Significance
Antibiotic resistance is fueled by antibiotic misuse and has become a major global health concern. The phenomenon warrants improved diagnostics that can more rapidly and efficiently elucidate information about the infectious agent to aid in establishing a more targeted and knowledge-based treatment regimen. This paper introduces a rapid antibiotic susceptibility test and automated data analysis algorithm that can, unlike traditional methods, deliver results on the same working day in an efficient and translatable manner for clinical use. This paper also introduces a method for direct urine testing that can help save days of diagnosis time. The platform is expected to promote more judicious use of antibiotics, thereby reducing the emergence of antibiotic resistance, lowering healthcare costs and ultimately saving lives.
Keywords: antibiotic resistance, nanoliter wells, antibiotic susceptibility testing, microfluidics, resazurin
Abstract
Antibiotic resistance is a major global health concern that requires action across all sectors of society. In particular, to allow conservative and effective use of antibiotics clinical settings require better diagnostic tools that provide rapid determination of antimicrobial susceptibility. We present a method for rapid and scalable antimicrobial susceptibility testing using stationary nanoliter droplet arrays that is capable of delivering results in approximately half the time of conventional methods, allowing its results to be used the same working day. In addition, we present an algorithm for automated data analysis and a multiplexing system promoting practicality and translatability for clinical settings. We test the efficacy of our approach on numerous clinical isolates and demonstrate a 2-d reduction in diagnostic time when testing bacteria isolated directly from urine samples.
Antibiotic/antimicrobial resistance (AMR) is among the leading global health concerns to date (1–3). Due to an overuse of antibiotics in medicine (1, 2) and agriculture (4, 5), antibiotic resistance mechanisms commonly emerge and threaten modern medicine by diminishing the utility of clinically relevant antibiotics. In 2014, infections with AMR were estimated to take the lives of over 700,000 people every year, and that number is expected to rise to 10 million people by the year 2050 (6). In addition, added and prolonged hospitalization due to AMR increases the cost of healthcare and imposes a large economic burden, one that is estimated to cost the United States $35 billion a year (2) and is expected to cost the world $100 trillion by the year 2050 (6). Antibiotic stewardship is an important approach used to conserve the utility of antibiotics and, in clinical settings, calls for the concomitant development of rapid diagnostics that can aid in providing improved antibiotic regimens (7–9). Not only can rapid diagnostics help guide proper antibiotic use but they also can be a critical determinant of patient outcomes. For patients with septic shock, for example, it is estimated that for every hour that effective antibiotic treatment is delayed survival rates drop by ∼7.6% (10).
Current methods for antimicrobial susceptibility testing (AST), a routine clinical test used to probe for resistant or nonresistant (susceptible) phenotypes of pathogens, typically require between 2 d to 1 wk from the time of taking the sample. Sample processing alone requires 24–48 h of incubation, and in the case of bacteremia and sepsis a blood culture incubation step is needed, which has a standard culture time of up to 5 d (11, 12). AST testing itself then takes an additional 8–24 h. During this time, to prevent the worsening of the condition of the patient, the clinician will often prescribe an antibiotic with a broad spectrum of activity in large doses to ensure its efficacy on the target pathogen. This approach facilitates the emergence of AMR in the clinic as well as damage the human microbiota (2, 6, 9).
Classical clinical AST methods rely on measuring growth inhibition of bacterial films or “lawns” on solid agar supplemented with growth medium in response to antibiotics diffusing from paper disks (Kirby–Bauer disk diffusion) or diffusing antibiotic gradients from strips (e.g., E-test). Although these methods are robust, they require at least 1 d of incubation and significantly lengthen the time to results. Liquid suspension-based methods such as the broth microdilution method are considered to be the gold standard for AST and are highly quantitative. In some cases, they can yield results sooner than solid-phase tests due to the increased growth rate capabilities of liquid-based media; however, these tests tend to be much more laborious. For this reason, automated liquid-phase testing (e.g., VITEK 2 by BioMerieux and Phoenix by Becton Dickinson) has become the popular choice in centralized laboratories today and uses algorithms that analyze the changing optical density of the samples due to bacterial growth. As a result, these methods offer improved speeds over solid-phase tests and can deliver results in 6–24 h (13). However, despite these advantages, current test speeds often leave results unused until the next working day. In addition, the test itself requires a pure bacterial culture that is derived from an overnight, culture-dependent plating step (solid-phase incubation). Current test speeds can be attributed to the sensitivity of optical density-based measurements, requiring massive proliferation to induce a detectable change. Genotypic AST, which has recently gained momentum, aims to deliver extremely rapid AST results (1–3 h) by probing for the presence of specific genetic sequences that are known to cause phenotypic resistance (14). The main drawback with this approach is that only known sequences associated with resistance can be targeted. Not only are there many more sequences that have yet to be elucidated (15), but new forms of resistance are out of reach.
Previous research efforts have resulted in a number of approaches to provide a faster phenotypic AST. More specifically, it has been shown that AST can be performed within a few hours by using single-cell optical imaging to monitor proliferation at different antibiotic conditions. Single-cell imaging has been coupled with various bacterial immobilization techniques such as agarose gel immobilization (16), dielectrophoresis-facilitated trapping (17), confined microchannels with electrokinetic loading (18), and valve-actuated filling of nanoliter wells (19). However, these methods require high-resolution optics and time-lapse investigation of multiple locations, making them expensive and complicated to multiplex and parallelize for high-volume clinical use. Other contributions for reducing AST time bypass the need to image individual cells while remaining sensitive by implementing other unique detection schemes such as sensitive changes in mass measured with a microchannel-embedded cantilever (20, 21), asynchronous magnetic bead rotation (22), or by speeding up cell growth with higher oxygen delivery using high-surface-to-volume-ratio microchannels (23). However, these schemes often involve complicated readouts and have not been demonstrated in a format that promotes automated data collection or multiplexing for different antibiotics or antibiotic concentrations. Methods that implement gradients of antibiotics (24, 25) vastly improve on multiplexing by simultaneously testing a range of antibiotic concentrations but require constant flow of antibiotics to hold the gradient profile, thereby limiting the number of simultaneous tests that can be performed.
Resazurin is a fluorescent dye that is minimally toxic and commonly used for cell viability assays. Cellular reduction potential causes an irreversible reaction of resazurin to resorufin, a reduced molecule exhibiting strong fluorescence characteristics unlike its unreduced counterpart. In a growth culture, this reaction occurs at a rate proportional to that of the aerobic respiration of cells in the medium (26). Due to the high sensitivity of fluorescent detection systems, resazurin has been used to monitor the viability of individual bacteria without the need for imaging individual cells, thereby bypassing the requirement for high-resolution optics and opening the doors to high-throughput scanning and parallelization. This sensitivity can aid in discerning between more minute changes in bacterial concentration or metabolic states of the culture compared with traditional optical density-based readouts. Boedicker et al. (27) used resazurin to achieve AST by single-cell viability tracking of stochastically confined bacteria in droplets. However, complex device setup, droplet handling, and data collection make it impractical for the large number of droplets needed for high-volume susceptibility profiling. Churski et al. (28) developed a novel microfluidic resazurin-based device, capable of performing AST within 3 h, that improves on sample preparation and data collection by implementing automated and preprogrammed droplet formation and sequential detection using an on-chip fiber optic. However, as in the work of Boedicker et al. (27), droplets are mobile, making their manipulation, handling, and tracking relatively complex. Storing of the droplets is nevertheless stationary but their indexing is encoded spatially within detachable tubing, making storage and data collection less practical for large sample numbers faced in hospitals. Weibull et al. (29) introduced a glass slide with a layer of silicon containing hundreds of nanoliter wells for rapid AST that offers simple loading, multiplexing, and imaging. Growth is observed using optical density, but an improvement in speed is attributed to the implementation of an algorithm that calculates the duration of the lag phase of bacterial growth. However, in this system the cultures within the wells are not completely isolated and can share molecules via diffusion, thereby reducing the independence of the cultures and limiting the multiplexing ability. Finally, previous research efforts mostly test only single or a few control strains of bacteria, making it difficult to estimate the characteristic time to results of the system when used with different bacterial types, which more resembles clinical-use cases.
