Significance
Ena/VASP proteins are tetramers that drive the processive elongation of actin filaments. Because Ena/VASP proteins are implicated in motility, embryogenesis, and cancer, it is mandatory to understand their molecular mechanism. The kinetic model of VASP-mediated filament elongation in solution and on beads revealed that VASP uses one arm for tethering to growing barbed ends while three GABs on other arms are used to capture and deliver actin monomers to growing filament tips. However, the underlying mechanism must be profoundly diverse because only in clustered arrays does VASP-mediated filament elongation become exceptionally processive and resistant against capping protein. Using VASP oligomerization mutants and total internal reflection fluorescence microscopy (TIRFM) imaging, we show now that, in clustered arrays, adjacent VASP tetramers synergize in the elongation of single filaments.
Keywords: actin, Ena/VASP, TIRF, cluster, formin
Abstract
Ena/VASP proteins act as actin polymerases that drive the processive elongation of filament barbed ends in membrane protrusions or at the surface of bacterial pathogens. Based on previous analyses of fast and slow elongating VASP proteins by in vitro total internal reflection fluorescence microscopy (TIRFM) and kinetic and thermodynamic measurements, we established a kinetic model of Ena/VASP-mediated actin filament elongation. At steady state, it entails that tetrameric VASP uses one of its arms to processively track growing filament barbed ends while three G-actin–binding sites (GABs) on other arms are available to recruit and deliver monomers to the filament tip, suggesting that VASP operates as a single tetramer in solution or when clustered on a surface, albeit processivity and resistance toward capping protein (CP) differ dramatically between both conditions. Here, we tested the model by variation of the oligomerization state and by increase of the number of GABs on individual polypeptide chains. In excellent agreement with model predictions, we show that in solution the rates of filament elongation directly correlate with the number of free GABs. Strikingly, however, irrespective of the oligomerization state or presence of additional GABs, filament elongation on a surface invariably proceeded with the same rate as with the VASP tetramer, demonstrating that adjacent VASP molecules synergize in the elongation of a single filament. Additionally, we reveal that actin ATP hydrolysis is not required for VASP-mediated filament assembly. Finally, we show evidence for the requirement of VASP to form tetramers and provide an amended model of processive VASP-mediated actin assembly in clustered arrays.
The highly coordinated spatial and temporal control of the assembly and disassembly of actin filaments is a major determinant of vital cellular processes such as endocytosis, cytokinesis, and cell migration (1–4). Specific protein assemblies, composed of various actin-binding proteins, operate in these processes to nucleate and elongate new actin filaments, arrange them into complex 3D arrays, and subsequently recycle them to replenish the G-actin pool (5). The only protein families known to date that actively drive the elongation of actin filaments by incorporation of actin monomers at growing barbed ends are formins and Ena/VASP proteins, albeit their modes of action are considerably different (6–9).
Ena/VASP proteins are a family of evolutionarily conserved proteins that are exclusively expressed in motile cells. In vertebrates, the family is composed of VASP, mammalian Enabled (Mena), and Ena-VASP-like (EVL) (10–12), whereas invertebrates such as Caenorhabditis elegans, Drosophila melanogaster, or Dictyostelium discoideum express only one variant. All of these proteins share a conserved architecture consisting of an N-terminal Ena/VASP homology 1 (EVH1) domain that binds FP4 motif-containing proteins for subcellular targeting (13). The central proline-rich region binds to the actin-binding protein profilin (PFN) and Src homology 3 (SH3) domains (14–16). The C-terminal EVH2 domain comprises the business end of the molecule and encompasses a WASP-homology 2 (WH2) G-actin–binding (GAB) domain (9, 15, 17), an F-actin–binding (FAB) domain (15, 17), and a short C-terminal coiled-coil region mediating tetramerization (18).
Comparable to formins, Ena/VASP proteins associate with growing barbed ends and accelerate filament elongation twofold to sevenfold (6, 9, 19, 20). However, contrary to formins, they deliver actin monomers directly to filament barbed ends by virtue of their GAB domains (6, 9, 19, 20). Moreover, PFN appears dispensable for VASP-mediated filament elongation in vitro (6, 20), albeit some studies have reported increased filament elongation rates in the presence of PFN (19, 21). Finally, because single Ena/VASP tetramers are not as processive as formins, they consequently only poorly protect barbed end growth from capping protein (CP) in solution (6, 19, 20). By contrast, when clustered in high density on surfaces, Ena/VASP proteins become highly processive and are virtually resistant to CP (6, 9).
On the basis of thermodynamic data and the determination of the association rate constants of G-actin to human and Dictyostelium GAB, and total internal reflection fluorescence microscopy (TIRFM), we previously developed a quantitative mathematical model of VASP-mediated filament elongation (9). It is derived from the “actoclampin” model of actin filament end-tracking proteins (22–24), in which processive and tethered elongation is achieved by alternating multivalent affinity-modulated interactions between the VASP tetramer and the filament barbed end. The model implies that, at steady state, one polypeptide chain of the VASP tetramer is bound to the terminal filament subunit, leaving a number (N) of free GABs to capture actin monomers from solution (concentration C) with rate constant kon. Captured monomers then either dissociate at rate constant koff or are transferred to the tip and are irreversibly incorporated with rate constant kt, so that the VASP-mediated filament elongation rate (rv),
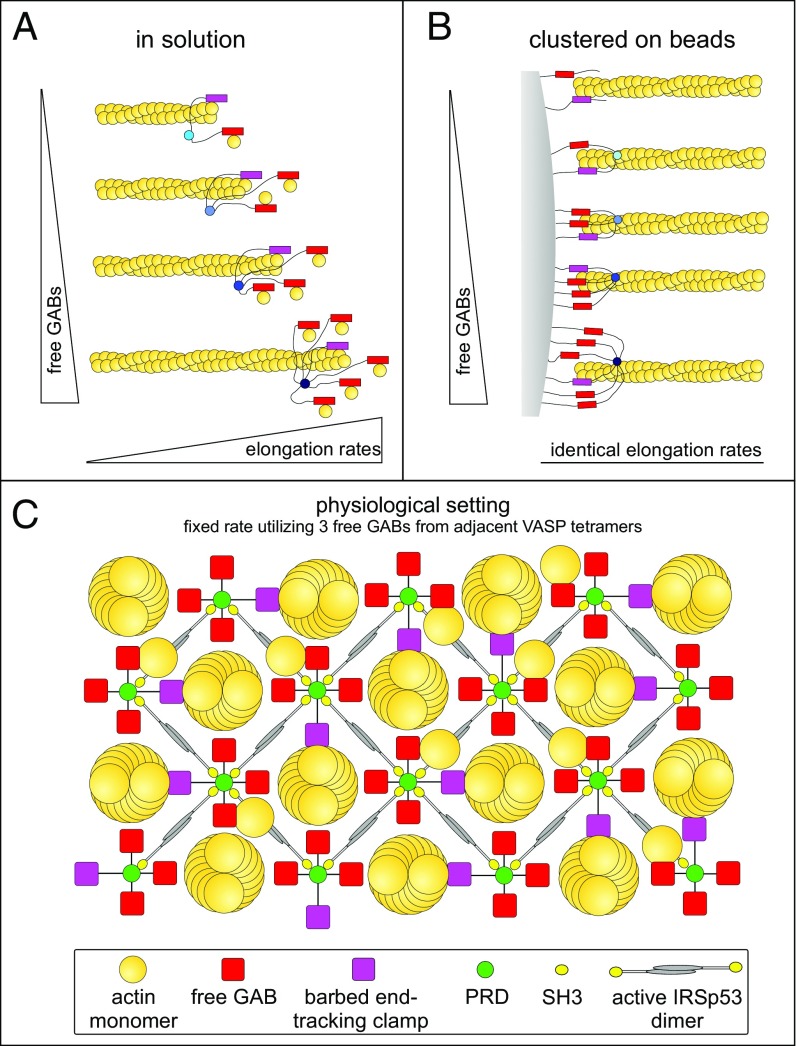
is correlated to the saturation of the GAB with actin monomers (9). Here, Kd = koff/kon is the equilibrium dissociation constant for monomer binding to GAB. Saturation of binding sites occurs for large C relative to Kd + kt/kon, such that rv ∼ ktN. Global fitting of the experimental rate data for filament elongation from beads coated with different VASP mutants with GAB domains of varying Kd yielded N = 3, formally suggesting that a single VASP tetramer operates at a filament tip during filament elongation. Indeed, processive filament elongation has been reported with single vertebrate and Drosophila Ena/VASP tetramers (19, 20). However, the marked differences in the context-specific processivity of VASP and its resistance to CP, indicated major differences in the underlying mechanisms. Here, we tested the model by analyses of VASP oligomerization mutants using single-color and multicolor TIRFM and show that the speed of VASP-mediated filament elongation directly correlates with N in solution. Strikingly, however, when clustered on beads, the elongation rate of these mutants was virtually identical revealing that, at a given actin monomer concentration, surface-tethered VASP operates at a fixed rate.
