Significance
Microtubules, essential in eukaryotic cells, are wide and hollow tubes that are formed through the polymerisation of αβ-tubulin proteins. Microtubules provide tracks for motor-driven cellular transport. Dynamic instability of microtubules, the stochastic change between slow growth and rapid shrinkage, is important during mitotic DNA segregation. Although microtubules were thought to be unique to eukaryotes, certain bacteria are exceptions because they possess BtubAB proteins that are closely related to tubulins and form hollow tubes. We demonstrate with the atomic structure of bacterial BtubAB “mini microtubules” that they share many features with eukaryotic microtubules, and that they exhibit dynamic instability. Our work highlights deep conservation of structure and dynamics between bacterial and eukaryotic microtubules, providing clues about their evolution.
Keywords: bacterial cytoskeleton, dynamic instability, BtubAB, cryoEM, subtomogram averaging
Abstract
Microtubules, the dynamic, yet stiff hollow tubes built from αβ-tubulin protein heterodimers, are thought to be present only in eukaryotic cells. Here, we report a 3.6-Å helical reconstruction electron cryomicroscopy structure of four-stranded mini microtubules formed by bacterial tubulin-like Prosthecobacter dejongeii BtubAB proteins. Despite their much smaller diameter, mini microtubules share many key structural features with eukaryotic microtubules, such as an M-loop, alternating subunits, and a seam that breaks overall helical symmetry. Using in vitro total internal reflection fluorescence microscopy, we show that bacterial mini microtubules treadmill and display dynamic instability, another hallmark of eukaryotic microtubules. The third protein in the btub gene cluster, BtubC, previously known as “bacterial kinesin light chain,” binds along protofilaments every 8 nm, inhibits BtubAB mini microtubule catastrophe, and increases rescue. Our work reveals that some bacteria contain regulated and dynamic cytomotive microtubule systems that were once thought to be only useful in much larger and sophisticated eukaryotic cells.
Eukaryotic microtubules are formed by the polymerization of αβ-tubulin protein heterodimers (1–4). Stabilized microtubules provide tracks for cellular cargo, and dynamic instability, the alternation between slow growth and fast shrinkage, is crucial for spindle function during mitosis (5). Although microtubules were thought to be absent from other organisms, it has been shown that certain bacterial Prosthecobacter species contain BtubAB proteins that are closely related to tubulin (6, 7) and that also form hollow tubes (8).