Herein, we present a system that combines the resazurin assay with a nanoliter well array containing lyophilized antibiotics within each well, forming a system with multiplexing potential that can be operated in a practical manner. Because well volumes are small and chemically isolated, the fluorescence buildup (i.e., metabolism) of a small number of bacterial cells can be detected for different antibiotic conditions simultaneously. The system provides rapid, same-day AST results requiring two orders of magnitude fewer reagents than current clinically available liquid-phase testing systems. Loading the sample can be easily achieved by hand, with a single-step injection using a conventional laboratory pipette, increasing its range of applications, such as for resource poor settings, by reducing its dependency on large and expensive accompanying machinery. In this paper, we demonstrate the efficacy of our assay by testing clinical isolates and directly comparing our results to those obtained in the clinic using conventional automated liquid testing. We also demonstrate the multiplexing potential of the system by introducing antibiotic lyophilization and array parallelization. We develop an algorithm for automatable analysis of results and production of susceptible/resistant (S/R) determinations. Finally, we investigate the potential for the stationary nanoliter droplet array (SNDA)–AST system to reduce sample preparation time for urinary tract infections (UTIs) by performing AST directly on bacteria harvested from clinical urine samples. By bypassing the solid-phase incubation step, up to 2 d of clinical diagnostic time can be saved using this system.
Description of the SNDA–AST System
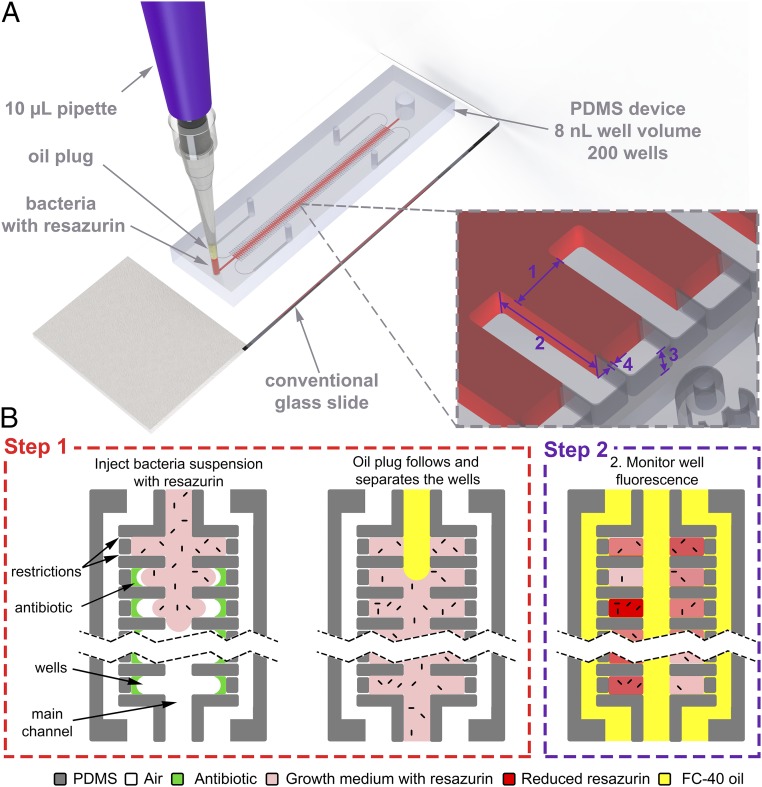
The method uses the SNDA (30) as a base platform for simple well loading/stationary droplet formation. The microfluidic device can interface with nearly any substrate, such as a conventional microscope glass slide, and can be loaded with a conventional 10-µL pipette. Each array consists of 200 wells of 8 nL, branching off a main channel (Fig. 1A). Well dimensions are 200 µm × 400 µm × 100 µm (width × length × height) and the main channel is 300 µm wide. Restrictions are 2–5 µm wide and serve as unique structures that are specifically designed to allow air in the wells to escape to surrounding channels, thus facilitating simple capillary-based filling. The system uses the standard AST cell concentration as well as growth medium (Mueller Hinton II, or MHII) to improve clinical translatability and interpretability using known standard breakpoints produced by organizations such as the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical & Laboratory Standards Institute. The well volume was set specifically so that the standard AST cell concentration () would produce an average of 4 cfu per well. This number was chosen specifically to balance three design considerations: (i) to use the minimal amount of bacteria possible to aid in the compatibility of future novel sample preparation schemes, (ii) to have large enough wells so that the standard AST testing concentration would result in a sufficient amount of bacteria per well to produce a predictable distribution of bacterial loading as governed by the Poisson distribution, and (iii) to maximize sample dispersion. Dispersion of the sample into hundreds of smaller nanoliter volumes serves to capture more cell–cell heterogeneity while lowering sample volumes. Because each well holds only 8 nL, each testing treatment requires <2 µL. In addition, sample dispersion increases the surface area-to-volume ratio of the cultures by an order of magnitude, thereby promoting increased gas transport and growth rates and faster AST times. Finally, sample dispersion can theoretically lower the rate of false-positive results by quarantining unrelated pathogens or contaminants to only a few number of wells.
Fig. 1.
Device design and principle of operation. (A) Illustration of SNDA–AST device on a conventional microscope slide being loaded with a conventional 10-µL pipette. The SNDA device consists of two rows of 8-nL wells connected by a main delivery channel. Each well contains ∼3-µm restrictions that allow the air to escape to two surrounding channels, thus facilitating simple capillary based filling. (Inset) 1, 200 µm; 2, 400 µm; 3, 100 µm; and 4, 2–5 µm. (B) Step 1: The SNDA-based AST device is loaded with a single-step injection of a two-plug formulation of the bacterial suspension with 10% resazurin followed by a plug of FC-40 oil. The purpose of the oil is to discretize the sample by isolating the wells while delivering oxygen and preventing evaporation. Step 2: The well fluorescence, indicating the level of metabolic activity occurring in the culture, is measured every 30 min and is proportional to the amount of bacteria/metabolism in the well. Bacteria are not drawn to scale.
Principle of Operation
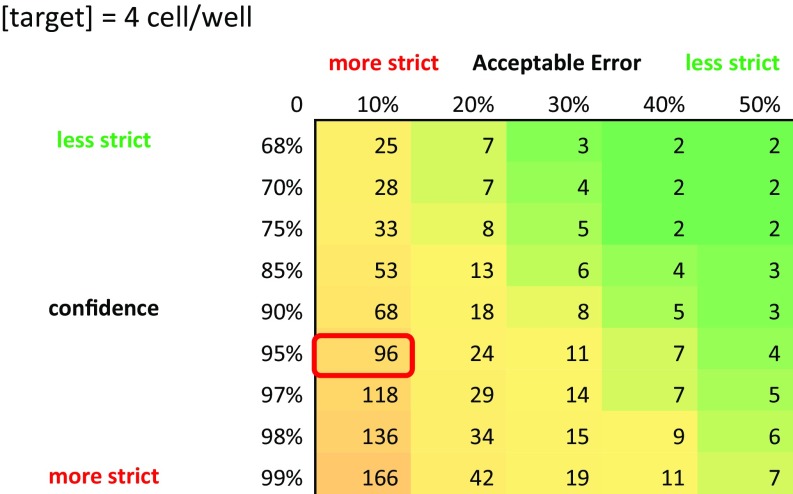
The sample is loaded with a single-step injection of a two-plug solution using a conventional laboratory micropipette (Fig. 1A). The first, or bottommost, plug is a ∼1.6 µL bacterial suspension of supplemented with 10% resazurin. The second, or topmost, plug contains ∼3 µL of FC-40 oil. The two-plug solution is achieved simply by aspirating the respective liquids sequentially. Low-pressure loading is enabled by restrictions in the well, allowing the volume of air in the wells to escape and be replaced by that of the liquid, making loading by hand possible using this system (Movie S1). After the first plug containing the bacteria flows in and fills the cavities of the reservoirs the oil plug follows and fills the main channel, thereby separating the wells with an immiscible barrier, effectively discretizing the sample (Fig. 1B, step 1). We specifically use fluorinated oil because it allows delivery of dissolved oxygen to the culture chambers while preventing evaporation of the well contents. In this work, an array is loaded for each testing treatment, and for each experiment a positive and negative control are included. We defined the positive control to be the bacterial testing suspension without the addition of any antibiotics and the negative control to be the bacterial testing suspension with the addition of sufficiently high amounts of antibiotics (one to two orders of magnitude higher in concentration to that of the testing concentration). We specifically chose this model for a negative control to account for any interactions between dead or nonproliferating bacteria in the system with resazurin that would not be present in a negative control composed simply of sterile growth medium with resazurin. For all experiments 100 wells were analyzed for each treatment (e.g., positive control, negative control, and ampicillin 8 mg/L) to account for nonuniform cell loading according to the Poisson distribution with λ = 4 cells per well, a 95% confidence level, and a 10% acceptable error (Fig. S1). Although only 100 wells are needed for analysis, in this work we used arrays containing 200 wells due to fabrication yield and loading aspects that seldom reduce the number of usable chambers. The fluorescence buildup within the wells indicates the level of metabolic activity occurring in the culture and is measured every 30 min for different antibiotic conditions to obtain results (Fig. 1B, step 2).