Results
Generation of Synthetic VASP Oligomerization Mutants.
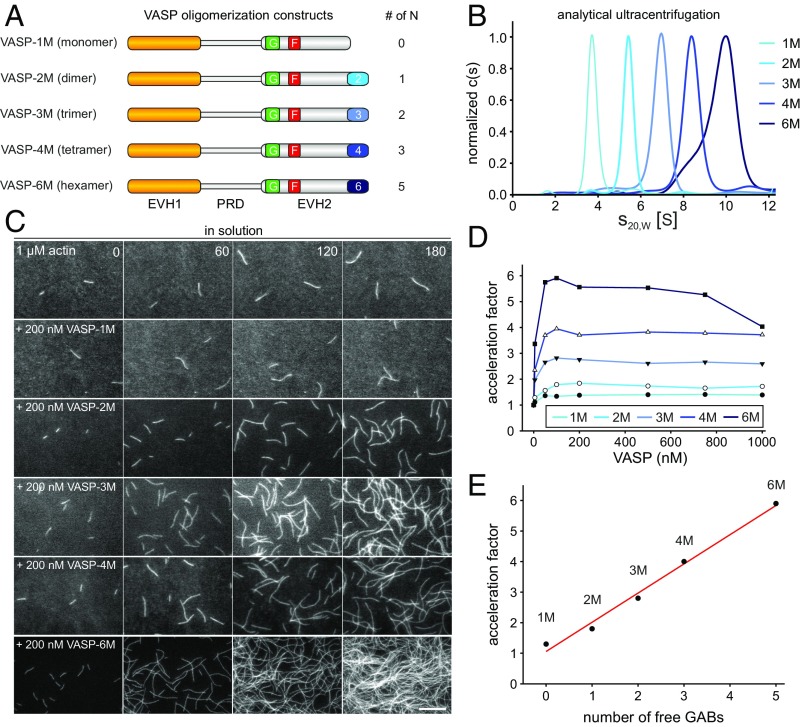
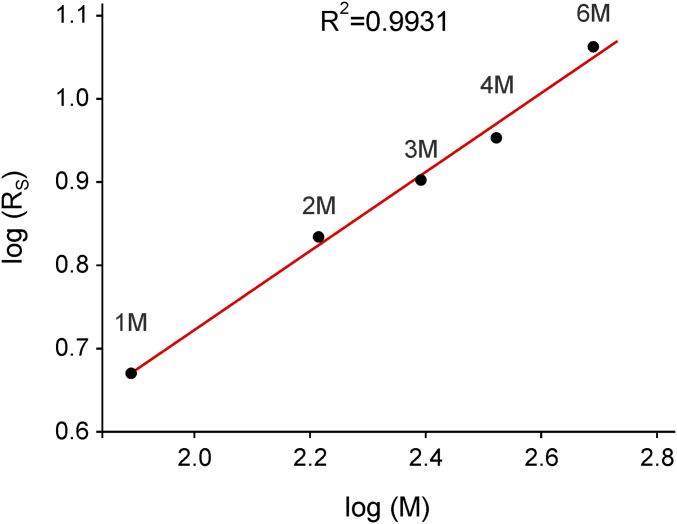
To thoroughly challenge the kinetic model, we first generated oligomerization mutants by replacing the C-terminal VASP coiled-coil domain that mediates tetramerization (18), by other oligomerization motifs (Fig. 1A and Table S1). For this reason, we decided to use both the natural as well as a mutated GCN4 leucine-zipper motif, which have been characterized as parallel coiled coils, forming dimers and trimers in solution, respectively (25). Moreover, we aimed to generate a higher order oligomerization mutant that exceeds the normal VASP tetramerization and used a synthetic coiled-coil hexamerization domain that forms a stand-alone, parallel six-helix bundle (26). These oligomerization mutants, designated VASP-2M, -3M, and -6M, were derived from the human VASP tetramer containing the high-affinity GAB from Dictyostelium (hVASP DGAB; here referred to as VASP-4M), because it facilitates biochemical analyses at the rather low actin concentrations in the subsequent TIRF assays (9). Moreover, we generated a monomeric VASP construct (VASP-1M) by deleting the C-terminal tetramerization motif. To study the oligomerization state of the VASP constructs, we expressed the proteins as fusions with the monomeric maltose-binding protein (MBP) and analyzed them at submicromolar or low micromolar concentration by analytical ultracentrifugation (AUC) in sedimentation velocity experiments (Fig. 1B). Sedimentation coefficients of the oligomers gradually increased from 4.0 S for VASP-1M over 5.7 S (VASP-2M), 7.3 S (VASP-3M), and 8.8 S (VASP-4M) to 9.9 S for VASP-6M. To check whether the increase in s value is consistent with the increase of the expected molar mass M of the oligomers, we calculated Stokes radii RS from the s values and plotted log(RS) versus log(M). As expected, a linear relationship was observed (Fig. S1). The parameters of this straight line are between those observed for globular and GdmCl-unfolded proteins (27). This behavior is not unexpected as the VASP fusion proteins consist both of globular domains (MBP and EVH1) and highly flexible regions (PRD and EVH2) (Fig. 1A). Thus, the results of the AUC experiments for the analyzed MBP-tagged VASP oligomerization mutants are in good agreement with predictions, confirming accurate and stable oligomerization of the constructs.
Fig. 1.
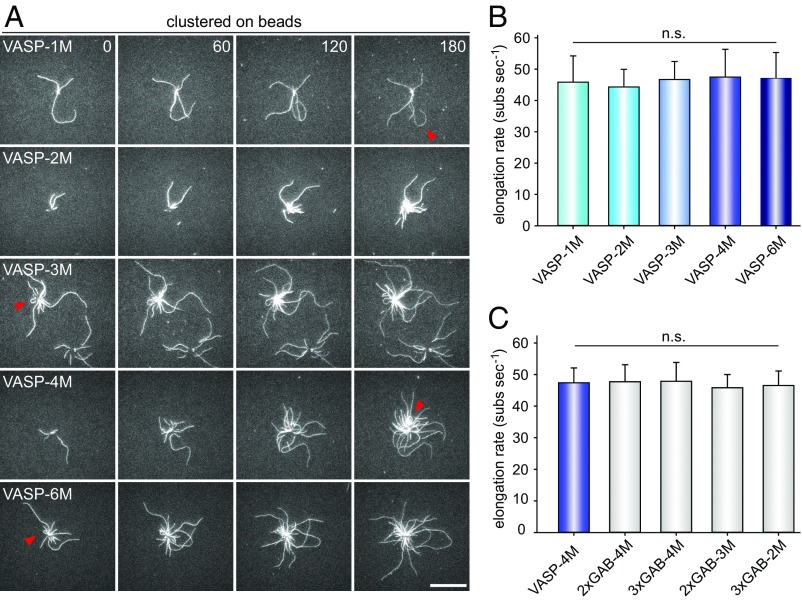
VASP-mediated filament elongation correlates directly with N in solution. (A) Schematic domain organization of the generated VASP-oligomerization mutants. Abbreviations: EVH1, Ena/VASP-homology domain 1; PRD, proline-rich domain; EVH2, Ena/VASP-homology domain 2 containing the G-actin-binding site (GAB) from Dictyostelium, the F-actin binding site (FAB), and the oligomerization domain. Abbreviations: F, FAB; G, GAB. (B) Sedimentation velocity analysis of the used VASP-oligomerization mutants fused to MBP. Differential sedimentation coefficient distribution analysis showed the proteins to sediment with s20°,w = 4.0 S (VASP-1M), 5.7 S (VASP-2M), 7.3 S (VASP-3M), 8.8 S (VASP-4M), and 9.9 S (VASP-6M) and is consistent with their predicted oligomerization status. (C) Time-lapse micrographs of TIRFM assays—corresponding to Movie S1—for determination of elongation rates. The actin (1 μM, 23% Alexa 488 labeled) was polymerized in the absence or presence of 200 nM of the indicated VASP construct in TIRF buffer, respectively. Time is given in seconds. (Scale bar, 20 µm.) (D) Dependence of the acceleration of actin-filament elongation on the oligomerization state of VASP. The elongation rates derived from TIRFM movies as shown in C were divided by the rate of spontaneous actin assembly to obtain the acceleration factors for each VASP oligomerization construct at the concentrations indicated. Concentrations are given in monomers. Fifteen actin control filaments and 30 filaments for all other data points in presence of VASP were measured. (E) Acceleration of VASP-mediated actin assembly at 100 nM VASP oligomer increased as a function of N (=total GABs − 1) as predicted by the kinetic model (9). Red line shows linear fit.