No other prokaryotic tubulin-like proteins, such as FtsZ or TubZ, form cylindrical protofilament-containing microtubules with a B lattice, an A-lattice seam, and dynamic instability (9). How related BtubAB filaments and microtubules are, and the consequences of binding of the third gene in the btub gene cluster (6, 10), have remained unclear.
We produced unmodified Prosthecobacter dejongeii BtubAB proteins (6) in Escherichia coli, where they are known to fold easily (7), in contrast to eukaryotic αβ-tubulin. As was found previously, when GTP is added to BtubAB, long, straight, and hollow tubes were observed by electron cryomicroscopy (cryoEM) (Fig. 1A) (7, 8). Our images showed gently twisting, apparently helical, filaments that are ∼14 nm wide.
Fig. 1.
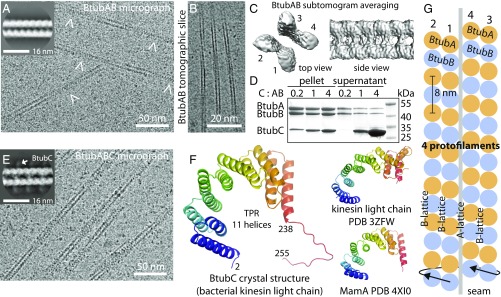
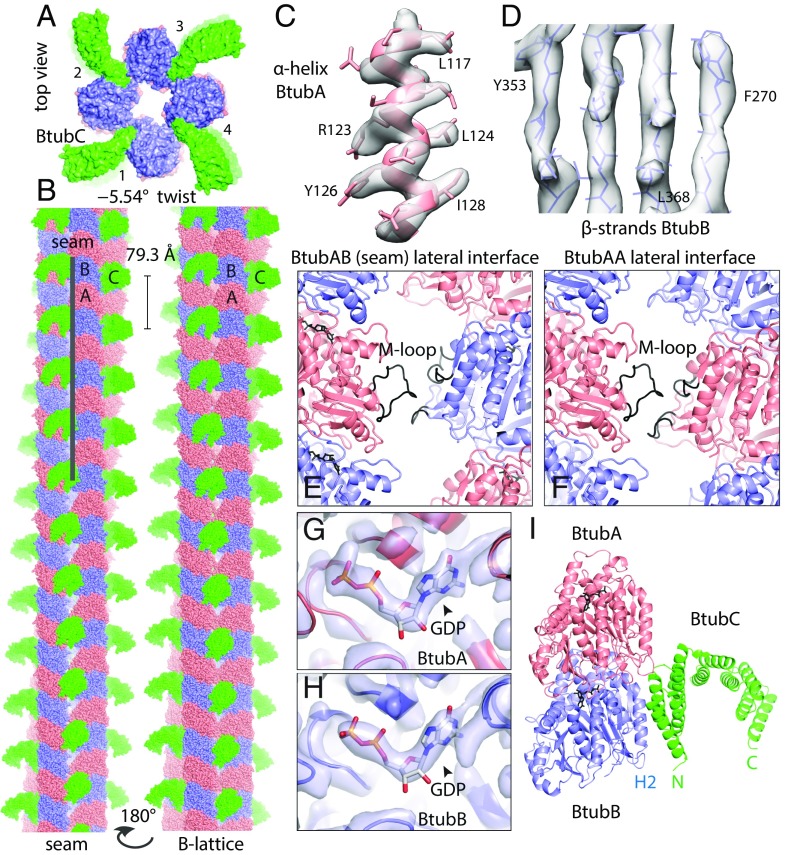
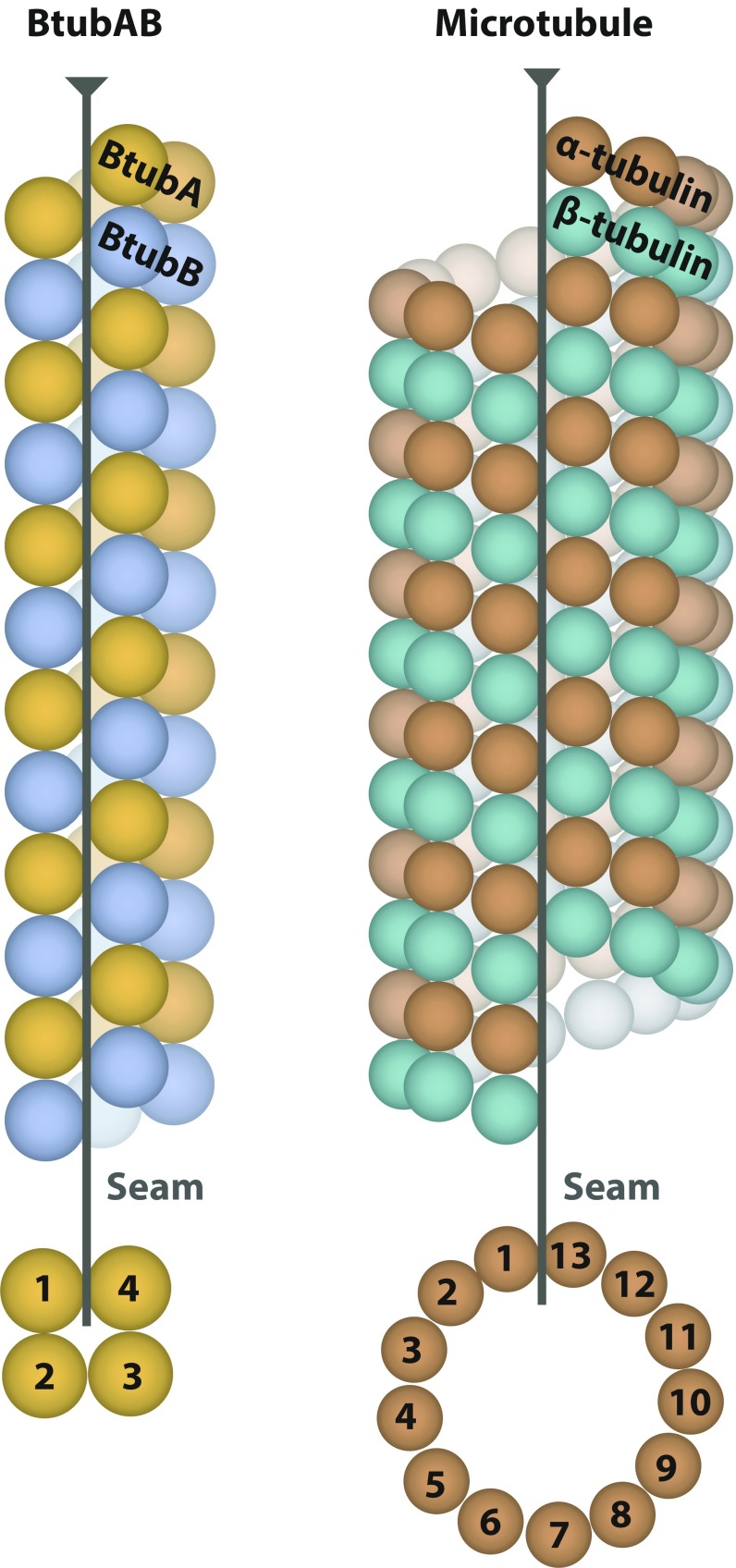
BtubAB form four-stranded mini microtubules that are decorated by BtubC. (A) Transmission cryoEM image of in vitro polymerized P. dejongeii BtubAB. The filaments show helicity as indicated by crossovers (arrowheads). (Inset) RELION (11) 2D class compatible with four protofilaments. (B) Section through cryoET tomogram of in vitro assembled BtubAB filaments. (C) Reference-free and unsymmetrised subtomogram averaged map (12) of BtubAB filaments showing four strands. (D) Pelleting assay showing that the third protein in the btub locus in P. dejongeii (6) binds stoichiometrically to BtubAB filaments. The protein has been renamed BtubC (formerly: BKLC, bacterial kinesin light chain). (E) Transmission cryoEM image of in vitro polymerized PdBtubABC. (Inset) RELION 2D class showing the binding of BtubC every 8 nm. (F, Left) Crystal structure of P. vanneervenii BtubC at 2.5 Å resolution. (F, Right) BtubC is as closely related to other TPR proteins such as MamA (45) as to the TPR domain of kinesin light chain (46). (G) Scheme summarizing the symmetry of BtubAB mini microtubules as deduced by cryoEM and cryoET: Four protofilaments with alternating filaments arrange into a hollow tube. The approximate helical parameters as deduced from the cryoET map in C dictate that the resulting structure must have a seam where lateral interactions change from B lattice (A-A and B-B) to A lattice (A-B).