Fig. S1.
A table summarizing calculations made using the Poisson distribution for different levels of acceptable error and confidence and a mean of bacteria per well. For our system, we chose to settle at an acceptable error of 10% in the concentration and be 95% confident that we have that concentration. This leads us to the conclusion that we have to average the results of ∼100 wells per treatment to achieve the desired level of accuracy.
Time to Results and Data Analysis
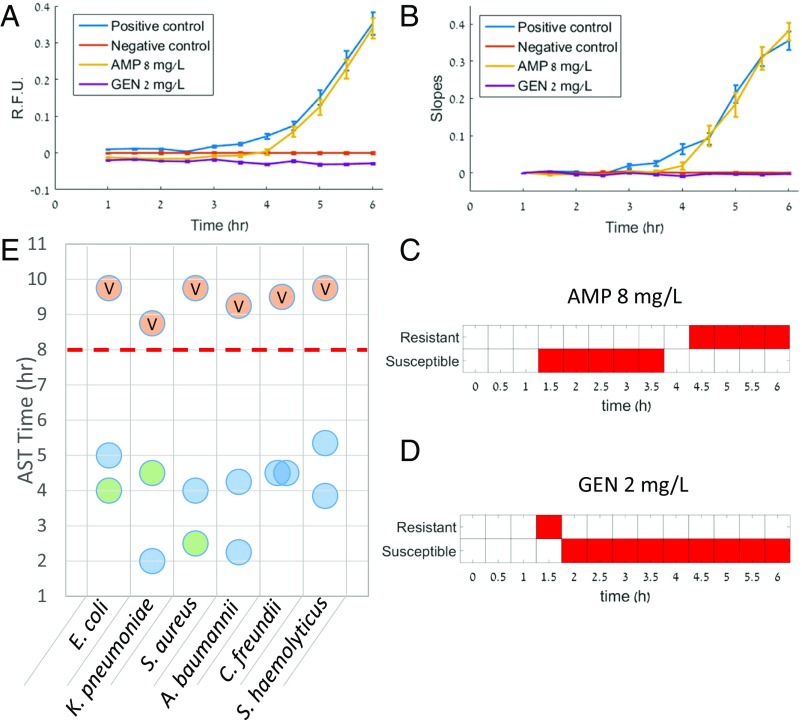
For automatable S/R determination we develop an algorithm that analyzes the kinetic growth curves of each well to make rapid determinations. After subtracting the data by the average rise profile of the negative control (Fig. 2A) we fit the rise profile of each well (100 wells per treatment) to both a linear and exponential model. Then, by comparing the goodness of the fit through the rms error, we keep only the best-fitting model for each specific well. The instantaneous slope of each fit is calculated analytically from the model equation and the slopes of the fits are averaged for each treatment at the last time point along the curve (Fig. 2B). This process is then repeated, and new fits are generated for each data point that is collected. For each experiment we used the difference in the average slopes of the positive and negative control to define a working range, Δ, for which we compare the slopes of the treatments for the S/R determinations. Treatments with slopes that were ≤30% of Δ were considered susceptible. Treatments with slopes that were ≥60% of Δ were considered resistant. These thresholds were optimized based on an existing dataset (Methods for more information). Slope comparisons between the treatments and the thresholds of the working range were performed with a one-tailed t test with significance level of thus allowing the method to use both the effect size (31) and P value for making determinations. In addition, to help avoid nonmeaningful determinations in the early stages of the growth curves, a logistical restriction was set so that determinations could only be made once there was a significant difference between the slopes of the positive and negative controls. We then visualize the results in the form of S/R determinations over time (Fig. 2 C and D) and consider the correct determination to be the one that the algorithm converges for sufficiently long times. The time to result is then defined as the time at which the algorithm made a decisive decision, that is, one that did not change for the remainder of the analysis period. Fig. 2 C and D and present the determination plots for treatments 1 and 2 from the dataset displayed in Fig. 2 A and B that produced resistant and susceptible determinations, respectively. We applied our determination assay and algorithm based on a “crude” minimum inhibitory concentration (MIC) of a single breakpoint concentration to a wide range of different microorganisms and antibiotic combinations and assessed the time to result. We then aggregated the result times from the different experiments to determine the characteristic time to results (Fig. 2E). Using this system, designations are evident in less than 5.5 h, with >80% of designations evident in 4.5 h or less, whereas the results on the same samples using the VITEK 2 took longer than the typical 8-h workday. We also found that our S/R determinations matched the determinations obtained in the clinic using the VITEK 2, as interpreted by EUCAST guidelines (32). Table 1 lists the 12 bacteria–antibiotic combinations that we tested and used for the time analysis. We designed our experiments in a manner that allowed us to test both gram-positive and -negative bacteria from different families as well as antibiotics from different mechanistic classes. In addition, we aimed to include both resistant and susceptible determinations in the analysis.
Fig. 2.
Time to S/R determination. (A) First, the fluorescent profile of each well is normalized to the average profile of the negative control by subtraction. Here, data for Klebsiella pneumoniae are used where treatments 1 and 2 were assigned resistant and susceptible determinations, respectively. Averages of each treatment are shown. Next, the normalized profile of each well is fit to both a linear or exponential model with an offset however only the ‟best” model is kept, using the root mean square error as a determiner. (B) Slope distributions are then obtained from the fits created for every time point. This is done iteratively after every data point collected using all data points collected up to that time point for the fits. Here the slopes of the fits created at the last time point are shown. (C and D) Finally, determinations are made for each treatment by categorizing the average slopes of the treatments using fixed fractional thresholds of that of the normalized positive control. We define the time to results as the time point at which the algorithm has presented a decisive determination (one that does not change for the remainder of the experiment). (E) A summary of AST times obtained for the clinical isolates studied in this work using the SNDA–AST system compared with the time to results obtained in the clinic for the same isolate sample using the VITEK 2 AST system. Blue circles represent susceptible determinations, green circles represent resistant determinations, and orange circles marked with a “V” present the results obtained by the VITEK 2 AST system in the clinic. A red dashed line illustrates the ending of a typical 8-h work day.
Table 1.
Experiments used to estimate the time to S/R determination of the SNDA–AST device
| Bacteria | Isolate | Source | SNDA–AST determination (time) | VITEK 2 (laboratory) determination (time) |
| E. coli | Clinical | Urine | AMP 8 mg/L – S (5.00 h) CIP 0.5 mg/L – R (4.00 h) | AMP – S CIP – R (9.75 h) |
| K. pneumoniae | Clinical | Urine | AMP 8 mg/L – R (4.50 h) GEN 2 mg/L – S (2.00 h) | AMP – R GEN – S (8.75 h) |
| Staphylococcus aureus | Clinical | BAL | PEN 0.125 mg/L – R (2.50 h) CIP 1 mg/L – S (4.00 h) | PEN – R CIP – S (9.75 h) |
| Staphylococcus haemolyticus | Clinical | Urine | CIP 1 mg/L – S(5.35 h) ERY 1 mg/L – S (3.85 h) | CIP – S ERY – S (9.75 h) |
| Acinetobacter baumannii | Clinical | Burn | CIP 1 mg/L – S (4.25 h) CST 2 mg/L – S (2.25 h) | CIP – SCST – S (9.25 h) |
| Citrobacter freundii | Clinical | Urine | CIP 1 mg/L – S (4.50 h) GEN 1 mg/L – S (4.50 h) | CIP – S GEN – S (9.50 h) |
For each experiment we list the type and source of the bacteria and provide a comparison between the results (determinations and time) obtained in the clinic with the VITEK2 and in our laboratory using the SNDA–AST system. In our experiments using the SNDA–AST system, breakpoint concentrations were used according to the EUCAST standards and S/R determinations were made accordingly. BAL, Bronchoalveolar lavage. Antibiotics: AMP, ampicillin; CIP, ciprofloxacin; CST, colistin; ERY, erythromycin; and GEN, gentamicin. Interpretation: R, resistant and S, sensitive.
