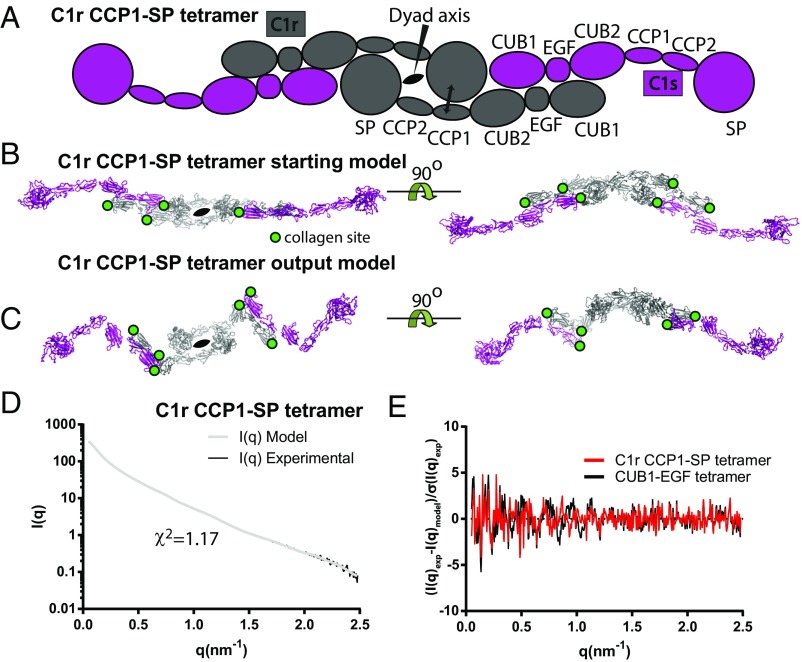
Arlaud et al. (1) raised concerns regarding our small-angle X-ray scattering (SAXS) and EM models of the unbound C1r2s2 tetramer (2). They reference studies supporting an interaction of two C1rs dimers via C1r CCP1–serine protease (SP) interactions. In response, we conducted refinements against our SAXS data by imposing distance restraints derived from C1r CCP1-CCP2-SP crystal structures (3, 4). The resulting models with central C1r dimers (Fig. 1) fit the SAXS data better (χ2 = 1.26 ± 0.07) than the models we described in ref. 2. The analysis that we presented in ref. 2 was therefore incomplete and should have considered C1r-mediated dimerization. We thank Arlaud et al. (1) for their remark. A representative output model is now deposited in the Small Angle Scattering Biological Data Bank, together with a model from our study (2).
Fig. 1.
SAXS rigid body modeling of free C1r2s2 tetramer assuming a central C1r dimer. (A) Schematic representation of the starting model for rigid body refinement with the imposed distance restraints indicated by a double-headed arrow. (B) Atomic start model. Visible collagen sites in C1r CUB1 and CUB2 and C1s CUB1 are marked with green circles. (C) Representative optimal output model after rigid body refinement with CORALXL. (D) Comparison of the scattering curve calculated from the model in C with the experimental curve. (E) Comparison of weighted residual plots for our original C1r2s2 output model (2) based on CUB1–EGF interactions and the C1r2s2 model based on C1r CCP1-SP contacts presented in C suggests similar resolution dependency for the fit between the experimental and model data for the two different output models.
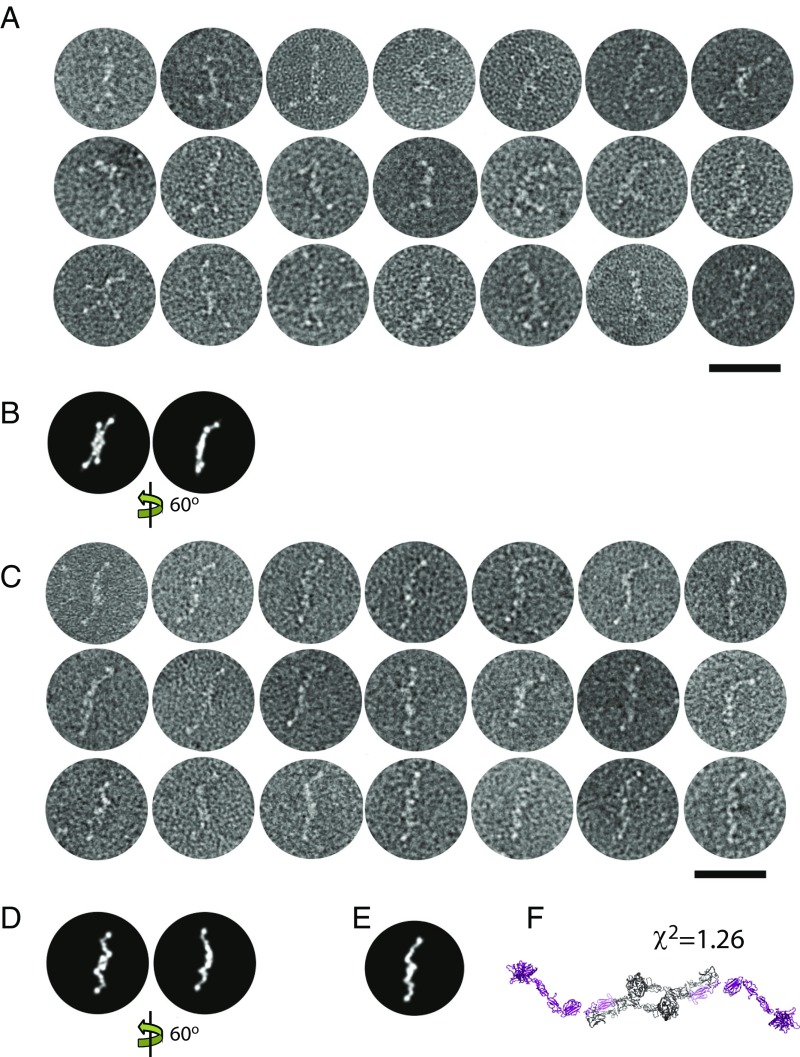
Negative stain (ns) images of free C1r2s2 (2) are not instrumental in excluding either of the models because many computed projections obtained by rotation of the SAXS models resemble each other at ns resolution levels (Fig. 2 B and D). Part of our ns images exhibit “split ends” (Fig. 2A), so this feature is not a result of averaging as proposed by Arlaud et al. (1). These images are not in agreement with the SAXS output model in Fig. 1. However, we also observed particles with simple ends, as in the study of Tschopp et al. (5) (compare Fig. 2C and averages 2 and 3 in figure 3 of ref. 2). Our SAXS models based on C1r CCP1-SP contacts are consistent with these images (Fig. 2 C–F), with best agreement observed for some nonrepresentative SAXS solutions with more extended C1s conformations (Fig. 2F). Further experiments are required to decide conclusively between the two possible structures of unbound C1r2s2.