Table S1.
Oligomerization domains used in this study
| Oligomerization motif | Amino acid sequence (S329-stop) |
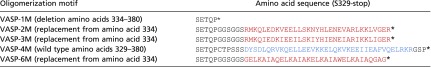 |
The tetramerization domain of wild-type human VASP is shown in blue letters. The oligomerization motifs of the mutants are depicted in red. Asterisk indicates end of sequence.
Fig. S1.
Analytical ultracentrifugation reveals a linear relationship of the Stokes radius on the expected molar mass for the different oligomeric forms of VASP in a log–log plot. Logarithm of the Stokes radii (circles) as calculated from the sedimentation coefficients from Fig. 1B plotted against the expected molar masses calculated from amino acid composition of the different oligomeric forms of VASP (VASP-1M, 77.7 kg/mol; VASP-2M, 163.9 kg/mol; VASP-3M, 246.4 kg/mol; VASP-4M, 332.8 kg/mol; and VASP-6M, 489.5 kg/mol). Linear regression (solid red line) resulted in a slope m = 0.474, an intercept b = −0.226, and a correlation coefficient R2 = 0.993.
VASP-Mediated Actin Filament Elongation Rate Directly Correlates with the Number of Free GABs in Solution.
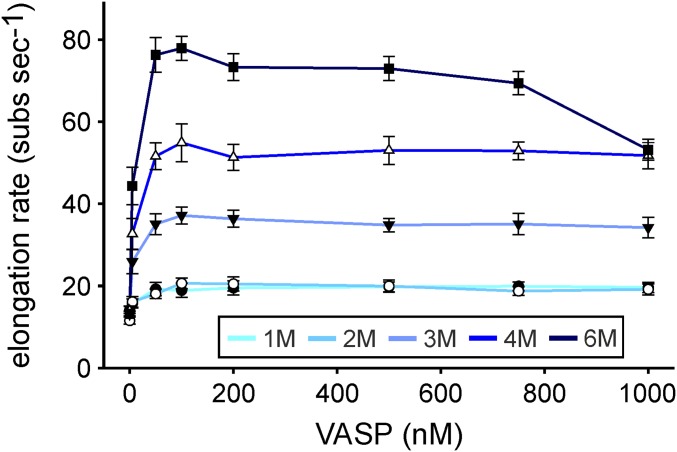
Next, we examined the effects of the mutants on VASP-mediated actin filament elongation by single-filament total internal reflection fluorescence microscopy (TIRFM) using Alexa 488-labeled actin in solution. In presence of MBP-tagged VASP-1M, actin filaments grew slightly faster as spontaneously polymerizing actin control filaments (Fig. 1C, Movie S1, and Fig. S2). This behavior is presumably caused by additional delivery of actin monomers by actin-loaded VASP-1M due its barbed-end binding capacity. All other mutants accelerated actin polymerization in a strictly concentration- and oligomerization state-dependent manner. The comparison of calculated acceleration factors (rate of VASP-mediated actin-filament elongation divided by the elongation rate of spontaneously growing control filaments) showed a maximum at 100 nM (Fig. 1D). This rate remained nearly constant up to 1 µM except for VASP-6M, where, presumably due to sequestration of actin monomers, the rate started to decrease at a concentration of 750 nM. Plotting of the acceleration factor at 100 nM oligomer against the number of free GABs (N) revealed a virtually perfect correlation (R2 = 0.99) as predicted by the kinetic model (Fig. 1E), even for the VASP hexamer. These findings therefore corroborated the notion that speed of VASP-mediated actin assembly is directly proportional to N. As expected, the line intercept at N = 0 (VASP-1M) is not equal to zero; rather, it is close the control elongation rate (acceleration factor of ∼1.0). This residual background rate that is close to the rate of the direct pathway for filament assembly suggests that monomers can still add directly to the filament tip from solution in parallel to the VASP-mediated pathway for monomer addition (9). Moreover, as mentioned above, the delivery of actin monomers by actin-loaded VASP-1M may additionally occur due to its barbed-end binding capacity.
Fig. S2.
Dependence of the oligomerization state of VASP on the acceleration of actin filament elongation. This figure corresponds to Fig. 1D but depicts the measured filament elongation rates instead of the acceleration factor.
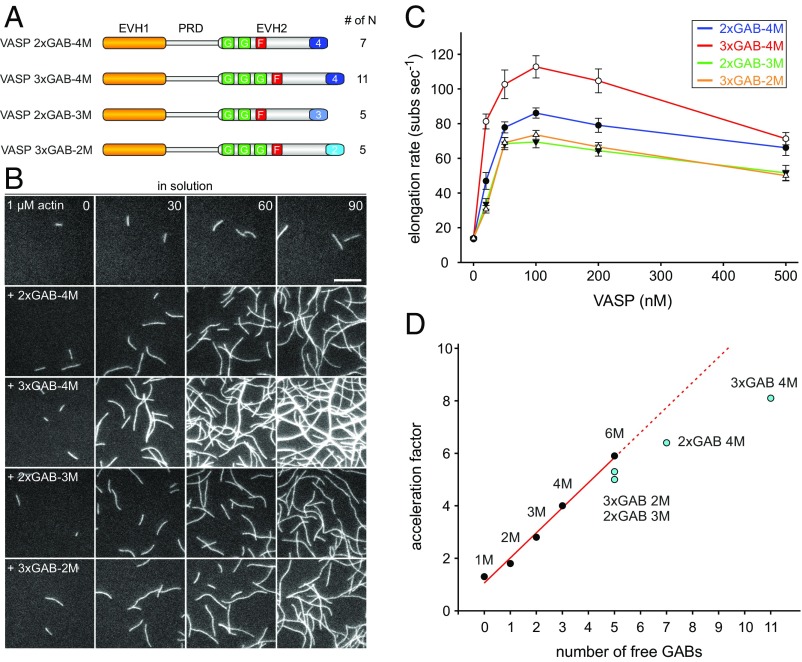
Analyses of VASP Mutants Containing Additional GABs in the Same Polypeptide Chain.
An alternative approach to vary the number of N is the creation of multiple GAB domains within the same EVH2 domain. Following this strategy, we engineered mutants containing two or three Dictyostelium GABs interspaced with flexible linker sequences of 44-aa residues, thus representing pseudo-VASP octamers (2×GAB VASP-4M) and pseudo-VASP dodecamers (3×GAB VASP-4M) with the wild-type tetramerization motif (Fig. 2A and Table S2). Additionally, we combined multiple GABs with different oligomerization motifs, yielding two distinct pseudo-VASP hexamers (2×GAB VASP-3M and 3×GAB VASP-2M, Fig. 2A). These proteins were again purified from bacterial extracts as MBP-fusion proteins and assayed by single-filament TIRFM with regard to their ability to stimulate actin assembly (Fig. 2B). These GAB mutants also promoted an increased filament elongation rate in a concentration-dependent manner and, similar to the oligomerization mutants, reached a maximal elongation rate at 100 nM (Fig. 2C). Albeit the acceleration factor increased with N again, in contrast to the rather fast pseudo-hexamers (2×GAB VASP 3-M and 3×GAB VASP-2M), the pseudo-octamer (2×GAB VASP-4M) and in particular the pseudo-dodecamer (3×GAB VASP-4M) significantly deviated from the theoretical values (Fig. 2D). Thus, engineering of too many GABs into VASP appears limiting to obtain the expected elongation rates predicted by the model, which in turn implies that the direct correlation of the elongation rate and N is either assured only in a certain range of N or that other steps of the reaction, such as the release of the bound FAB from the penultimate subunit, become rate limiting at high N values.
Fig. 2.