Antibiotic Incorporation via lyophilization and Multiplexing
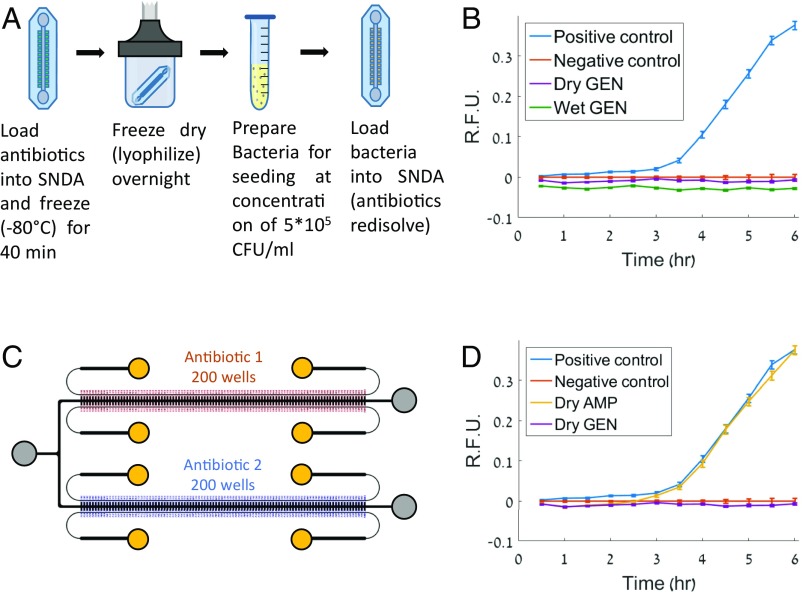
We developed a lyophilization scheme to incorporate freeze-dried antibiotics within the wells of the SNDA before sample loading to improve ease of use, practicality, and multiplexing. To lyophilize antibiotics within the SNDA, before the AST experiment, we loaded liquid antibiotics at the target concentration in the SNDA and separated the wells with air instead of oil. Next, we froze the arrays at −80 °C for 40 min and subsequently placed them into vacuum chambers for overnight lyophilization in a lyophilizer machine (Fig. 3A). After lyophilization, these devices had dried antibiotics within the wells and were then ready to be used for AST experiments and loaded with a bacterial suspension using the normal loading procedure mentioned previously. Using this method we observed similar kinetic growth profiles with freeze-dried antibiotics compared with those obtained using the equivalent “wet” antibiotics that we supplemented before loading (Fig. 3B).
Fig. 3.
Antibiotic incorporation via lyophilization and multiplexing. (A) Workflow illustration for antibiotic loading and lyophilization. (B) Comparison of the standard “wet” and dried gentamicin at the breakpoint concentration using E. coli with 8 mg/L ampicillin showing that the dried antibiotic has efficacy similar to that of the standard “wet” counterpart. (C) Schematic of parallel SNDA–AST device used for multiplexing. A bacterial sample can be loaded into the device and tested against two different types of antibiotics simultaneously. Inlet/outlet reservoirs are indicated by gray and yellow circles. Yellow circles specifically indicate oil reservoirs. (D) Demonstration of sensitivity testing of two different dried antibiotics simultaneously at breakpoint concentrations using the parallel SNDA–AST device, confirming that this isolate of E. coli is resistant to ampicillin and susceptible to gentamicin. Error bars in all graphs present 95% CIs on the mean.
To demonstrate the concept of multiplexing through parallelization we designed a device of two parallel and interconnected arrays for which two different antibiotics can be lyophilized within the wells of each array (Fig. 3C). Using this system, we could investigate the susceptibility of Escherichia coli to gentamicin and ampicillin simultaneously and confirm that our determinations matched the expected determinations (Fig. 3D) that were produced by the VITEK 2 for the same strain.
Same-Day Detection and AST for UTIs
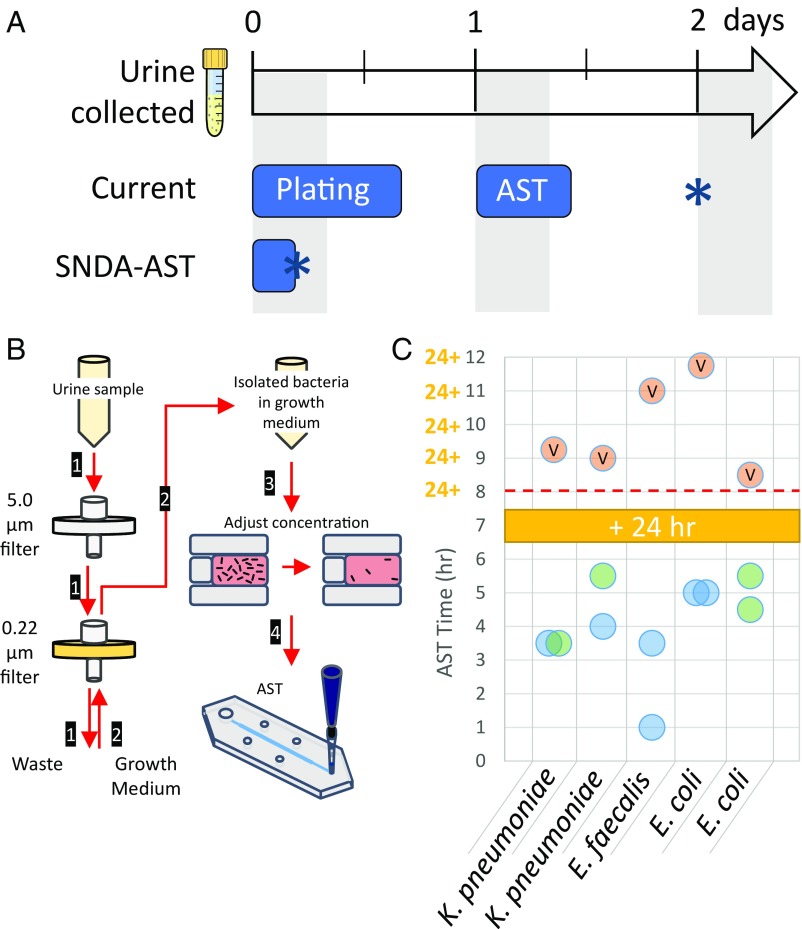
UTIs are among the most common type of bacterial infections dealt with in the clinic and pose a significant healthcare burden (33). After a urine sample is collected, current clinical protocols dictate the need to culture the sample on an agar-growth medium to first confirm the presence of bacteria in the sample, isolate the bacteria from the urine, and identify the purity of the culture (34, 35). This step is necessary because most urine cultures for suspected UTIs yield negative results (do not contain a significant concentration of bacteria). However, urine cultures require 18–24 h of incubation, and only after that can AST testing begin, thereby greatly lengthening the diagnostic time. Here, we investigated the ability to perform rapid AST directly on bacteria collected from fresh urine samples to skip the plating and incubation steps and provide same-day AST results. Because current plating and AST methods only yield results the next working day, we hypothesized that using the SNDA–AST system directly on bacteria harvested from urine samples can save ∼2 d of clinical diagnostic time (Fig. 4A).
Fig. 4.
Same-day AST for UTIs. (A) Timeline comparing the current clinical workflow (Top) to the SNDA–AST workflow (Bottom). In the current workflow, lengthy plating and AST steps cause the next step to be initiated only the next working day (shaded regions), causing AST results (asterisk) to be delivered in 2 d. With the SNDA–AST setup, the entire workflow can be performed and results (asterisk) can be delivered the same day. (B) Illustrated schematic of bacteria isolation protocol developed for clinical urine samples. (1) The urine sample is directed through a 5-µm filter to trap WBC and large debris followed by a 0.22-µm filter to trap bacteria. (2) Growth medium is directed through the 0.22-µm filter in the opposite direction to flush out trapped bacteria and the suspension is collected in a separate vial. (3) The bacterial concentration is adjusted via microscope observation. (4) The sample is then supplemented with 10% resazurin and loaded into the SNDA–AST device for analysis. (C) A summary of AST times obtained for the clinical urine samples studied in this work using the SNDA–AST system compared with the time to results obtained in the clinic for the same isolate sample using the VITEK 2 AST system. Blue circles represent susceptible determinations, green circles represent resistant determinations, and orange circles marked with a “V” present the results obtained by the VITEK 2 AST system in the hospital. A red dashed line indicates the assumed 8-h work day, showing that, unlike results from the VITEK 2, results from the SNDA–AST system can be used the same day.