When performing 2D classification of filament segments in RELION (11), classes emerged that were not compatible with the previous suggestion that BtubAB tubes contain five protofilaments (8) (Fig. 1A, Inset). To determine the protofilament number of the filaments unambiguously, we performed electron cryotomography (cryoET) (Fig. 1B and Movie S1) with subsequent reference-free subtomogram averaging (12) (Fig. 1C). It is clear from the tomograms and from the subtomogram average (Movie S1 and Fig. 1C) that the tubes contain four protofilaments, and this arrangement is compatible with the 2D class average from cryoEM (Fig. 1A, Inset). The discrepancy we found with previous estimations of the BtubAB protofilament number, 4 versus 5, might reflect differences in conditions (in vitro versus in vivo) or could be related to the low resolution currently attainable with the method of electron tomography of entire cells as was used previously (8).
Approximate helical parameters were deduced by manual inspection of the subtomogram-averaged map (Fig. 1C) and were used to obtain a 4.2-Å map of the filaments through iterative helical real space reconstruction (13) in RELION (Fig. S1). Refined helical parameters were: twist −90.7° and rise 10.4 Å. These symmetry operators averaged BtubA and BtubB map regions and, hence, indicated that the overall structure is not helical, as explained in Fig. 1G. Applying the twist and rise four times around the tube means that the nature of the subunits must change from BtubA to B or B to A. This is a well-known feature of eukaryotic microtubules, named the seam, where the otherwise B-lattice (A-A and B-B lateral contacts) changes into A lattice, with A-B contacts (14, 15).
Fig. S1.
CryoEM density map (4.2 Å) of PdBtubAB filaments. BtubA and BtubB are similar in sequence and structure so they cannot be distinguished and, hence, the map was fully symmetrised, averaging all subunits together (Materials and Methods). Because of the symmetrization, the M loop that reaches over from BtubA (only) to the next protofilament has half occupancy in this map and is only visible at much lower contour level because the corresponding loop in BtubB does not fold into the same conformation and does not contact the neighboring protofilament. Beyond this observation, BtubAB filaments, without BtubC, are similar in appearance and helical parameters to BtubABC filaments (when transformed to a different symmetry setting). Refined helical parameters were twist −90.7° and rise 10.4 Å.
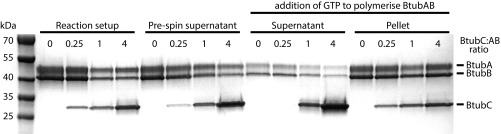
The alternating arrangement of BtubAB subunits means, to obtain a correct reconstruction and structure, only helical symmetry along one protofilament must be applied (twist ∼ –5.6°, rise ∼80 Å), and the reconstruction algorithm must be able to distinguish BtubA and BtubB. Because BtubA and BtubB are extremely similar in structure (7), we needed to add a feature to the BtubAB filaments that could mark the 8-nm heterodimer repeat along each protofilament. Our attention turned to the third gene in the btub gene cluster, bacterial kinesin light chain, bklc (6), or btubC, as explained below. Using pelleting assays we found that P. dejongeii BtubC, expressed in E. coli and purified with a tag, binds to BtubAB filaments in stoichiometric fashion (Fig. 1D and Fig. S2) as was also reported elsewhere, recently (10). Adding more BtubC beyond the number of BtubAB heterodimers in the filaments did not significantly increase the amount of BtubC that spun down. Encouraged by this we performed again cryoEM with subsequent 2D classification, clearly showing that BtubC binds every 8 nm, to each BtubAB heterodimer, on the outside of the filaments (Fig. 1 E and E, Inset).
Fig. S2.
Pelleting assay showing stoichiometric binding of PdBtubC to PdBtubAB filaments. Replicate experiment to the one shown in Fig. 1D. Fourfold excess of BtubC to BtubAB (molar, C to AB heterodimer) leads to roughly the same amount of BtubC in the pellet compared with equimolar quantities. Excess BtubC protein remains in the supernatant and does not spin down, indicating a specific interaction of BtubC with BtubAB heterodimers in the filaments.
Because BtubC shows sequence similarity to the TPR domain of eukaryotic kinesin light chain (that does not bind to microtubules), we solved the crystal structure of BtubC from Prosthecobacter vanneervenii BtubC (76% sequence identity to P. dejongeii BtubC) at 2.5-Å resolution by mercury single anomalous diffraction (SAD) (Fig. 1F and Table S1). The structure shows the expected curved TPR repeat fold, containing 11 helices and a C-terminal tail beyond residue 238. When comparing the BtubC structure to all known protein structures, it became clear that BtubC is as similar to TPR domains from eukaryotic kinesin light chains as to many other TPR domains such as MamA (Fig. 1F, Right). Given the organization of btubABC into a likely operon (6, 16), the fact that BtubC binds to BtubAB filaments (10) and the misleading appearance of the term kinesin in its previous BKLC name, we decided to rename the protein BtubC.
Table S1.