Additional GABs in the same polypeptide chain of VASP are less efficient to accelerate filament elongation in solution at higher N values. (A) Schematic domain organization of generated chimeric VASP mutants containing additional Dictyostelium GABs in the EVH2 domain. (B) Time-lapse micrographs of TIRFM assays for determination of elongation rates. The actin (1 μM, 23% Alexa 488 labeled) was polymerized in absence or presence of 200 nM of the indicated VASP construct in TIRF buffer, respectively. Time is given in seconds. (Scale bar, 5 µm.) (C) Comparison of the actin-filament elongation rates derived from TIRFM movies as shown in B in the presence of different VASP mutants at the concentrations indicated. Concentrations are given in monomers. Data correspond to means ± SD. Fifteen actin control filaments and 30 filaments for all other data points in presence of VASP were analyzed. (D) Comparison of the acceleration factors of VASP oligomerization construct and VASP constructs containing multiple GABs at 100 nM (relating to monomer concentration). Red line shows linear fit. Note less augmented filament elongation rates with VASP constructs containing multiple GABs within the same EVH2 domain.
Table S2.
Complete sequences of VASP constructs encompassing additional GABs
| VASP construct | Complete amino acid sequence |
 |
The high-affinity GAB domain of Dictyostelium VASP is shown in red. The flexible 44-aa linkers are depicted in blue. The FAB domain is highlighted in magenta. Asterisk indicates end of sequence.
VASP-Mediated Filament Elongation on Beads Is Constrained to a Fixed Rate.
Of note, in the physiological context, VASP mainly operates in clusters beneath the plasma membrane, for instance at the tips of filopodia and lamellipodia (28). Moreover, clustering is key to drive resilient and long-lasting filament elongation in presence of CP (6, 9). Thus, we asked how altered VASP oligomerization or the presence of multiple GABs would affect filament growth after clustering of the mutants on derivatized beads. However, as opposed to adsorption of the proteins in random orientations on polystyrene beads, which results in the formation of undefined geometries, we aimed to attach VASP in more physiological arrays. Thus, we mimicked clustering and appropriate orientation of the molecules by covalent attachment of N-terminal SNAP-tagged VASP constructs to the surface of benzylguanine-derivatized beads. Because processive, VASP-mediated filament elongation on beads is largely resistant to CP, whereas the latter efficiently abrogates growth of spontaneously polymerizing actin filaments in solution, thereby aiding analyses (6), we monitored filament growth on the beads by TIRFM in the presence of 40 nM CP. All oligomerization mutants promoted processive filament elongation as evidenced by frequent buckling of the growing actin filaments (Fig. 3A and Movie S2). Intriguingly, however, all analyzed oligomerization mutants, including VASP-6M carrying two additional GABs (N = 5) compared with the wild-type control, reached virtually the same, maximal elongation rate of about 46 ± 2.4 subs per second (mean ± SD) in the presence of 1 µM actin (Fig. 3B). The immobilized mutants harboring multiple GABs exhibited the same behavior as evidenced by the measured filament elongation rates in the range of 46 subs per second (Fig. 3C). These data therefore revealed a fundamental difference between the underlying mechanisms of VASP-mediated actin filament elongation in solution versus VASP cluster-driven filament growth on a surface.
Fig. 3.
Clustered VASP promotes processive filament elongation at a fixed rate irrespective of its oligomerization state. (A) Actin assembly of 1 µM actin (23% Alexa 488 labeled) in 1× TIRF buffer in the presence of 40 nM CP on 2 µm Ø benzylguanine beads derivatized with the VASP constructs indicated. Red arrows heads highlight processively growing, buckling filaments with their barbed ends attached to the bead surface. Time is given in seconds. (Scale bar, 20 µm.) (B and C) Determination of the elongation rates from TIRFM time-lapse movies corresponding to Movie S2 revealed a single, fixed rate for all tested VASP constructs. For each mutant, the fastest filaments growing on coated beads were analyzed. Data correspond to means ± SD. n = 30. n.s., nonsignificant. Color coding as in Fig. 1.
Hydrolysis of Actin-Bound ATP Is Not Required for VASP Activity.
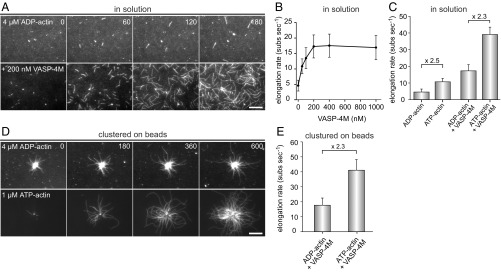
Another factor that could affect processive barbed-end elongation of Ena/VASP protein is the utilization of free energy obtained by ATP hydrolysis of newly incorporated actin subunits. In fact, the formin mDia1 has been reported to require PFN and ATP hydrolysis for processive barbed-end elongation (29), although these findings could not be confirmed by others (30). Thus, to examine a potential dependence on actin ATP hydrolysis for Ena/VASP proteins, we prepared ADP-G-actin and monitored the elongation rates in the absence or presence of VASP-4M in solution and on beads. First, we compared the rates of spontaneous actin assembly from either ATP- or ADP-charged monomers (Fig. 4A). The control filaments grew from 1 µM ATP-G-actin with 11.6 ± 2.4 subs per second (mean ± SD), whereas their growth rate dropped to 4.6 ± 1.8 subs per second with 4 µM ADP-G-actin (Fig. 4B). VASP-4M is known to accelerate barbed-end growth about threefold to fourfold (9). When VASP-4M was added, the actin polymerization rate in the presence of ADP-actin increased in a concentration-dependent manner to a maximum of 17.7 ± 3.7 subs per second at 200 nM VASP-4M in solution (Fig. 4 A and B). Thus, albeit the growth rate from ADP-G-actin was approximately diminished by one-half, VASP-4M still stimulated an almost fourfold increase compared with the growth rate of the control filaments. In case of VASP-derivatized beads, we could only compare VASP-mediated actin assembly from either ATP- or ADP-actin. In this assay, we observed processive filament elongation in presence of both nucleotides as evidenced by the buckling of the filaments, and the growth rate of VASP-4M–generated filaments from ADP-G-actin was again diminished by about one-half to 17.7 ± 3.7 subs per second, which is consistent with the reduction of filament growth in solution. These data therefore do not indicate a requirement of actin ATP hydrolysis for processive VASP-mediated filament assembly from actin monomers.
Fig. 4.
VASP does not require actin ATP hydrolysis of terminal subunits for filament elongation. (A) TIRF images of 4 µM ADP-actin (23% Alexa 488 labeled) polymerized in 1× ADP-TIRF buffer in the absence (Top) or presence (Lower) of 200 nM VASP-4M. Time is given in seconds. (B) Elongation rates of ADP-actin in the presence of increasing concentrations of VASP-4M in solution. (C) Comparison of elongation rates of ADP-actin or ATP-actin in absence or presence of 200 nM VASP-4M. (D) TIRFM images of 4 µM ADP-actin (Top) and 1.0 µM ATP-actin (Lower) polymerized in presence of VASP-4M–coated microspheres and 40 nM CP. (E) Comparison of elongation rates from ADP-G-actin and ATP-G-actin in the presence VASP-4M–derivatized beads. Note similar decrease in presence of ADP-actin for spontaneous and VASP-mediated actin assembly. (A and D) Time is given in seconds. (Scale bars, 20 µm.) (B, C, and E) Data correspond to means ± SD. Fifteen actin control filaments and 30 filaments for all other data points in presence of VASP-4M were analyzed.
Profilin Does Not Accelerate VASP-Mediated Actin Assembly.