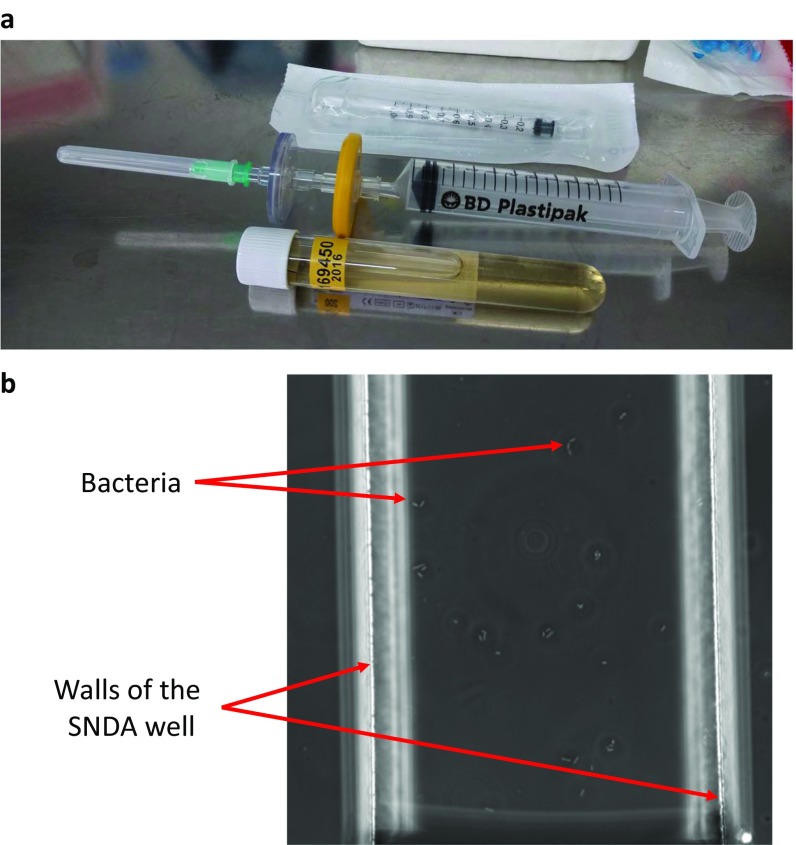
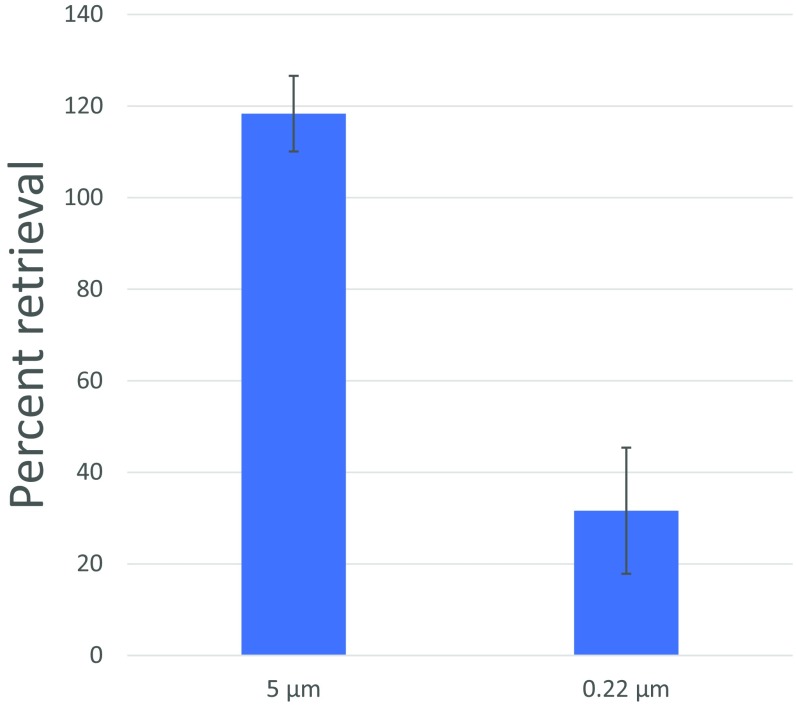
Performing AST directly on urine samples requires that the urine be used as a growth medium. This can alter growth kinetics and thus affect determinations and their compatibility with breakpoint standards for interpretation. In addition, extraneous components in the urine sample could cause inaccurate testing. Instead, we opted to use a series of carefully designed filtering steps to isolate the bacteria directly from clinical urine samples into our growth medium of choice. First, the sample is filtered sequentially through a series of a 5-µm filter followed by a 0.22-µm filter (Fig. 4B and Fig. S2A). The 5-µm filter traps WBC and large debris that may be present in the sample and the 0.22-µm filter traps the bacteria. Second, the standard AST growth medium is passed through the opposite direction of the 0.22-µm filter only, consequently flushing out the bacteria that were trapped during the first filtering step. The bacteria that are removed during this process are kept for use in AST. The ratio of volumes of the urine sample to the volume of the growth medium used is roughly 5:1, which consequently concentrates the bacteria five times in the growth medium compared with the load in the original urine sample. Because positive urine samples of patients with UTI generally contain more than 105 cfu/mL bacteria, this ensures that there will be a high-enough concentration to perform AST accurately when taking into account the estimated efficiency of the filtering scheme (Fig. S3). Also, the SNDA–AST system requires only a few tens to hundreds of microliters of a solution and urine samples are generally collected in volume of ∼9 mL, and thus sufficient sample volume is most likely not a concern with this system. Next, because AST requires a specific bacterial concentration, the sample is adjusted to the correct concentration by manually probing the average number of bacteria per well (Fig. S2B) in the SNDA and diluting accordingly. Finally, AST is performed using the SNDA–AST using the same methodology mentioned earlier. If developed, perhaps an image analysis program can perform counting before AST automatically and also give insight into the gram classification of the bacteria with some level of confidence.
Fig. S2.
(A) An image of the filtration setup used to isolate the bacteria from clinical urine samples where the sample is drawn through the needle and through the series of filters from left to right. (B) A microscope image of bacteria in a single well of the SNDA as seen during the counting step required for adjusting the cell concentration for subsequent AST.
Fig. S3.
Retrieval efficiency of the filtration process. Here we quantify the retrieval efficiency of the filtration process used to produce the data in Fig. 4 on clinical urine samples. A laboratory-grown culture of E. coli was used at an initial concentration of 5 × 105 cells per mL as determined by optical density and a standard curve. n = 4 repetitions for each measurement. Error bars represent 95% CIs on the mean. Results were quantified by plating the bacterial suspensions before and after the respective filtering steps using the Drigalski technique. Percent values are calculated with respect to the bacterial concentration before each specific step.
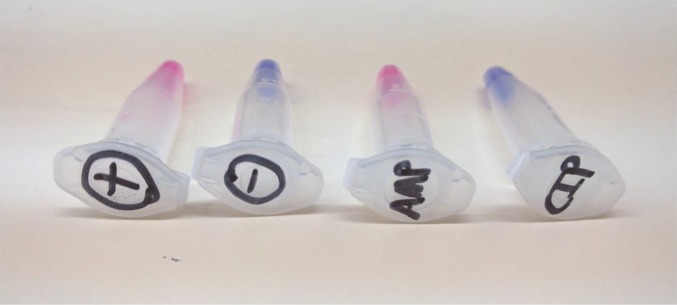
We next aggregated the time to results of 10 bacteria–antibiotic combinations from five clinical urine samples using this system (Fig. 4C). We observed times similar to those found in our time analysis with the clinical isolates shown in Fig. 2B. This allowed us to validate our hypothesis that this method is capable of shaving up to 2 d off the clinical diagnostic timeline for UTIs. Table 2 summarizes the experiments performed and results obtained on clinical urine samples for the time analysis of AST for UTIs. We found our determinations to match the determinations obtained using the VITEK 2 in the clinic, with the exception of two cases (single determinations in Exp. 2 and 4). For these discrepancies, the results obtained by a bulk control ran in parallel did match the VITEK results (Fig. S4), indicating that there was no problem regarding the accuracy of the resazurin assay itself, but rather that a delayed proliferation cycle was present. For these experiments, we hypothesize that this delayed growth cycle is most likely attributed to a pronounced lag phase associated with the bacteria coming directly from patient urine samples which, for the work described in this paper, were stored in a refrigerator overnight before use.