Crystallography and cryoEM data
| Sample | Prosthecobacter vanneervenii PvBtubC EMTS derivative | Prosthecobacter vanneervenii PvBtubC native | Prosthecobacter dejongeii PdBtubABC |
| NCBI database ID | CAP16140.1 | CAP16140.1 | AA012155.1, AA012159.2, this work |
| Constructs | 1–256-KLHHHHHH | 1–256-KLHHHHHH | A: 1–473, no tag, B: 1–426 no tag, C: MGSSHHHHHH-SSGLLPRGSH-1–257 |
| Method | Crystallography SAD | Crystallography | CryoEM, |
| 0.5 mM EMTS | helical reconstruction | ||
| Data collection | |||
| Beamline/microscope | ESRF ID30B | ESRF ID29 | FEI Polara G2, Falcon III |
| Wavelength/energy | 1.006 Å | 0.977170 | 300 kV |
| Crystal/helical | |||
| Space/point group | H32 | H32 | |
| Cell, Å | 120.0, 120.0, 239.1 | 119.7, 119.7, 239.4 | |
| Twist/rise | −5.54°, 79.31 Å | ||
| Data | |||
| Resolution, Å | 2.8 | 2.5 | 3.6 |
| Completeness*, % | 100 (100) | 99.4 (100) | |
| Multiplicity* | 11.8 (11.3) | 4.9 (5.2) | |
| (I)/σ(I)* | 17.8 (2.4) | 10.8 (2.7) | |
| Rmerge* | 0.102 (1.036) | 0.108 (0.674) | |
| Rpim* | 0.044 (0.467) | 0.054 (0.330) | |
| CC1/2 | 0.999 (0.726) | 0.996 (0.690) | |
| Anomalous correlation | 0.24 (0.0) | ||
| Mercury sites | 4 | ||
| Images, pixel size | 6105, 1.34 Å | ||
| Defocus range, dose | −1.5 to −3 µm, 40 e/Å2 | ||
| Helical segments | 257,661 | ||
| Refinement | |||
| R/Rfree† | 0.186/0.237 | 0.32 | |
| Models | 2 chains/ASU: | 1 BtubAB heterodimer: A residues 3–435, B 2–37; 46–273;281–426 | |
| Residues 2–255, | 0.014 | ||
| 92 waters | |||
| Bond length rmsd, Å | 0.015 | ||
| Bond angle rmsd, ° | 1.716 | 1.556 | |
| Favored‡, % | 100 | 99.5 | |
| Disallowed‡, % | 0 | 0 | |
| MOLPROBITY score | 95th percentile | 100th percentile | |
| PDB/EMDB IDs | 5O01 | 5O09, EMD-3726 |
Values in parentheses refer to the highest recorded resolution shell.
Five percent of reflections were randomly selected before refinement.
Percentage of residues in the Ramachandran plot (PROCHECK “most favored” and “additionally allowed” added together).
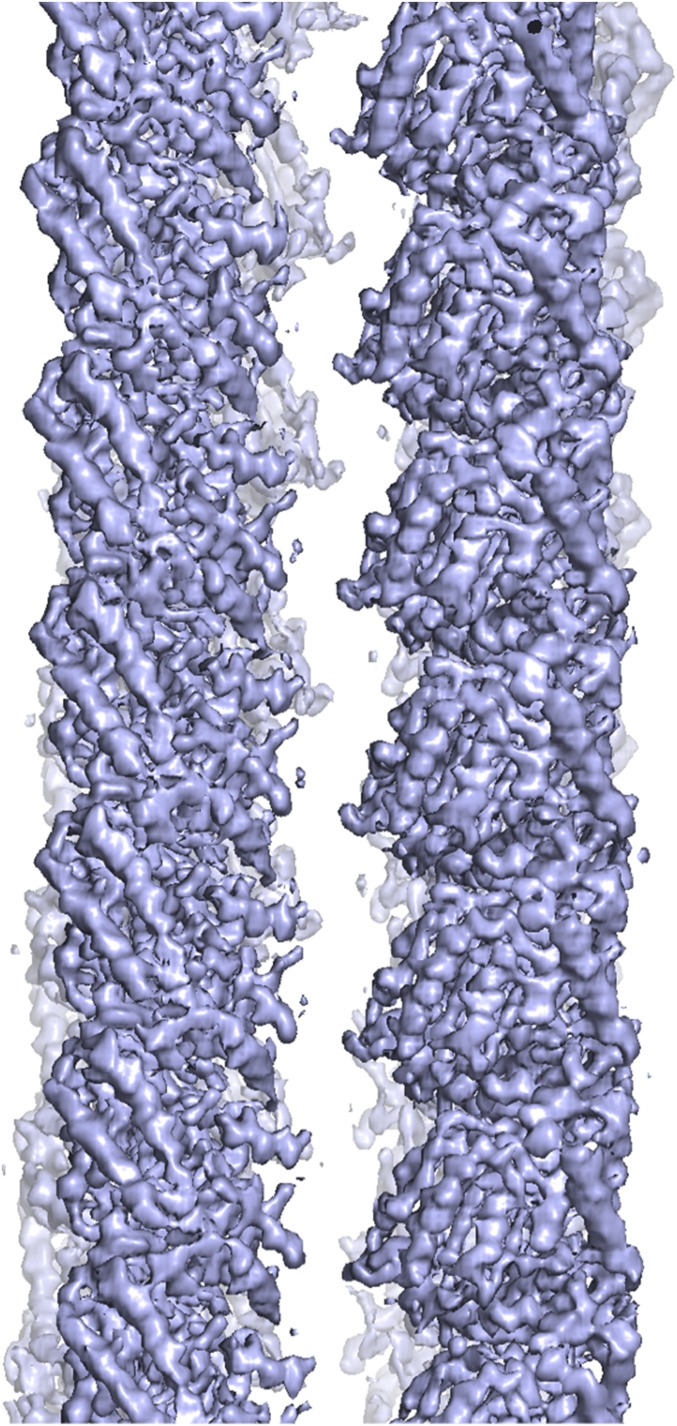
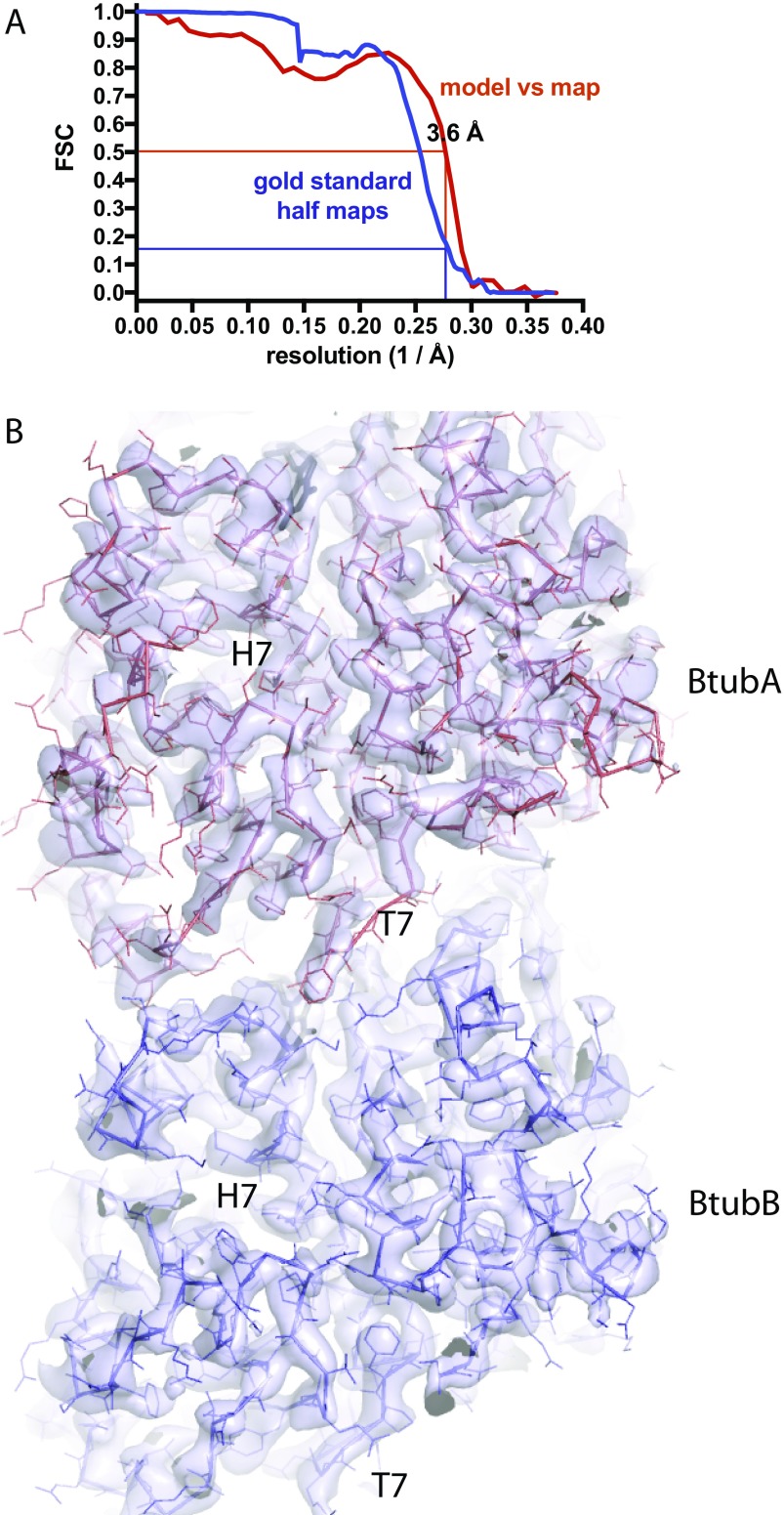
We performed iterative helical real space reconstruction in RELION from BtubABC filament cryoEM images as depicted in Fig. 1E (Table S1, refined helical parameters twist –5.54°, rise 79.3 Å). The resulting density map at 3.6 Å [as determined by gold standard Fourier shell correlation (FSC), Fig. S3A] was of excellent quality, allowing the fitting and all-atom refinement of previous crystal structures of BtubA and BtubB (7) against the best part of the filament density (Fig. 2 C and D, Fig. S3B, and Movie S2). Manual assignment of BtubA and BtubB to their corresponding map regions was enabled by many side chain differences between the otherwise similar structures (Fig. S4). Density was less good for BtubC and its structure was from a different organism (Pv versus Pd, Table S1), so we fitted the BtubC crystal structure as a rigid body only. Expanding symmetry yielded a complete atomic model of the BtubABC filaments (Fig. 2 A and B). The structure contains four protofilaments that arrange into a gently twisting hollow tube. Along three protofilament interfaces, lateral contacts form a B-lattice with A-A and B-B contacts (Fig. 2B, Right), whereas along the seam, A-B contacts form an A lattice (Fig. 2B, Left). The protofilaments are ∼90° rotated against each other. The contacts between protofilaments are exclusively mediated by a loop that has been named M-loop in tubulin because it facilitates microtubule formation through a similar contact (Fig. 2 E and F) (3). The M loop in BtubB is not ordered in our cryoEM map and is unlikely to form a contact, leaving only contacts made by BtubA's M-loop to bind the filaments together. The contacts made along the A (seam) and B lattices are similar, despite containing different partners. A superposition of M loops holding microtubules and BtubAB mini microtubules together shows segments of similar size and shape but also that the contacts are quite different at the secondary structure and side chain level. The differences at secondary structure and side chain level probably enable a change in interprotofilament angle from 27.7° in 13-protofilament microtubules to 90° in BtubAB mini microtubules (Fig. S5). In mini microtubules, both BtubA and BtubB are GDP-bound as deduced by inspecting the cryoEM density map (Fig. 2 G and H), in contrast to tubulin, which forms a stable heterodimer because α-tubulin is always bound to nonexchangeable GTP. BtubC binds to the outside of the BtubAB filaments (Fig. 2A) and contacts predominantly BtubB (Fig. 2I), including at the seam. The N-terminal helices of BtubC contact BtubB, but there are some minor additional contacts to the other subunits surrounding the interface. The structure of BtubABC mini microtubules is best appreciated by inspecting Movie S3.
Fig. S3.
(A) BtubABC cryoEM Fourier shell correlation (FSC) of the half maps and atomic model. The blue curve describes the FSC as calculated by RELION during postprocessing, comparing the two gold standard half maps that have been kept separate for the entire procedure before they are combined. Filament segments were kept separate at the filament level to avoid overfitting through the inclusion of identical parts of the original images in the two half sets because filament segments overlap. A spherical mask was used for the determination of a sharpening B factor and half map FSC during postprocessing only, and covered one complete BtubAB