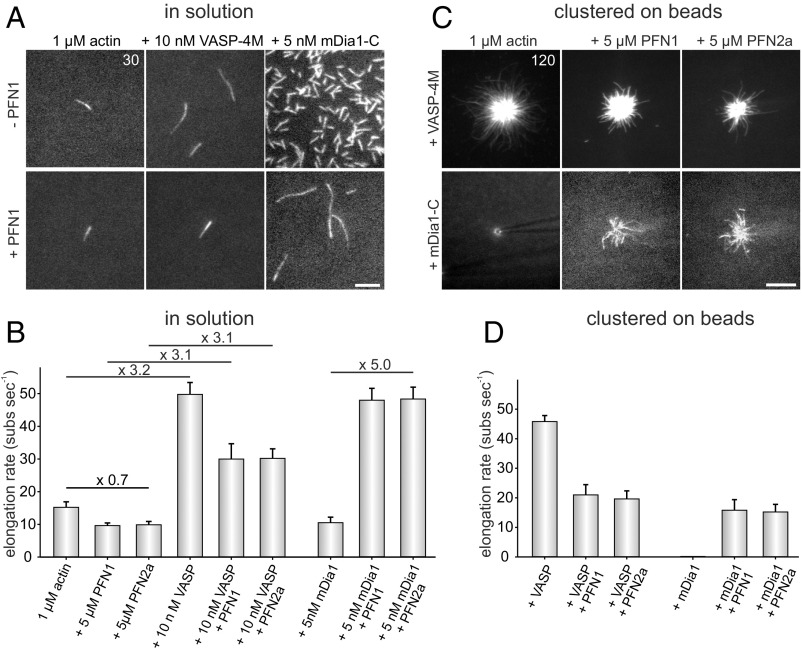
The requirement of PFN for VASP-mediated actin filament assembly is still controversial. Although some laboratories found that PFN does not accelerate VASP-mediated filament elongation in vitro (6, 20), others reported on increased filament elongation rates in the presence of PFN (19, 21). To conclusively clarify this issue, we analyzed filament elongation of VASP-4M by TIRF imaging in the absence or presence of different human PFN isoforms and compared it side-by-side to filament elongation driven by a C-terminal fragment of the formin mDia1 (mDia1-C) in solution and after clustering on beads. In the presence of 5 µM PFN1 or PFN2a, the actin filaments grew about 30% slower (9.6 ± 0.8 and 9.8 ± 1.0 subs per second) compared with spontaneously polymerizing actin control filaments (15.2 ± 1.7 subs per second) in solution (Fig. 5 A and B). The filament elongation rate increased to about 49.7 ± 3.6 subs per second by addition of 10 nM VASP-4M in the absence of PFN and dropped to ∼30 subs per second in the presence of VASP-4M in combination with either PFN1 or PFN 2a (30.0 ± 4.7 and 30.2 ± 2.9 subs per second). Thus, although VASP can clearly utilize PFN-actin as a substrate, the elongation rates were noticeably slower in the presence of both PFN isoforms, whereas the approximately threefold increase of VASP-mediated actin assembly was virtually unaffected by PFN. By contrast, addition of PFN1 and PFN2a to mDia1-C–generated filaments resulted in fivefold increased elongation rates with 47.9 ± 3.6 and 48.3 ± 3.6 subs per second), because formins rely on profilin binding to capture PFN-actin from solution, whereas VASP may use its PRD and its GABs to recruit monomers (6, 9, 15). Similar behaviors in filament elongation for VASP and mDia1 were also observed on beads, excluding the possibility that PFN is required to boost VASP-mediated filament elongation in clustered arrays (Fig. 5 C and D). These data therefore clearly demonstrate that, as opposed to formins, PFN does not accelerate but rather diminishes VASP-driven filament barbed-end elongation. The impaired growth of VASP-elongated filaments and of actin control filaments in presence of PFN is most likely caused by the necessity to remove PFN from the terminal filament subunit after barbed-end delivery before incorporation of the next monomer into growing filament ends.
Fig. 5.
Profilin does not accelerate VASP-mediated filament elongation but is required to increase speed of formin-driven actin assembly. (A) Time-lapse micrographs of single TIRFM assays for determination of elongation rates in solution. The actin (1 μM, 23% Alexa 488 labeled) was polymerized in the absence or presence of either 5 µM PFN1, 10 nM VASP-4M, or 5 nM mDia1-C in TIRF buffer, respectively. Time is given in seconds. (Scale bar, 5 µm.) (B) Quantification of the elongation rates in solution derived from TIRFM experiments as shown in A. Bars represent means ± SD from at least three independent experiments. (C) Time-lapse micrographs of single TIRFM assays for determination of elongation rates in clustered arrays on beads. The actin (1 μM, 23% Alexa 488 labeled) was polymerized in absence or presence of either 5 µM PFN1 or PFN2a in TIRF buffer, respectively using SNAP-tag derivatized VASP-4M or mDia1-C beads. The spontaneous assembly of actin was inhibited by addition of 40 nM CP. No filament growth from mDia1-C–derivatized beads in the absence of PFN was observed. Time is given in seconds. (Scale bar, 10 µm.) (D) Quantification of the elongation rates on beads derived from TIRFM experiments as shown in C. Bars represent means ± SD from at least three independent experiments.
VASP Processivity Increases with Higher Oligomerization.
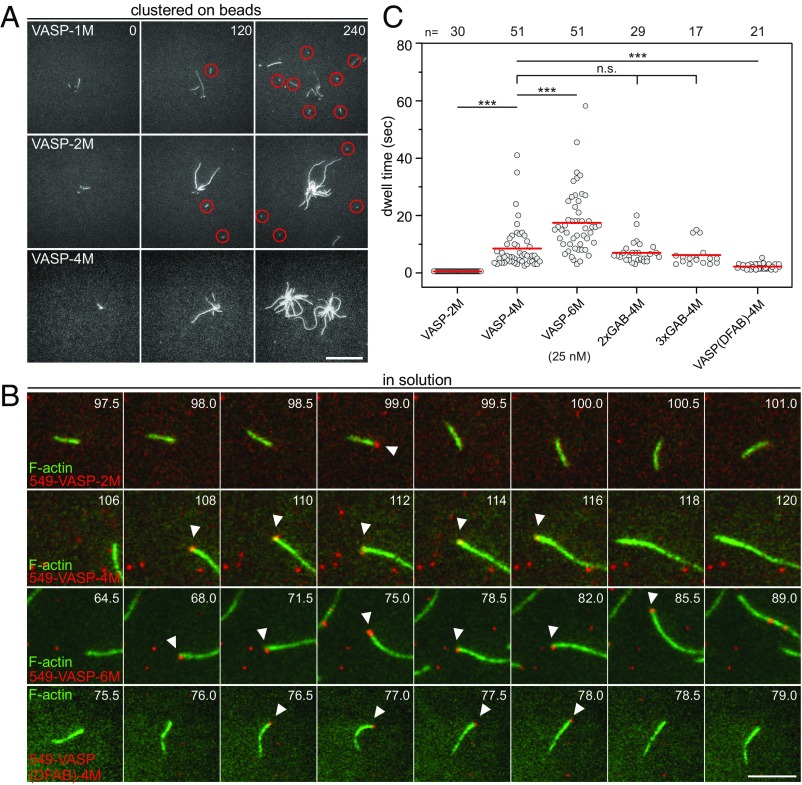
Finally, we asked why VASP is a tetramer, given the fact that even monomeric VASP-1M is capable of reaching maximum elongation rates when clustered on beads. Of note, with VASP-2M, and even more evident with VASP-1M–coated beads, we observed many short actin filaments in the vicinity of the beads (Fig. 6A). Because the growth of spontaneously nucleated actin filaments in solution is virtually completely suppressed by the presence of 40 nM CP in these experiments, the small filaments must have been initially generated on the surface of VASP-coated beads, and were subsequently released into solution. As the short filaments were not observed with VASP-4M (Fig. 6A), these findings further suggested that an increased oligomerization correlates with a more stable attachment of growing actin filament-barbed ends to the bead surface. Because it is technically not possible to monitor barbed-end growth and attachment of individual filaments on the bead surface, we explored the association of VASP oligomerization mutants with filament-barbed ends during processive filament elongation. For this reason, we labeled purified SNAP-tagged VASP constructs with SNAP-Surface 549 dye and measured their processivity in solution using multicolor TIRFM (Fig. 6B and Movie S3). Tetrameric VASP-4M remained associated with the tip of a growing actin filament for about 8.5 ± 7.7 s (mean ± SD), whereas the dwell time of hexameric VASP-6M more than doubled to 17.7 ± 10.7 s (Fig. 6C). By contrast, dimeric VASP-2M reproducibly appeared at the filament tip for only one frame in the time-lapse movies recorded at 2 Hz, meaning that the dwell time was less than 0.5 s. Moreover, the dwell time was not affected by additional GAB motifs as in 2×GAB VASP-4M and 3×GAB VASP-4M, showing that these constructs were virtually equally processive as VASP-4M. Finally, we tested processivity of a tetrameric VASP construct encompassing the lower-affinity FAB (Kd = 1.0 µM) from Dictyostelium instead of the higher affinity human FAB (Kd = 0.1 µM) (9). In excellent agreement with previous calculations (9), VASP-(DFAB)-4M remained in average only 1.6 ± 0.8 s at growing filament tips (Fig. 6 B and C). Thus, the run length of VASP and the stable association of growing actin filament-barbed ends to a VASP-coated surface are mainly dependent on the oligomerization state and the affinity of the filament side binding FAB domain.