Table 2.
Experiments used to assess the efficacy and time to results of the SNDA–AST assay performed on bacteria extracted directly from clinical urine samples
| Exp. | Organism | SNDA–AST determination (time) | VITEK 2 (laboratory) determination (time) |
| 1 | K. pneumoniae | AMP 8 mg/L – R (3.5 h) CIP 0.5 mg/L – S (3.5 h) | AMP – R CIP – S (9.25 h) |
| 2 | K. pneumoniae | AMP 8 mg/L – R (5.5 h) CIP 1 mg/L – S (4.0 h) | AMP – R CIP – R (9.0 h) |
| 3 | Enterococcus faecalis | AMP 8 mg/L – S (1.0 h) CIP 4 mg/L – S (3.5 h) | S – AMP S – CIP (11.0 h) |
| 4 | E.coli | AMP 8 mg/L – S (5.0 h) CIP 0.5 mg/L – S (5.0 h) | R – AMP S – CIP (11.75 h) |
| 5 | E. coli | AMP 8 mg/L – R (4.5 h) CIP 0.5 mg/L – R (5.5 h) | R – AMP R – CIP (8.5 h) |
For each experiment we list the type of bacteria and provide a comparison between the results (determinations and time) obtained in the clinic with the VITEK2 on isolates after plating and in our laboratory using the SNDA–AST system on bacteria directly from the urine samples. Antibiotic concentrations were chosen to be the breakpoint according to the EUCAST standards and S/R determinations were made accordingly. Antibiotics: AMP, ampicillin and CIP, ciprofloxacin. Interpretation: R, resistant and S, sensitive.
Fig. S4.
Bulk colorimetric assay performed in parallel with the same sample used for AST with the SNDA–AST. This example is from the E. coli tested in Table 2 in Exp. 4. From left to right: Positive control, negative control, ampicillin 8 mg/L, and ciprofloxacin 0.5 mg/L. The result with ampicillin determined using this bulk colorimetric assay matches that of the VITEK 2 on the same isolate performed in the clinic, although does not match the determination produced by the SNDA–AST. We hypothesize that this is most likely due to a pronounced lag phase associated with the bacteria that were extracted directly from clinical urine samples that were refrigerated overnight before testing.
Discussion
A few hours could mean the difference between same-day and next-day results. Because the general working day is approximately 8 h long, AST systems that approach or exceed this amount of time to yield results are more likely to allow appropriate treatment to be administered only the next working day.
Our results from testing 12 bacteria–antibiotic combinations suggest that same-day AST results are possible using the SNDA–AST system. The use of resazurin as a reporter allows for higher sensitivity for detecting bacterial cell viability compared with that with optical density-based systems. Furthermore, because bacterial isolation via plating takes an additional day, simple sample preparation methods, such as the urine filtration method demonstrated here, can help reduce the diagnostic timeline by a number of days via directly transferring the bacteria from the sample to a usable and standard format for AST. Our results from testing 10 bacteria–antibiotic combinations from clinical urine samples showed AST times similar to those obtained from the standard isolate protocol with two exceptions that did not produce rapid determinations. Both exceptions were associated with a delayed growth cycle beyond the observation period (false negative determining S instead of R), and a closer look at the data revealed slight evidence of signal growth during the observation period that may be attributed to metabolizing bacteria in the lag phase that did not undergo significant proliferation (Fig. S5). These findings suggest that further improvements to the analysis algorithm (e.g., through the use the absolute fluorescence intensity values rather than only the slopes) may be of value. For positive/negative urine sample detection we believe that an automated system based on image analysis can be developed and may also be able to give insight into the gram classification of the organism. Regarding identification, a separate method may be used for. Although MS is commonly used for identifying isolate colonies it is not yet commonly performed on urine samples directly, although it is possible (35).
Fig. S5.
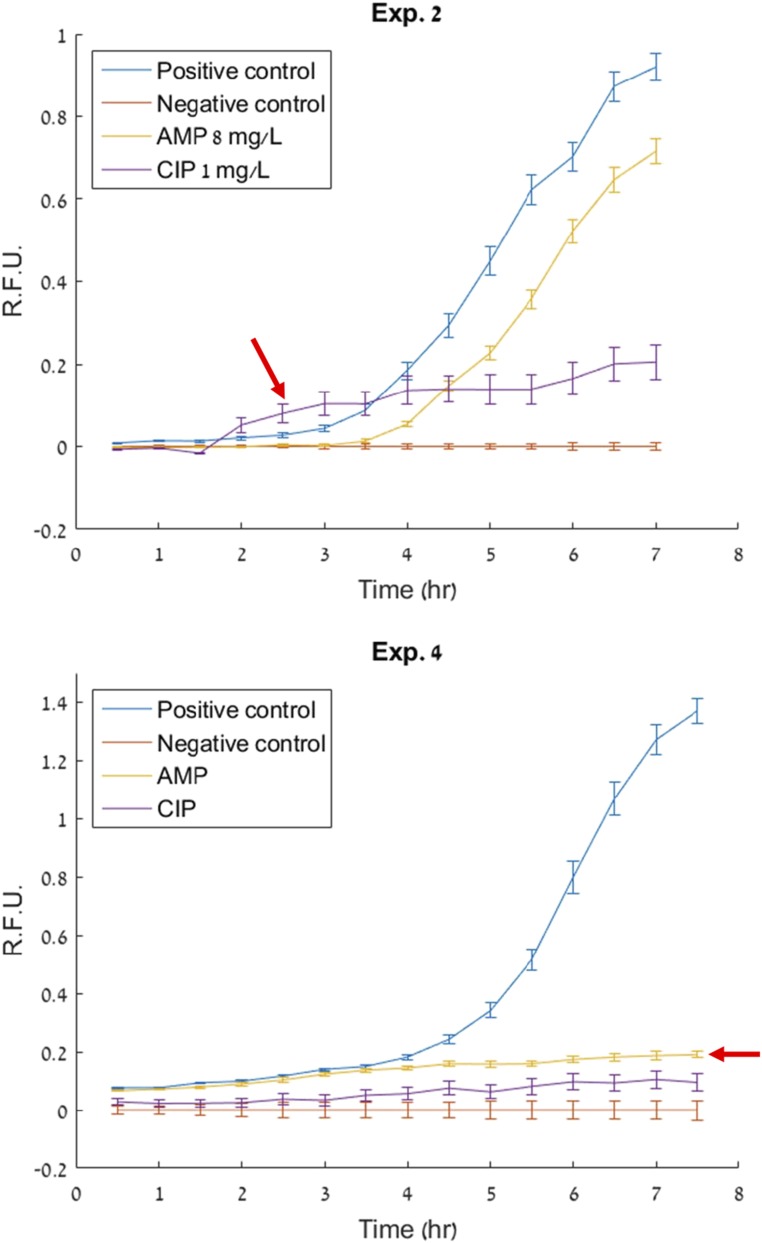
Data from two discrepancies in the urine analysis studies summarized in Table 2. Evidence of a slight signal rise compared with the negative control (red arrows) can be seen for the treatments that were incorrectly determined susceptible [ciprofloxacin (CIP) in Exp. 2 and ampicillin (AMP) in Exp. 4]. This rise may be due to metabolism from live bacteria that are not undergoing significant proliferation due to a lag phase. These data suggest that an improvement to the analysis algorithm that uses absolute fluorescence intensity rather than slope to make determinations can be of value. R.F.U., relative fluorescence units.