heterodimer (but not BtubC, which had weaker density and was not refined). The mask was softened at the edges with an eight-pixel raised cosine edge. The red curve describes the FSC of a map generated from the one used to refined BtubAB heterodimer (omitting BtubC, both half maps averaged), cut around the atomic model, against the final atomic model. Using the suggested resolution estimates at FSC 0.5 for the model vs. map and FSC 0.143 for the two gold standard half maps, identical values of 3.6 Å were obtained. (B) PdBtubABC filament cryoEM density map at 3.6 Å resolution. Final map used for model building and refinement with the refined atomic models for BtubA (red) and BtubB (blue) superimposed. See also Movie S2.
Fig. 2.
BtubABC mini microtubule cryoEM structure at 3.6 Å resolution. (A) View along the filament axis showing the four protofilaments end-on and also the protruding BtubC subunits in green. (B, Left) BtubABC filaments. PdBtubA is in red, PdBtubB in blue, and PvBtubC in green. The view shown highlights the seam (A lattice, A-B lateral contacts). (B, Right) Rotated by 180°, showing the B lattice (A-A and B-B lateral contacts). Helical parameters: twist = –5.54°, rise = 79.3 Å. (C and D) CryoEM density map with fitted atomic model. (E) Close-up of the only lateral contact holding the protofilaments together. The contact is related to the equivalent contact in eukaryotic microtubules, with the M loop (3) of BtubA (residues ∼280–290) reaching over to contact residues around 90 and also 55–65. Shown here is the seam contact (BtubA M loop reaching over to BtubB). Note that the M loop in BtubB is not ordered. (F) Same as in E, but B-lattice contact, BtubA contacting BtubA. (G) Close-up of density in the nucleotide binding pocket in BtubA, which is best fitted with GDP. (H) Pocket in BtubB, also GDP-bound. (I) Cartoon plot of the interaction of BtubC with the BtubAB heterodimer. BtubC predominantly binds to BtubB. See also Movie S3.
Fig. S4.
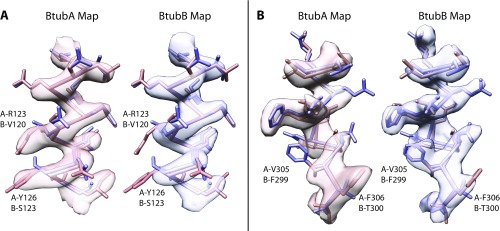
Assignment of BtubA and BtubB to their corresponding map regions. Because BtubA and BtubB are similar in structure (7), we needed to use BtubC, which only binds to each BtubAB heterodimer once to keep BtubA and BtubB separate during the cryoEM helical reconstruction. We then needed to determine which of the monomers in the map are BtubA and BtubB. The subunit assignment was done based on the excellent quality of the 3.6 Å resolution map, which enabled the unambiguous assignment through inspection of many map regions. A and B show two such map regions, superimposed with both the BtubA and BtubB model from the crystal structure, fitted into the map with CHIMERA (28). BtubA map and model are in red, blue for BtubB.
Fig. S5.