Fig. 6.
Dependence of VASP oligomerization and multiple GABs on processivity. (A) Assembly of 1 µM actin (23% Alexa 488 labeled) in 1× TIRF buffer in the presence of 40 nM CP on 2 µm Ø benzylguanine beads derivatized with the SNAP-tagged VASP constructs indicated. Red circles indicate regions with short capped filaments that detached from the beads. Time is given in seconds. (Scale bar, 20 µm.) (B) The actin (1 μM, 23% Alexa 488 labeled) was polymerized in 1× TIRF buffer in the presence of 25 nM SNAP-Surface 549-labeled VASP constructs indicated. White arrowheads indicate single VASP molecules (red) surfing on growing filament barbed ends (green). Note different timescales. Time is given in seconds. (Scale bar, 5 µm.) (C) Dot plot summarizing barbed-end dwell times of the tested VASP constructs. Red lines indicate mean values. n, sample size.
Discussion
Our kinetic model implies that, at high actin-to-VASP ratios, the rate of VASP-mediated filament elongation is proportional to the product of the transfer rate kt and the number of free GABs (N) (9). Because N was obtained by fitting, we experimentally tested the model by variation of VASP oligomerization through deletion or substitution of the natural tetramerization motif with alternative oligomerization motifs leading to VASP-1M, -2M, -3M, and -6M to obtain VASP variants with lower and higher oligomerization states compared with the wild type. In excellent agreement with model predictions, in TIRFM experiments, we determined a linear correlation of VASP-mediated filament elongation rates with the number of N in solution (Figs. 1E and 7A). Conspicuously, insertion of two or even three GABs, interspaced with 44-aa flexible linker sequences, into the same polypeptide also markedly increased the elongation rates compared with the oligomers containing a single GAB, albeit the rates were slightly lower compared with the VASP constructs harboring the GABs on distinct polypeptide chains. For instance, although 100 nM VASP-6M promoted actin filament growth with 77.9 ± 2.9 subs per second (mean ± SD) (Fig. S2), the pseudo-VASP hexamers 2×GAB VASP-3M and 3×GAB VASP-2M promoted growth at the same concentrations with 69.4 ± 3.4 and 73.6 ± 2.5 subs per second, respectively (Fig. 2D). Thus, two or even three adjacent GABs loaded with actin monomers ahead of the FAB appear similar effective in filament end-tracking and delivering actin subunits to growing barbed ends compared with the natural GAB-FAB architecture in the VASP-6M hexamer, although the sequence of events during monomer delivery by the 2×GAB and 3×GAB constructs is currently unclear. By contrast, the elongation rates promoted by the pseudo-VASP octamer (2×GAB VASP-4M) with 86.1 ± 3.0 subs per second and in particular of the pseudo-VASP dodecamer (3×GAB VASP-4M) with 112.7 ± 6.4 subs per second markedly dropped below the theoretical values of 104.4 and 161.1 subs per second, respectively. Because the saturation of the three GABs in the pseudo-hexamer and pseudo-VASP dodecamer with actin monomers are expected to be identical, we assume that, at high N values, the diminished increase in elongation rates promoted by the pseudo-octamer and dodecamers are most likely attributable to another, and as yet concealed, rate-limiting step in the reaction. Formally, this effect could be caused by either the release of the end-tracking GAB-FAB clamp from the trailing end or by steric hindrance due to competition of too many G-actin–loaded GABs for access to the leading barbed end during monomer delivery. However, given the decreased dwell times with VASP-(DFAB)-4M harboring the lower-affinity Dictyostelium FAB, we clearly favor the first possibility. Consistently, this construct also promotes faster filament elongation compared with VASP-4M (9).
Fig. 7.
Context-dependent mechanisms of VASP-mediated filament elongation. (A) The rates of VASP-mediated actin filament elongation in solution are directly proportional to the number of free GABs (N). (B) The rates of VASP-mediated actin filament elongation after clustering on beads are fixed, irrespective of the oligomerization state or the number of additional GABs, and correspond to the rate of the canonical VASP tetramer using three free GABs (N = 3) as predicted by the kinetic model (9). Because after dense clustering, even monomeric VASP-1M can drive processive filament elongation at the same rate, the three free GABs must be provided by distinct VASP molecules. The constructs harboring additional GABs within the same polypeptide chain were omitted for reasons of clarity. Clustering of constructs is not shown for reasons of clarity. (C) Scheme for the proposed cellular arrangement of VASP and accessory proteins in the paracrystalline lattice beneath the plasma membrane. Top view is shown. Upon macroscopic clustering, mediated by multivalent interaction of the two C-terminal SH3 domains of IRSp53 and the four PRD regions within the VASP tetramer, the spacing of neighboring VASP molecules becomes close enough to reach a sufficiently high density of VASP at the surface to trigger long-lasting processive filament elongation in the presence of CP using three free GABs from neighboring VASP tetramers.
Previously, we have shown that individual filaments elongated from VASP-coated beads at roughly the same rates as with VASP in solution, suggesting that the number of participating GABs is a fixed value in both conditions (9). A hitherto-unresolved question was related to the finding that monomeric Dictyostelium VASP [N = 0 (6)] clustered on beads displayed a similar elongation rate as the tetrameric wild-type protein. Although this behavior may be formally explained by mimicking tetramerization by dense clustering, it did not exclude the possibility that N no longer correlates to the filament elongation rate when VASP is clustered on a surface. Thus, we repeated the bead assays with all VASP oligomerization mutants. However, again, the measured elongation rates of processive actin-filament elongation for all oligomerization constructs including the fast hexameric VASP-6M and the pseudo-oligomerization mutants were virtually identical with about 46 subs per second (Figs. 3B and 7B). Because the growth rate remained constant and was not affected by the number of the GABs, either by oligomerization or the quantity of GABs per monomer, it is therefore evident that presumably due to geometrical constrains the elongation rate on a surface is limited to a defined number of contributing GABs (N = 3). This scenario is supported by sustained processive growth on beads even in the presence of high concentrations of CP (6), showing that the filament barbed-end region is permanently in close contact with bead-derivatized VASP, presumably restricting the number of GABs, which have access to the growing barbed end. In contrast, in solution, the barbed ends of growing filaments are freely accessible for VASP oligomers resulting in filament elongation rates proportional to N, if N does not exceed too high values.
In the physiological context, Ena/VASP proteins operate mainly, if not exclusively, on a surface such as in the tips of protruding filopodia or lamellipodia (28). Based on our data, we therefore propose the “paracrystalline lattice model” of VASP-mediated actin assembly in clustered surface arrays. According to this model, each functional unit in the lattice is composed of four adjacent VASP tetramers that synergize in the elongation of a central actin filament (Fig. 6C). At any time, one end-tracking module from one VASP tetramer is tethered to the filament barbed end region while the GAB domains of three other VASP tetramers are available for monomer delivery. However, the free energy of ATP hydrolyses appears not to be required for release of the end-tracking clamp, thereby acting as a passive actoclampin motor (23, 31), although this finding does not rule out a potential role of ATP hydrolysis in facilitating VASP-mediated filament assembly from PFN-actin at concentrations found in vivo. The units in the lattice are symmetric and overlapping, so that each VASP tetramer can tether or deliver actin monomers to four neighboring actin filaments. This concept is not only compatible with the actin filament geometry in cross-sections of filopodia (32), but also with previous work on Drosophila Ena, which accelerates filament elongation about threefold by processively tracking the barbed end (20). A single Ena tetramer can also simultaneously track two adjacent barbed ends in a Fascin–cross-linked bundle, but interestingly, the elongation rate of both filaments then drops by one-half, demonstrating that individual Ena/VASP tetramers can deliver actin monomers to neighboring actin filaments. Moreover, as opposed to formins, which have to compensate the torsional twist of growing filaments by rotation within the membrane (33), due to the distinct mechanism, Ena/VASP proteins are not expected to build up any torsional forces and therefore constitute an ideal machinery to elongate tightly cross-linked actin architectures such as filopodia.
