Abstract
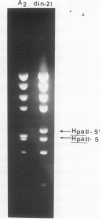
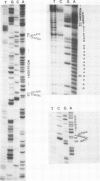
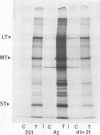
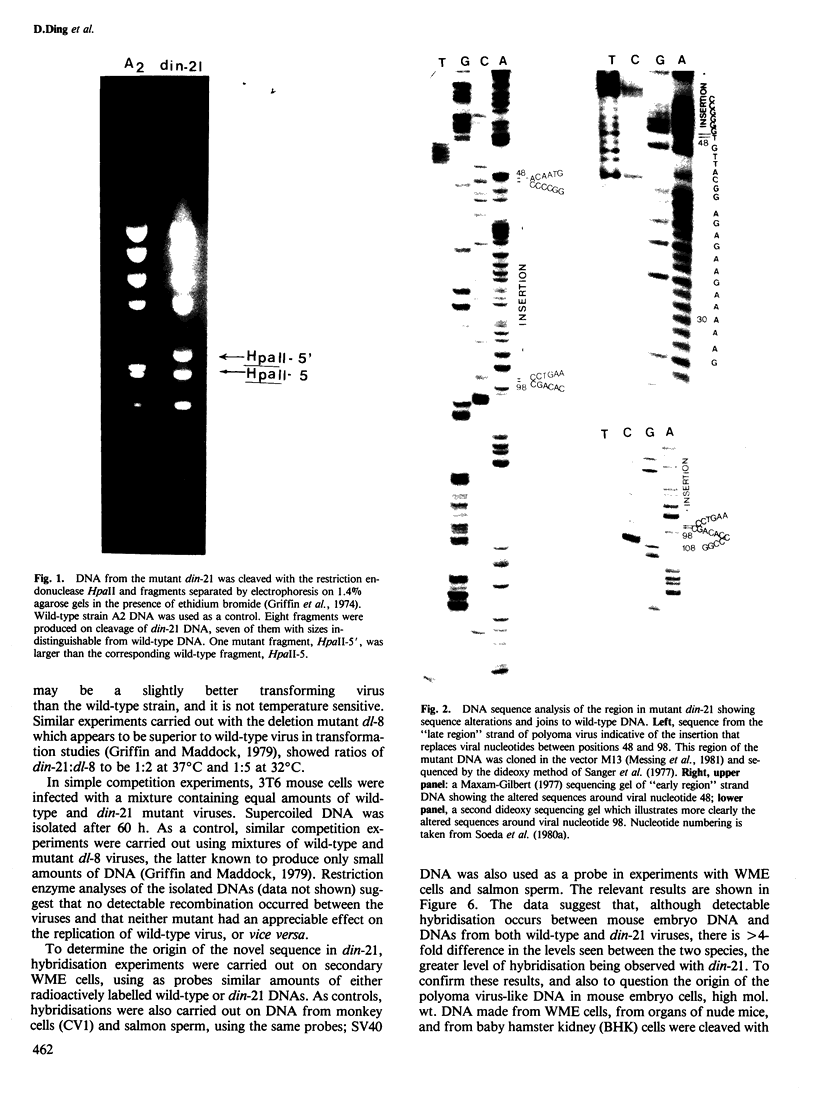
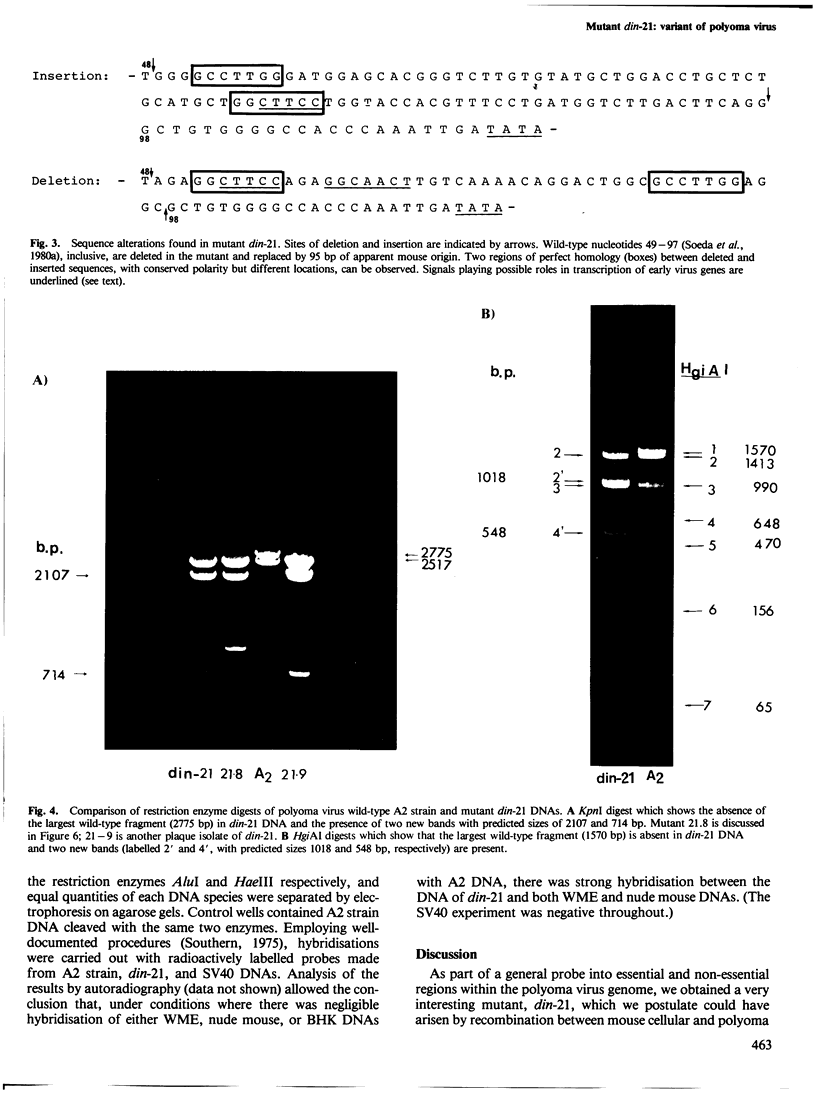
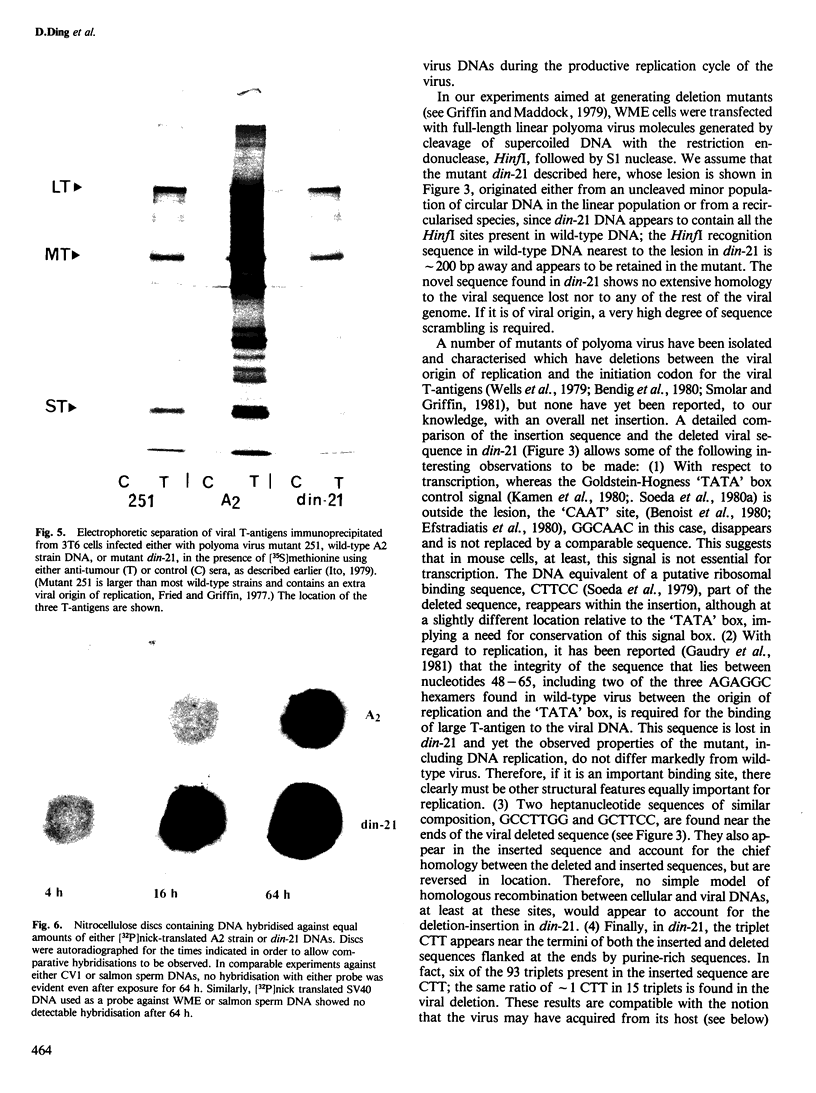
An unusual non-defective mutant of polyoma virus with an anomalously large genome, designated din-21, has been isolated. The viral chromosome lacks 49 base pairs of the putative control region between the origin of replication and the initiation codon for the early proteins, the T-antigens. In their stead , 95 base pairs, with limited homology to the deleted sequence and apparently of mouse origin, have been inserted. The primary sequence of the insert DNA has been determined and some of the biological properties of the mutant examined. It transforms rat-1 cells slightly better than wild-type virus and grows slightly less well in lytically infected mouse cells. It does not interfere with the growth of wild-type polyoma virus. The properties of this mutant suggest that it is a natural isolate of mouse cells. The mutant was presumably generated by reciprocal recombination between polyoma DNA and mouse host DNA. This could be associated with the integration of a viral DNA sequence into the host chromosome during the viral replicative cycle.