The use of resazurin not only allows for more sensitive bacterial cell viability detection than optical density-based systems but it also maintains the ability to quickly image well intensities (scan) and obtain results without high-magnification imaging of bacterial cells themselves. Overall, this allows for higher throughput, parallelization of sample processing, and decreased cost and complexity. In this study, a 4× objective was used for scanning 20 wells per image, allowing for four arrays of an assay to be scanned in ∼24 s. Scanning four antibiotic concentrations for a panel of 10 antibiotics (40 treatments) would then take ∼4 min. Because the time interval between scanning the same sample is 30 min, seven of these multiplexed tests could be scanned by the same instrument simultaneously. Regardless, signal intensity was plentiful in our setup. Using PrestoBlue, an N.A. of 0.1, and a CCD camera with 5 × 5 binning, we were able to image using <40-ms exposure times. For comparison, a standard 1× objective with an N.A. of 0.04 will produce ∼40% the signal intensity of the previous objective, making data acquisition still possible using slightly higher exposure times and can allow for scanning four times as many samples simultaneously for high-throughput applications. Also, using an LED-based illumination source can obviate the need for a mechanical shutter and furthermore decrease the scanning time by half. Finally, the number of wells analyzed per treatment drastically changes with the allowable acceptable error and confidence according to the Poisson distribution (Fig. S1) and thus can be an important parameter to optimize. For example, for the same confidence level, a 10% reduction in the allowable error can reduce the number of wells per treatment from ∼100 to ∼25. Consequently, this method can be expanded to testing a multitude of concentrations per antibiotic for a full panel of antibiotics. Our analysis method can then be adopted to solve for a more full MIC to produce more accurate S/R determinations.
The work presented here investigated six clinical isolates and five urine samples and in some cases, such as with enterococcus (which is notorious for having a relatively long doubling time), times to designations were interestingly short. This could possibly be due to the ability of resazurin to monitor cell viability through metabolism and independently of proliferation. Also, our sample preparation steps including filtering and concentration adjustment, which took 30–50 min, could have possibly reduced the lag time during the subsequent AST test for these bacteria associated with the refrigerated urine samples. Nonetheless, for AST isolated nanowells have the advantage of capturing bacterial heterogeneity more accurately and can possibly help reduce false positives/false negatives by confining or quarantining contaminations or unexplained phenomena to individual wells. Finally, the low reagent consumption of the system, <1 µL per test treatment, compared with hundreds of microliters to milliliters per test treatment for conventional methods, can drastically help lower costs and laboratory space requirements while potentially improving reliability due to a decreased quantity requirement of antibiotics, which may be produced from different batches where potency can vary. Perhaps most importantly, in consequence of using low reagent volume, the SNDA–AST system requires less than three orders of magnitude fewer bacteria, which opens the door to further shorten sample preparation, notably incubation, especially with samples containing a relatively low pathogen load. We note that the principle of direct bacterial isolation from patient samples and testing using the SNDA–AST system can possibly be applied to other sample types as well, although sample preparation techniques would likely differ for each sample type given the difference in pathogen load and biological background complexity.
Methods
Device Fabrication.
SNDA–AST devices were made from polydimethylsiloxane (PDMS) (Silgard 184; Dow Corning) using a silicon mold fabricated using deep reactive ion etching. For this, silicon-on-insulator wafers were used with a device layer of 100 µm and a BaO layer that served as an etch-stop to create flat channel profiles. PDMS was mixed with a 1:10 curing agent to polymer base ratio (by mass) and cured for 4 h in a 70 °C convection oven.
Device Loading/Operation.
The inlet of the SNDA–AST device was made using a 1-mm biopsy punch and a 10-µL pipette was used for the injection with conventional matching pipette tips. To create the two-plug solution, 3 µL of FC-40 oil was first aspirated with the pipette set to 3 µL. Next, the pipette tip containing the 3 µL of oil was dipped into the sample containing the bacteria and 1.6 µL was aspirated by turning the pipette set wheel from 3.0 to 4.6 µL. The SNDA–AST device was loaded by first loading the sample with a single step two-plug injection using a conventional laboratory pipette. Before incubation, the secondary channels were also filled with oil to help prevent evaporation. A video demonstration can be seen in Movie S1. Loading of the parallel SNDA–AST devices was achieved using the shared inlet and the same aforementioned loading protocol.
Antibiotic Susceptibility Experiments.
In all experiments, MHII (90922; Sigma-Aldrich), a standard AST growth medium, was used. Antibiotic solutions were prepared from powdered stocks and used either the same day or frozen in aliquots at −20 °C for no more than 30 d. Bacterial suspensions were prepared by inoculating a colony from a fresh streak plate into MHII and vortex mixing for 60 s. For all experiments, the standard AST bacterial concentration of ∼ was estimated using optical density measurements (NanoDrop ND-1000) that were calibrated previously with corresponding traditional plate counts.
In each experiment one array was loaded for each testing treatment. We defined the positive control to be the bacterial testing suspension without the addition of any antibiotics and the negative control to be the bacterial testing suspension with the addition of sufficiently high amounts of antibiotics (one to two orders of magnitude higher in concentration to that of the testing concentration). The antibiotic types used for the negative control were chosen based on their broad spectrum of activity. We specifically chose this model for a negative control to account for any interactions between dead or nonproliferating bacteria in the system with resazurin that would not be present in a negative control composed of simply sterile growth medium with resazurin.
For all experiments not involving lyophilized antibiotics (clinical isolates and urine samples) the bacterial suspension was supplemented with 10% resazurin solution (PrestoBlue; Molecular Probes) and supplemented with the appropriate amounts of antibiotics before loading into the SNDA–AST device. A one-step injection of a two-plug solution containing the bacterial suspension and FC-40 oil was used as mentioned previously. Each experiment used four SNDA–AST devices placed (positive control, negative control, and two treatments) in parallel on a one-well plate filled with ∼30% water to create a humid chamber during imaging.
Imaging was performed with an inverted epifluorescent microscope (Zeiss Axiovert 200M) equipped with an incubation chamber (Pecon xl-3 s1 054) set to 37 °C, a CCD camera (Zeiss Axiocam MRm), and a 120-W mercury arc lamp (Excite 120Q). All images were taken using a 4× (Zeiss Achroplan, N.A. 0.10) objective and at 40× total magnification, which allowed for monitoring the fluorescence of 20 wells per image (two rows of 10 wells). For each experiment, each position was imaged every 30 min for 7 h. Although the start time for each experiment was recorded as the moment the resazurin was added to the bacterial suspension and not when the imaging began. In addition, in each experiment, the bacterial suspension with resazurin was used to perform a “bulk” experiment similar to that of the broth microdilution method to verify the results of the algorithm with the colorimetric results of the bulk assay that were obtained the following morning. Resazurin and its reduced version, resorufin, are known to have colorimetric properties as well, which allows results to be interpreted in colorimetrically at the expense of sensitivity and assay speed. We found the results of the colorimetric bulk assay to match the results of the SNDA–AST determinations with two exceptions, one of which is presented in Fig. S4. Antibiotic concentrations were chosen according to version 6 of the EUCAST breakpoints (32).
Data Analysis and S/R Determination.
The fluorescence of the medium within the wells was calculated by averaging the pixel intensities in a given rectangular region which accounts for ∼50% of the well area in the image. A custom MATLAB script was used to analyze the images with high throughput as well as determine the S/R detection times. For all experiments, 100 wells were analyzed for each treatment (e.g., positive control, negative control, and ampicillin 8 mg/L) to account for nonuniform cell loading according to the Poisson distribution with λ = 4 cells per well, a 95% confidence level, and a 10% acceptable error (Fig. S1). The data shown are normalized by subtraction to the negative control to account for the natural reduction of Resazurin in this setup. Determinations were produced using the method mentioned previously in the results section. The S/R thresholds were optimized based on this dataset to produce the most rapid determinations that still matched those produced by the VITEK. Although we found these threshold to be rather insensitive to change, because a 10% change in the resistant threshold changed determination times by an average of ∼20 min.
Antibiotic Lyophilization Within the SNDA–AST Device.
A liquid suspension of antibiotics at the target concentration was prepared and loaded into the SNDA–AST device and sheared with air using the aforementioned single-step loading method of the two-plug solution consisting of a plug of bacterial suspension followed by a plug of air. The device was then incubated for 40 min at −80 °C and subsequently placed in a glass jar for overnight lyophilization (FreeZone; Labconco). Reconstitution with a bacterial suspension containing 10% resazurin was performed using the normal SNDA–AST loading protocol for each experiment.