Long-lasting processivity of Ena/VASP proteins in the presence of physiological concentrations of CP requires dense clustering of the neighboring VASP tetramers in the range of 10 nm (6). Notably, clustering of receptors or of other membrane proteins is a widespread concept in biology and has been shown to be driven by phase transitions through interactions of multivalent proteins (34). Although lamellipodin and the inverse Bar-domain (IBAR)-protein insulin receptor substrate of 53 kDa (IRSp53) have been both implicated in clustering of VASP (16, 35), the oligomerization state of lamellipodin is not yet clearly established. By contrast, IRSp53 is a Cdc42-regulated antiparallel dimer that contains C-terminal SH3 domains that interact with proline-rich regions of Ena/VASP proteins (36, 37). Moreover, IRSp53 has been shown to cluster VASP by dynamic light scattering (16). Consistently, VASP constructs lacking the PRD fail to form IRSp53-induced clusters (16). Thus, currently, IRSp53 appears the best candidate to drive clustering of VASP through multivalent SH3–PRD interactions downstream of Cdc42 signaling in cell protrusions (16). The utilization of a tunable adapter molecule such as IRSp53 provides several advantages. After its activation by Cdc42, the antiparallel IRSp53 dimers will impose a defined spacer geometry to the lattice that is stable and close enough to trigger processive filament elongation in cross-bridged VASP arrays. Moreover, after inactivation of Cdc42, IRSp53 is expected to return to its autoinhibited state, leading to termination of filament growth after dissociation of the macroscopic VASP clusters, a behavior commonly observed in retracting lamellipodia (28).
Finally, by analyzing processivity of VASP oligomerization mutants, we can provide a rational explanation for the necessity of the natural Ena/VASP tetramerization. We found that the run length of VASP in solution was dependent on the oligomerization state and hence the number as well as the affinity of the FABs. Whereas dimeric VASP-2M remained associated for less than 0.5 s, tetrameric VASP-4M remained associated to growing barbed ends for about 8.5 s, a value similar to the 9 s reported for the single Drosophila Ena tetramer (20). Of note, we previously calculated the dwell time for Dictyostelium DdVASP to about 1–2 s (9), a value experimentally confirmed in this study. However, the dissociation constant of the Dictyostelium FAB (∼1.0 µM) is about an order of magnitude lower compared with the human FAB (0.1 µM) (9). Given the fact that DdVASP accelerates filament elongation sevenfold compared with only onefold to twofold of human VASP (6), it is therefore likely that the tighter interaction of human VASP with growing barbed ends results in increased processivity at the cost of a reduced elongation rate at the rather low actin concentration used in the TIRF assay. Thus, there may be an optimum affinity for processivity, but in all cases the affinity must be low enough to allow unbinding and advancement on the growing tip, yet high enough to facilitate tip binding and processivity. For long-lasting filament elongation, allowing incorporation of thousands of monomers in presence of CP, VASP must reliably shield growing filament barbed ends by close-fitting contact to prevent dissociation of filaments from the elongation machinery. As opposed to tetrameric VASP-4M and hexameric VASP-6M, the nonprocessive or poorly processive VASP constructs VASP-1M and VASP-2M were much less efficient in tightly tethering growing filaments on beads in the presence of CP. We therefore assume that a certain number of FABs is required for sustainable growth. Although the hexameric VASP could potentially provide even better attachment to the filaments, we show that, when clustered on beads, the number of contributing arms is limited to four (one tethering and three delivering monomers). Thus, the tetramer was presumably selected as an ideal actin polymerase during evolutionary development.
Materials and Methods
Constructs.
The construct for the expression of hVASP GST-tagged full-length human VASP (residues 1–380) containing the high-affinity GAB from D. discoideum (hVASP-DdGAB) has been described (9). It was used here as the VASP wild-type reference (VASP-4M) because it facilitates analyses of TIRFM experiments as shown previously (9, 16). All other VASP constructs were derived from this plasmid. VASP-1M (amino acids 1–333) was amplified as a BamHI–SalI fragment using VASP-4M as a template and inserted in the same sites of pGEX-6P1-SNAP, which allows the purification of GST-SNAP-tag fusions and subsequent labeling of the SNAP-tag with fluorophores as well as oriented immobilization of the proteins on benzyl-guanine–derivatized beads (16). For the other VASP oligomerization mutants, first a 5′ 945-bp BamHI–SalI fragment was amplified from VASP-4M cDNA using the forward primer 5′-GCGGGATCCATGAGCGAGACGGTCATCTGTTCCA-3′ and the reverse primer 5′-GCGGTCGACTTACCATGGTCTCCGCACAGATTCACTCTGGGC-3′. The reverse primer was additionally used to introduce a unique NcoI site as a silent mutation in the codon encoding proline 309 by changing the coding sequence from CCC to CCA. The fragment was inserted into pGEX-6P1-SNAP, yielding pGEX-SNAP-VASP-N. The remaining 3′ sequences containing the wild-type tetramerization domain of VASP or of variants substituting the wild-type tetramerization domain of VASP with GCN4 dimerization and trimerization coiled-coil motifs (25, 38) or of a synthetic hexamerization coiled-coil domain (26) in the backbone of the VASP 3′ end were synthesized from commercial sources (GenArt) and inserted in the NcoI and Sal sites of plasmid pGEX-SNAP-VASP-N yielding pGEX-SNAP-VASP-2M, -3M, -4M, and -6M (Table S1 for more details). Similarly, constructs containing one or two additional GABs interspaced with flexible 44-residue linkers (Table S1 for details) were synthesized as PstI–NcoI fragments and inserted into the same sites of the respective oligomerization mutants yielding pGEX-SNAP-VASP 2×GAB-4M and pGEX-SNAP-VASP 3×GAB-4M as well as pGEX-SNAP-VASP 2×GAB-3M and pGEX-SNAP-VASP 3×GAB-2M. Human VASP containing the Dictyostelium GAB and FAB was excised from pGEX-6P1 hVASP DGAB/FAB motifs (9) and inserted into the same sites of pGEX-SNAP to yield VASP-(DFAB)-4M. For experiments in solution and AUC experiments, VASP-1M, -2M, -3M, -4M, and -6M constructs were excised with BamHI–SalI from the pGEX-SNAP plasmids and inserted into pMAL-c2X (New England Biolabs), resulting in the addition of an N-terminal MBP-tag. For expression of SNAP-tagged murine mDia1-C containing the last three polyproline regions followed by the complete C-terminal sequences (amino acids 701–1,225), the corresponding coding sequence was amplified from mDia1 cDNA using the primers 5′-CGCGTCGACTTGCCTGGAGGACCAGGACTG-3′ and 5′-CTGGTTGGCCGTGCAAGCTAAGCGGCCGCGCG-3′ and inserted into the SalI/NotI sites of pGEX-SNAP. Human PFN2a cDNA was synthesized by commercial sources (Genart) and inserted as an NcoI–XhoI fragment into expression plasmid pET15b (Novagen). All constructs were verified by sequencing.
Protein Purification.
