1097/00006676-199904000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Kaiser N, et al. Psammomys obesus, a model for environment-gene interactions in type 2 diabetes. Diabetes. 2005;54:S137–S144. doi: 10.2337/diabetes.54.suppl_2.s137. [DOI] [PubMed] [Google Scholar]
- 4.Kalman R, Ziz E, Galila L, Shafrir E. The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents. Elsevier; London: 2012. Sand rat; pp. 1171–1190. [Google Scholar]
- 5.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard J, et al. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 7.Bürglin TR. A comprehensive classification of homeobox genes. In: Duboule D, editor. A Guidebook to Homeobox Genes. Oxford Univ Press; Oxford: 1994. pp. 25–71. [Google Scholar]
- 8.Miller CP, McGehee RE, Jr, Habener JF. IDX-1: A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 10.Brooke NM, Garcia-Fernàndez J, Holland PWH. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- 11.Ashizawa S, Brunicardi FC, Wang XP. PDX-1 and the pancreas. Pancreas. 2004;28:109–120. doi: 10.1097/00006676-200403000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Servitja JM, Ferrer J. Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia. 2004;47:597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- 13.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 14.Leibowitz G, et al. IPF1/PDX1 deficiency and β-cell dysfunction in Psammomys obesus, an animal With type 2 diabetes. Diabetes. 2001;50:1799–1806. doi: 10.2337/diabetes.50.8.1799. [DOI] [PubMed] [Google Scholar]
- 15.Vedtofte L, Bödvarsdóttir TB, Karlsen AE, Heller RS. Developmental biology of the Psammomys obesus pancreas: Cloning and expression of the Neurogenin-3 gene. J Histochem Cytochem. 2007;55:97–104. doi: 10.1369/jhc.6A7073.2006. [DOI] [PubMed] [Google Scholar]
- 16.Gustavsen CR, et al. The morphology of islets of Langerhans is only mildly affected by the lack of Pdx-1 in the pancreas of adult Meriones jirds. Gen Comp Endocrinol. 2008;159:241–249. doi: 10.1016/j.ygcen.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 18.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 19.Schwitzgebel VM, et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab. 2003;88:4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- 20.Thomas IH, et al. Neonatal diabetes mellitus with pancreatic agenesis in an infant with homozygous IPF-1 Pro63fsX60 mutation. Pediatr Diabetes. 2009;10:492–496. doi: 10.1111/j.1399-5448.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pessia E, et al. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol Evol. 2012;4:675–682. doi: 10.1093/gbe/evs052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratnakumar A, et al. Detecting positive selection within genomes: The problem of biased gene conversion. Philos Trans R Soc Lond B Biol Sci. 2010;365:2571–2580. doi: 10.1098/rstb.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwitzgebel VM. Many faces of monogenic diabetes. J Diabetes Investig. 2014;5:121–133. doi: 10.1111/jdi.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 27.Chen YC, Liu T, Yu CH, Chiang TY, Hwang CC. Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS One. 2013;8:e62856. doi: 10.1371/journal.pone.0062856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hron T, Pajer P, Pačes J, Bartůněk P, Elleder D. Hidden genes in birds. Genome Biol. 2015;16:164. doi: 10.1186/s13059-015-0724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seroussi E, et al. Identification of the long-sought Leptin in Chicken and Duck: Expression pattern of the highly GC-rich avian Leptin fits an autocrine/paracrine rather than endocrine function. Endocrinology. 2016;157:737–751. doi: 10.1210/en.2015-1634. [DOI] [PubMed] [Google Scholar]
- 30.Luo R, et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 32.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller O, Kollmar M, Stanke M, Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27:757–763. doi: 10.1093/bioinformatics/btr010. [DOI] [PubMed] [Google Scholar]
- 34.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsik CG, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.