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Thomas T., Folk W. R. Regulatory mutants of polyoma virus defective in DNA replication and the synthesis of early proteins. Cell. 1980 Jun;20(2):401–409. doi: 10.1016/0092-8674(80)90626-1. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Gaudray P., Tyndall C., Kamen R., Cuzin F. The high affinity binding site on polyoma virus DNA for the viral large-T protein. Nucleic Acids Res. 1981 Nov 11;9(21):5697–5710. doi: 10.1093/nar/9.21.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Björck E., Bjursell G., Lindahl T. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J Virol. 1981 Oct;40(1):11–19. doi: 10.1128/jvi.40.1.11-19.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E. Fine structure of polyoma virus DNA. J Mol Biol. 1977 Dec 5;117(2):447–471. doi: 10.1016/0022-2836(77)90137-1. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Katinka M., Yaniv M., Vasseur M., Blangy D. Expression of polyoma early functions in mouse embryonal carcinoma cells depends on sequence rearrangements in the beginning of the late region. Cell. 1980 Jun;20(2):393–399. doi: 10.1016/0092-8674(80)90625-x. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Nilsson M. G. Multiple free viral DNA copies in polyoma virus-transformed mouse cells surviving productive infection. J Virol. 1977 Jun;22(3):646–653. doi: 10.1128/jvi.22.3.646-653.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Berg P. Does simian virus 40 DNA integrate into cellular DNA during productive infection? J Virol. 1978 Nov;28(2):475–489. doi: 10.1128/jvi.28.2.475-489.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekikawa K., Levine A. J. Isolation and characterization of polyoma host range mutants that replicate in nullipotential embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1100–1104. doi: 10.1073/pnas.78.2.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolar N., Griffin B. E. DNA sequences of polyoma virus early deletion mutants. J Virol. 1981 Jun;38(3):958–967. doi: 10.1128/jvi.38.3.958-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Maruyama T., Arrand J. R., Griffin B. E. Host-dependent evolution of three papova viruses. Nature. 1980 May 15;285(5761):165–167. doi: 10.1038/285165a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Türler H. Interaction of polyoma and mouse DNAs. IV. Time course and extent of integration of polyoma DNA into mouse DNA during lytic infection. J Virol. 1977 Aug;23(2):272–285. doi: 10.1128/jvi.23.2.272-285.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells R. D., Hutchinson M. A., Eckhart W. Isolation and characterization of polyoma virus genomes with deletions between the origin of viral DNA replication and the site of initiation of translation in the early region. J Virol. 1979 Nov;32(2):517–522. doi: 10.1128/jvi.32.2.517-522.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]