Fig. 2.
Ns EM of selected particles of the free C1r2s2 tetramer. Raw images from the dataset in our study (2) are shown without averaging. (Scale bar: 50 nm.) (A) Particles with split ends. (B) Two computed projection images of the SAXS model our study (2). (C) Particles with simple ends. (D) Projection images of the SAXS model shown in Fig. 1. (E and F) Projection image of a SAXS model with more extended C1s domains compared with the SAXS model in Fig 1. Note that the SAXS models in both figures are symmetrized twofold.
However, although our analysis of unbound C1r2s2 in our study (2) is incomplete, this fact does not change our conclusions regarding the structure of nonactivated C1 and its consequential intermolecular activation mechanism. The CUB1-EGF tetramerization of the C1r2s2 inside the C1 complex assumed by us (2) is consistent with models developed by Brier et al. (6) and Venkatraman Girija et al. (7) featuring the CUB1-EGF-CUB2 domains of C1r and C1s located in one central plane, consistent with the fact that both C1r CUB domains, but only the C1s CUB1 domain, contain a binding site for C1q collagen stems (8, 9). The CCP1-CCP2-SP domains were suggested to be tucked inside the C1q collagen stems in nonactivated C1, and Arlaud et al. (1) reference an EM study in favor of this model (10). However, the micrographs presented in that study (10) and a related contemporary study (11) do not prove a central location of the C1r and C1s SP domains. Our SAXS rigid body modeling of the nonactivated C1 complex placed the C1r and C1s SP domains at the periphery independent of their initial position, and this location was supported by our ab initio SAXS modeling (2). Consistently, our ns EM class averages showed up to 10 protruding spherical domains in agreement with six C1q globular heads and four SP domains from C1r and C1s located in the periphery of C1. If the SP domains were tucked in the center of nonactivated C1, a maximum of six peripheral domains would be observed.
Footnotes
The authors declare no conflict of interest.
References
- 1.Arlaud GJ, Gaboriaud C, Ling WL, Thielens NM. Structure of the C1 complex of complement. Proc Natl Acad Sci USA. 2017;114:E5766–E5767. doi: 10.1073/pnas.1703977114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortensen SA, et al. Structure and activation of C1, the complex initiating the classical pathway of the complement cascade. Proc Natl Acad Sci USA. 2017;114:986–991. doi: 10.1073/pnas.1616998114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budayova-Spano M, et al. The crystal structure of the zymogen catalytic domain of complement protease C1r reveals that a disruptive mechanical stress is required to trigger activation of the C1 complex. EMBO J. 2002;21:231–239. doi: 10.1093/emboj/21.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kardos J, et al. Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r. Mol Immunol. 2008;45:1752–1760. doi: 10.1016/j.molimm.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Tschopp J, Villiger W, Fuchs H, Kilchherr E, Engel J. Assembly of subcomponents C1r and C1s of first component of complement: Electron microscopic and ultracentrifugal studies. Proc Natl Acad Sci USA. 1980;77:7014–7018. doi: 10.1073/pnas.77.12.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brier S, et al. Mapping surface accessibility of the C1r/C1s tetramer by chemical modification and mass spectrometry provides new insights into assembly of the human C1 complex. J Biol Chem. 2010;285:32251–32263. doi: 10.1074/jbc.M110.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatraman Girija U, et al. Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc Natl Acad Sci USA. 2013;110:13916–13920. doi: 10.1073/pnas.1311113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bally I, et al. Identification of the C1q-binding sites of human C1r and C1s: A refined three-dimensional model of the C1 complex of complement. J Biol Chem. 2009;284:19340–19348. doi: 10.1074/jbc.M109.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips AE, et al. Analogous interactions in initiating complexes of the classical and lectin pathways of complement. J Immunol. 2009;182:7708–7717. doi: 10.4049/jimmunol.0900666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strang CJ, Siegel RC, Phillips ML, Poon PH, Schumaker VN. Ultrastructure of the first component of human complement: Electron microscopy of the crosslinked complex. Proc Natl Acad Sci USA. 1982;79:586–590. doi: 10.1073/pnas.79.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poon PH, Schumaker VN, Phillips ML, Strang CJ. Conformation and restricted segmental flexibility of C1, the first component of human complement. J Mol Biol. 1983;168:563–577. doi: 10.1016/s0022-2836(83)80302-7. [DOI] [PubMed] [Google Scholar]