AST on Urine Samples.
Anonymous discarded clinical urine samples were obtained from Rambam Hospital, Haifa, Israel with the consent of the Rambam IRB from patients that were confirmed to have UTIs by the hospital. Samples were refrigerated overnight until they were confirmed positive by the hospital and transferred to our laboratory for testing. Our AST assay was performed on these clinical samples using methods described above. Filtering of the urine samples (Fig. S2A) and replacing the urine with fresh medium was performed at a ratio of 5:1 urine:medium, which led to a 5× concentration of the bacteria in the urine sample to account for losses during the filtration scheme. Depending on the sample, clogging of the 0.22-µm filter sometimes occurred before being able to reach a concentration ratio of 5:1. It is unclear whether the bacteria themselves or the pyuria (pus) was responsible for clogging the filters. However, if clogging did occur, we simply filtered as much urine as permissible due to clogging (<5 mL) and found that there was still substantially enough bacteria for testing in these cases. Adjusting the cell concentration after bacterial isolation via filtering was achieved using a phase-contrast microscopy analysis of the bacterial sample in the SNDA. Because wells of the SNDA contain a known volume of liquid, the average number of bacteria per well were counted manually for ∼20 wells and the concentration of the sample was determined (Fig. S2B). After appropriate dilution, the sample was counted again in the same way to confirm the correct concentration before initiating the experiment. Total sample preparation time, including the time for filtering and concentration adjustment, took 30–50 min during our experiments in the laboratory.
Supplementary Material
Acknowledgments
We thank Dr. Ayala Cohen for her generosity and time involving statistical consultations, Dr. Marc Assous for fruitful conversations; Ariel Szklanny and Maria Salameh for their assistance; Galia Ben David for helping with regulations and ascertaining materials; the Micro and Nano Fabrication Unit at Technion for providing the capabilities and facilities needed for fabrication, and the Russell Berrie Nanotechnology Institute for their support. This work was supported by a Kamin Grant from the Israel Innovation Authority and the I-CORE Israel Centers of Research Excellence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703736114/-/DCSupplemental.
References
- 1.Gelbrand H, Miller-Petrie M, Pant S. 2015. The state of the world’s antibiotics 2015 (Center for Disease Dynamics, Economics & Policy, Washington, DC)
- 2.CDC, US Department of Health and Human Services 2013 Antibiotic resistance threats in the United States, 2013 (CDC, Atlanta). Available at www.cdc.gov/drugresistance/threat-report-2013.
- 3.WHO 2015. Worldwide country situation analysis: Response to antimicrobial resistance (WHO, Geneva)
- 4.Elliott K. 2015. Antibiotics on the farm: Agriculture’s role in drug resistance (Center for Global Development, London)
- 5.Carmen Cordova AK. 2015. FDA’s efforts fail to end misuse of livestock antibiotics (Natural Resources Defense Council, New York)
- 6.O’Neill J. 2014. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance (HM Government, London)
- 7.Dryden MS, Cooke J, Davey P. Antibiotic stewardship–More education and regulation not more availability? J Antimicrob Chemother. 2009;64:885–888. doi: 10.1093/jac/dkp305. [DOI] [PubMed] [Google Scholar]
- 8.Okeke IN, et al. Diagnostics as essential tools for containing antibacterial resistance. Drug Resist Updat. 2011;14:95–106. doi: 10.1016/j.drup.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Kerremans JJ, et al. Rapid identification and antimicrobial susceptibility testing reduce antibiotic use and accelerate pathogen-directed antibiotic use. J Antimicrob Chemother. 2008;61:428–435. doi: 10.1093/jac/dkm497. [DOI] [PubMed] [Google Scholar]
- 10.Kumar A, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 11.Kuper KM, Boles DM, Mohr JF, Wanger A. Antimicrobial susceptibility testing: A primer for clinicians. Pharmacotherapy. 2009;29:1326–1343. doi: 10.1592/phco.29.11.1326. [DOI] [PubMed] [Google Scholar]
- 12.Opota O, Croxatto A, Prod’hom G, Greub G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin Microbiol Infect. 2015;21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Machen A, Drake T, Wang YFW. Same day identification and full panel antimicrobial susceptibility testing of bacteria from positive blood culture bottles made possible by a combined lysis-filtration method with MALDI-TOF VITEK mass spectrometry and the VITEK2 system. PLoS One. 2014;9:e87870. doi: 10.1371/journal.pone.0087870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schofield CB. Updating antimicrobial susceptibility testing: Methods. Clin Lab Sci. 2012;25:233–239. [PubMed] [Google Scholar]
- 15.Nseir S, Povoa P. Multipathogen real-time PCR system adds benefit for my patients: No. Intensive Care Med. 2015;41:531–533. doi: 10.1007/s00134-014-3607-y. [DOI] [PubMed] [Google Scholar]
- 16.Choi J, et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci Transl Med. 2014;6:267ra174. doi: 10.1126/scitranslmed.3009650. [DOI] [PubMed] [Google Scholar]
- 17.Peitz I, van Leeuwen R. Single-cell bacteria growth monitoring by automated DEP-facilitated image analysis. Lab Chip. 2010;10:2944–2951. doi: 10.1039/c004691d. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, et al. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal Chem. 2013;85:3971–3976. doi: 10.1021/ac4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan R, et al. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens Bioelectron. 2013;49:118–125. doi: 10.1016/j.bios.2013.04.046. [DOI] [PubMed] [Google Scholar]
- 20.Etayash H, Khan MF, Kaur K, Thundat T. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat Commun. 2016;7:12947. doi: 10.1038/ncomms12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knudsen SM, von Muhlen MG, Schauer DB, Manalis SR. Determination of bacterial antibiotic resistance based on osmotic shock response. Anal Chem. 2009;81:7087–7090. doi: 10.1021/ac900968r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinn I, et al. Asynchronous magnetic bead rotation (AMBR) biosensor in microfluidic droplets for rapid bacterial growth and susceptibility measurements. Lab Chip. 2011;11:2604–2611. doi: 10.1039/c0lc00734j. [DOI] [PubMed] [Google Scholar]
- 23.Chen CH, et al. Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal Chem. 2010;82:1012–1019. doi: 10.1021/ac9022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, et al. Gradient microfluidics enables rapid bacterial growth inhibition testing. Anal Chem. 2014;86:3131–3137. doi: 10.1021/ac5001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou Z, et al. Time lapse investigation of antibiotic susceptibility using a microfluidic linear gradient 3D culture device. Lab Chip. 2014;14:3409–3418. doi: 10.1039/c4lc00451e. [DOI] [PubMed] [Google Scholar]
- 26.González-Pinzón R, Haggerty R, Myrold DD. Measuring aerobic respiration in stream ecosystems using the resazurin-resorufin system. J Geophys Res Biogeosci. 2012;117 doi: 10.1029/2012JG001965. [DOI] [Google Scholar]
- 27.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churski K, et al. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip. 2012;12:1629–1637. doi: 10.1039/c2lc21284f. [DOI] [PubMed] [Google Scholar]
- 29.Weibull E, et al. Bacterial nanoscale cultures for phenotypic multiplexed antibiotic susceptibility testing. J Clin Microbiol. 2014;52:3310–3317. doi: 10.1128/JCM.01161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shemesh J, et al. Stationary nanoliter droplet array with a substrate of choice for single adherent/nonadherent cell incubation and analysis. Proc Natl Acad Sci USA. 2014;111:11293–11298. doi: 10.1073/pnas.1404472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan GM, Feinn R. Using effect size-or why the P value is not enough. J Grad Med Educ. 2012;4:279–282. doi: 10.4300/JGME-D-12-00156.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.European Committee on Antimicrobial Susceptibility Testing 2016 Breakpoint tables for interpretation of MICs and zone diameters, Version 6.0 (European Committee on Antimicrobial Susceptibility Testing, Basel). Available at www.eucast.org.
- 33.Kumar S, Dave A, Wolf B, Lerma EV. Urinary tract infections. Dis Mon. 2015;61:45–59. doi: 10.1016/j.disamonth.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Leber AL, editor. Clinical Microbiology Procedures Handbook. 4th Ed ASM; Washington, DC: 2016. [Google Scholar]
- 35.Ferreira L, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:2110–2115. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.