VASP constructs were expressed as GST-SNAP– or MBP-tagged fusion proteins in Escherichia coli host Rosetta 2 (Novagen). Expression was induced with 0.75 mM isopropyl-β-d-thiogalactopyranoside at 21 °C for 12 h. The bacteria were harvested and lysed by freeze–thaw and ultrasonication in lysis buffer containing 30 mM Hepes, pH 7.4, 150 mM KCl, 2 mM EDTA, 1 mM DTT, 5% (vol/vol) glycerol, and 2 units/mL Benzonase. The proteins were subsequently purified from bacterial extracts by affinity chromatography using glutathione-conjugated agarose 4B (Macherey-Nagel) or amylose high-flow resin (New England Biolabs) using standard procedures. The GST-tag was cleaved off by PreScission protease (GE Healthcare), and the GST tag absorbed on fresh glutathione-conjugated agarose and the proteins in the flow through separated by size-exclusion chromatography (SEC) with a HiLoad 26/600 Superdex 200 or Superose 6 controlled by an Äkta Purifier System. The expression and purification of both profilins by poly-l-proline affinity chromatography was performed as described for PFN1 (6). The purification of human heterodimeric CP CAPZ has been described (6). The purified proteins were stored at −20 °C in storage buffer [30 mM Hepes, pH 7.4, 150 mM KCl, 1 mM DTT, and 60% (vol/vol) glycerol] for later measurements. Ca2+-ATP actin was purified from rabbit skeletal muscle as described (39), stored in G buffer (5 mM Tris⋅HCl, pH 8.0, and 0.2 mM CaCl2, 0.5 mM DTT, 0.2 mM ATP), and labeled on Cys374 with Alexa 488 maleimide (Life Technologies) for TIRFM assays. ADP-actin was freshly prepared by polymerizing 23% Alexa 488-labeled G-ATP-actin in 1× KMEI buffer containing 10 mM imidazole, pH 7.0, 1 mM MgCl2, 1 mM EGTA, and 50 mM KCl in the presence of 15 units/mL hexokinase and 5 mM glucose. The obtained F-ADP-actin was sedimented at 100,000 × g for 2 h at 4 °C. The pellet was resuspended in 5 mM Tris⋅HCl, pH 7.8, containing 1 mM DTT, 100 µM CaCl2, 100 µM ADP, 15 units/mL hexokinase, and 5 mM glucose, and incubated on ice for 1 h and subsequently applied to SEC in the same buffer using a HiLoad 26/600 Superdex 75 column using an Äkta Purifier System yielding ADP-G-actin.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were carried out in a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge at 35,000 rpm in 12-mm double sector centerpieces filled with 400-µL sample using an An-50 Ti rotor (Beckmann Coulter). Concentration profiles were measured using the UV absorption scanning optics and data acquisition software ProteomeLab XL-I GUI 6.0 (firmware 5.7). MBP-tagged VASP-1M, VASP-2M, VASP-3M, and VASP-4M were examined at a concentration of 0.6 µM at 10 °C and 230 nm in 10 mM Hepes, pH 7.5, 50 mM KCl, 0.1 mM DTT, and 2 µg/mL PMSF. MBP-tagged VASP-6M was examined at 4.3 µM, 4 °C, and 280 nm in 30 mM Hepes, pH 7.4, 0.15 M KCl, 2 mM EDTA, 0.5 mM DTT, 4 µg/mL PMSF, and 5% (vol/vol) glycerol. Data were analyzed using a model for diffusion-deconvoluted differential sedimentation coefficient distributions implemented in SEDFIT (40). Extinction coefficients at 280 nm, partial specific volumes, molar masses, densities, and viscosities were calculated from amino acid or buffer compositions, respectively, with the program SEDNTERP (41) and were used to correct experimental s values to s20,w. Concentrations are given in monomers throughout the text.
TIRFM.
For the in vitro TIRFM solution experiments, the MBP-tagged VASP constructs were prediluted to the indicated concentrations in 1× ATP-TIRF buffer [20 mM imidazole, pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 20 mM β-mercaptoethanol, 0.5 mM ATP, 15 mM glucose, 2.5 mg⋅mL−1 methylcellulose (4,000 cP), 20 µg⋅mL−1 catalase, and 100 µg⋅mL−1 glucose oxidase] or 1× ADP-TIRF buffer (20 mM imidazole, pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 20 mM β-mercaptoethanol). For the TIRFM bead assays, O6-benzylguanine-derivatized beads with a diameter of 2 µm were used (New England Biolabs). Eighty microliters of the bead solution was used for a single coating reaction as described previously (42), and after equilibration in coating buffer (20 mM Hepes, pH 7.4, 150 mM KCl, 1 mM EDTA, 1 mM DTT, and 1% BSA), 120 µL of purified SNAP-tagged VASP constructs diluted to 1 mg⋅mL−1 each were added, and the mix was incubated overnight on a rotating wheel at 4 °C. Unbound SNAP-tagged VASP was removed by three washing steps with coating buffer. The immobilized SNAP-VASP constructs were stored on ice and analyzed within 2 d. Two microliters of VASP-coated beads were used for each TIRFM experiment. For two-color TIRF imaging, SNAP-tagged VASP constructs were labeled with SNAP-Surface 549 (BG-Dy 549) following the protocol of the manufacturer (New England Biolabs), and unbound dye was removed by 2-mL Zeba desalting spin columns (Thermo Scientific). The TIRFM assays were initiated by addition of ATP-G-actin (1 µM final concentration, 23% Alexa 488 labeled) or ADP-G-actin (4 µM final concentration, 23% Alexa 488 labeled) and by flushing of the mixtures into mPEG-silane (Mr 2,000) (Laysan Bio)-precoated flow chambers. Images were captured with a Nikon Eclipse TI-E inverted microscope equipped with a TIRF Apo 100× objective at 0.5-s intervals with exposure times of 40 ms by Ixon3 897 EMCCD cameras (Andor) for at least 10 min. The pixel size corresponded to 0.159 µm. The elongation rates of filaments were determined by manual tracking of growing barbed ends using ImageJ software. At least 30 filaments were measured from three independent movies per condition for the experiments in presence of the VASP constructs and 10 filaments from two independent movies in the actin control experiments.
Statistical Analysis.
Statistical analysis was performed using SigmaPlot 11.2 software (Systat Software). Statistical significance of differences between nonnormally distributed populations was determined by the Mann–Whitney U test. When data fulfilled the criteria of normality (Shapiro–Wilk test) and equal variance (Levene’s test), statistical differences were analyzed with a two-tailed, unpaired Student t test. Statistical differences are reported as ***P < 0.001 and n.s. as not significant.
Supplementary Material
Acknowledgments
We thank Annette Breskott, Lidia Litz, and Kristina Borst for technical assistance and all members of the group for fruitful discussions. This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants FA 330/9-1 and FA 330/10-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.B.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703145114/-/DCSupplemental.
References
- 1.Bugyi B, Carlier M-F. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 4.Krause M, Gautreau A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 5.Le Clainche C, Carlier M-F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 6.Breitsprecher D, et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesarone MA, Goode BL. Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol. 2010;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20:217–225. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitsprecher D, et al. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 2011;30:456–467. doi: 10.1038/emboj.2010.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 11.Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell Biol. 2003;13:386–392. doi: 10.1016/s0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 12.Sechi AS, Wehland J. ENA/VASP proteins: Multifunctional regulators of actin cytoskeleton dynamics. Front Biosci. 2004;9:1294–1310. doi: 10.2741/1324. [DOI] [PubMed] [Google Scholar]
- 13.Ball LJ, et al. Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J. 2000;19:4903–4914. doi: 10.1093/emboj/19.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-l-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- 15.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Disanza A, et al. CDC42 switches IRSp53 from inhibition of actin growth to elongation by clustering of VASP. EMBO J. 2013;32:2735–2750. doi: 10.1038/emboj.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- 18.Kühnel K, et al. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci USA. 2004;101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci USA. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasic L, Kotova T, Schafer DA. Ena/VASP proteins capture actin filament barbed ends. J Biol Chem. 2008;283:9814–9819. doi: 10.1074/jbc.M710475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickinson RB, Purich DL. Clamped-filament elongation model for actin-based motors. Biophys J. 2002;82:605–617. doi: 10.1016/S0006-3495(02)75425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickinson RB, Caro L, Purich DL. Force generation by cytoskeletal filament end-tracking proteins. Biophys J. 2004;87:2838–2854. doi: 10.1529/biophysj.104.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purich DL. Actoclampin (+)-end-tracking motors: How the pursuit of profilin’s role(s) in actin-based motility twice led to the discovery of how cells crawl. Biophys Chem. 2016;209:41–55. doi: 10.1016/j.bpc.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 26.Zaccai NR, et al. A de novo peptide hexamer with a mutable channel. Nat Chem Biol. 2012;7:935–941. doi: 10.1038/nchembio.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 28.Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- 29.Romero S, et al. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 31.Dickinson RB. Models for actin polymerization motors. J Math Biol. 2009;58:81–103. doi: 10.1007/s00285-008-0200-4. [DOI] [PubMed] [Google Scholar]
- 32.Aramaki S, Mayanagi K, Jin M, Aoyama K, Yasunaga T. Filopodia formation by crosslinking of F-actin with fascin in two different binding manners. Cytoskeleton (Hoboken) 2016;73:365–374. doi: 10.1002/cm.21309. [DOI] [PubMed] [Google Scholar]
- 33.Mizuno H, et al. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 34.Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife. 2014;3:e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen SD, Mullins RD. Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. Elife. 2015;4:e06585. doi: 10.7554/eLife.06585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krugmann S, et al. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 37.Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: Crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 38.O’Shea EK, Rutkowski R, Stafford WF, 3rd, Kim PS. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- 39.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 40.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 41.Laue MT, Shah BD, Rigdeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. Computer-aided interpretation of analytical sedimentation data for proteins; pp. 90–125. [Google Scholar]
- 42.Winterhoff M, Brühmann S, Franke C, Breitsprecher D, Faix J. Visualization of actin assembly and filament turnover by in vitro multicolor TIRF microscopy. Methods Mol Biol. 2016;1407:287–306. doi: 10.1007/978-1-4939-3480-5_21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.