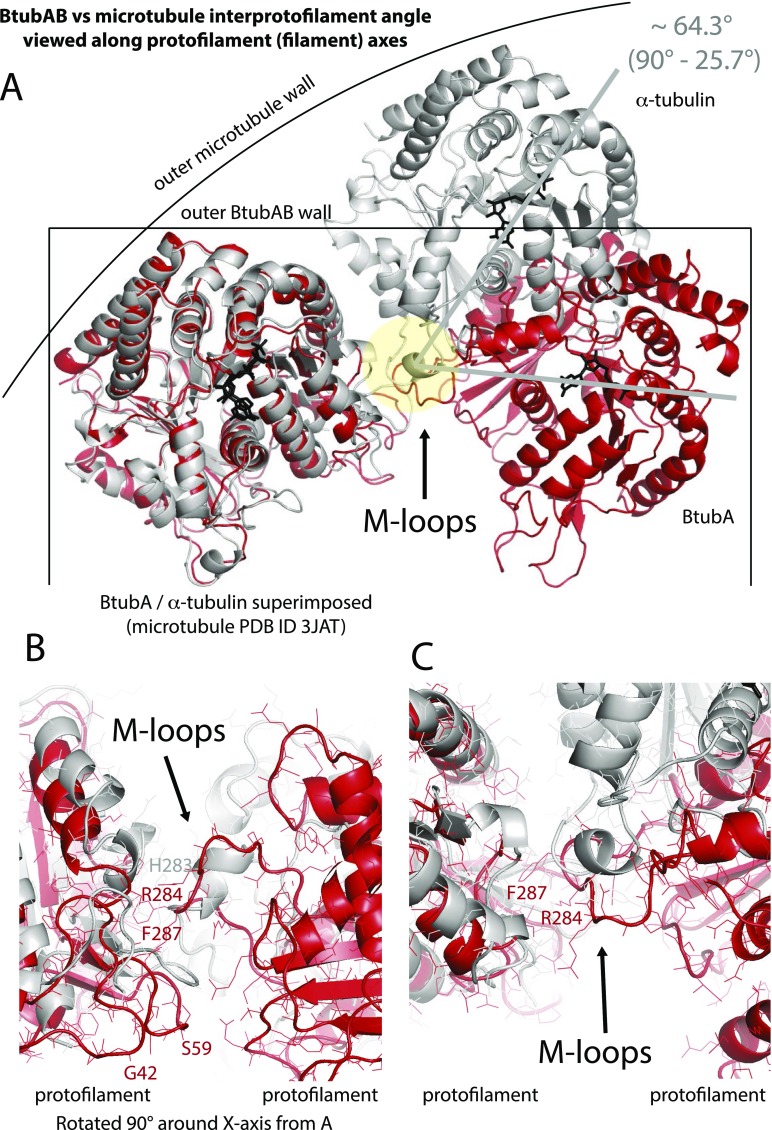
Comparison of interprotofilament interfaces in microtubules and BtubAB minimicrotubules. (A) Superposition of BtubA (red) and α-tubulin (gray) when in 4-protofilament BtubAB mini microtubules and 14-protofilament microtubules (PDB ID code 3JAT) (4). The protofilaments are rotated by ∼26° almost perfectly around the M loop, the structure in both polymers that facilitates protofilament contacts. (B and C) Detailed view of the same BtubAB/α-tubulin superposition showing the M loops. Although in roughly the same place, secondary structures are different and there is no obvious similarity of the contacts at the residues level. Divergent evolution probably changed the entire contact so to enable different interprotofilament angles, leading to 4 instead of more than 10 protofilaments. C is rotated 90° around the x axis with respect to B.
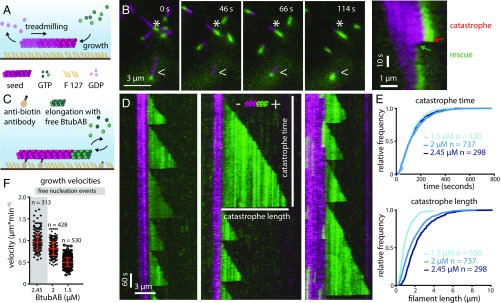
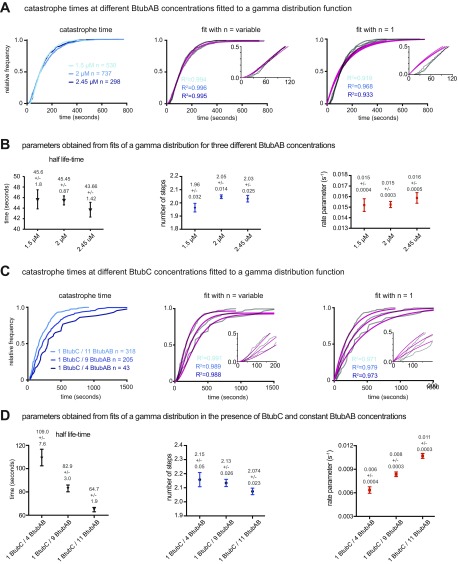
Because the BtubAB filament structure showed strong similarities to eukaryotic microtubules, despite being very different in diameter, we investigated the dynamic properties of BtubAB by total internal reflection fluorescence (TIRF) fluorescence microscopy. The experimental setups are shown in Fig. 3 A and C and discussed in Materials and Methods. In single-color TIRF experiments, upon addition of GTP, BtubAB filaments grew into long, rigid filaments with little bundling propensity, but also showed directed movement of smaller filaments (Movie S4). The movement could be completely abolished by the addition of GTP-γ-S, a slowly hydrolysable GTP analog (Movie S5). A two-color experiment highlighted that the movement was caused by treadmilling, the addition of subunits at one end (the plus end) and loss of subunits at the other end (the minus end) (Fig. 3B and Movies S6 and S7). In this experiment, both magenta and green filaments were dynamic (GTP added), but the magenta filaments were grown first, after which green subunits were added, so that green filaments were seeded on the older, magenta filaments. Filaments disassembled from the magenta (old) end only, marking the minus ends, whereas growth appeared exclusively at the green ends, marking the plus ends. We noted that sometimes filaments would rapidly depolymerize from the plus ends (Fig. 3B, Right and Movie S7), possibly indicating dynamic instability, one of the hallmarks of eukaryotic microtubule behavior (5). Dynamic instability is the rapid, or catastrophic, disassembly of a cytomotive filament when the protective cap of triphosphate nucleotide-bound subunits has disappeared because nucleotide hydrolysis within the filament caught up with filament growth, reaching the terminal subunits at the plus end. So we performed another dual-color experiment, this time starting with magenta seeds that had been stabilized by adding GMPCPP, another slowly-hydrolysable GTP analog (Fig. 3D and Movie S8). Because the magenta seeds were not dynamic, long runs of green filament extensions at the plus ends could be observed. The green extensions displayed many rounds of catastrophe and repolymerization, termed rescue. The rate of growth was 0.38 µm⋅min−1⋅µM−1, which is only slightly outside the reported range of mammalian microtubule growth rates of 0.20–0.33 µm⋅min−1⋅µM−1 (17, 18). We analyzed the times between catastrophe events and the extension length at which catastrophe occurred and plotted these distributions in cumulative plots (Fig. 3E). As expected, catastrophe time did not depend on protein concentration because it is a zero order event, but catastrophe length varied with BtubAB concentration because of the change in growth velocities (Fig. 3F). Generally, the shape of the distribution of catastrophe times may deviate from one caused by a simple exponential decay if more than one step is needed to cause catastrophe. In that case, the reaction kinetics of catastrophe lead to an aging process, in which older filaments are more likely to experience catastrophe (19). When performing this analysis on our data, it became clear that BtubAB filament catastrophe time distributions were best fitted with a gamma function describing such behavior (Fig. S6 A and B), resembling complex dynamic behavior of eukaryotic microtubules for which catastrophes are best described as a multistep process.
Fig. 3.
BtubAB mini microtubules show dynamic instability. (A) Sketch of TIRF microscopy setup used for B. Elongation of Atto655-labeled BtubAB filaments (GTP seeds, magenta) upon addition of Atto488-labeled BtubAB monomers, also in GTP. (B) Image series showing treadmilling filaments (asterisks). Seeds grow at the plus ends (green extension), whereas the opposing minus ends shrink (shortening of magenta seeds). Filaments display catastrophic shortening and regrowth from the plus ends only (arrowheads), indicating dynamic instability. The kymograph (Right) highlights treadmilling and dynamic instability. (C) Sketch of TIRF microscopy setup used for D. Stable GMPCPP seeds (magenta) were elongated by green BtubAB in GTP. (D) Kymographs of three separate BtubAB filaments showing dynamic instability from their plus ends. Extensions show a sawtooth pattern because filaments undergo several cycles of growth and rapid shrinkage, called catastrophe. (E) Graphs showing cumulative distributions of catastrophe times and lengths (indicated by bars in the middle kymographs in D) for three different concentrations (n = 3 experiments). (Upper) Catastrophe times do not change with concentration and follow a gamma distribution, revealing that catastrophe is a multistep process (Fig. S6). (Lower) Catastrophe length increases with BtubAB concentration. (F) Scatter plot of growth velocities versus BtubAB concentration (n = 3, 2.45 µM: n = 2). As expected, growth rate increases with concentration. Filaments grow at 0.96 ± 0.18 µm⋅min−1, mean ± SD; 0.81 ± 0.16 µm⋅min−1 and 0.5 ± 0.1 µm⋅min−1 for 2.45 µM, 2 µM, and 1.5 µM BtubAB, respectively. The growth velocities correspond to the observed changes in catastrophe length (E), meaning catastrophe is independent of growth rate.
Fig. S6.
Analysis of BtubAB catastrophe times and gamma function fitting parameters obtained for different BtubAB and BtubC concentrations. Related to Fig. 3. (A) Cumulative distribution of catastrophe times fitted to a gamma function (Materials and Methods). A multistep process where n > 1 is compared with a single-step process (n = 1) obtained by fitting a gamma distribution describes catastrophe of BtubAB filaments best over a range of concentrations. The goodness of the fits for different n (magenta) is given as the mean R2 value for each condition (color coded) in the corresponding graph. (B) BtubAB concentrations have only small effects on mean lifetimes, number of steps and rate parameters that have to take place before catastrophe. Values are given above each data point, and error bars represent 95% confidence intervals. (C) Experimentally obtained distribution of catastrophe times for different ratios of BtubC to BtubAB, for which the concentration of BtubAB was kept constant at 2 µM. Catastrophe is still a multistep process with n = 2 steps as seen for BtubAB alone. Again, fits for different n (magenta) are evaluated by R2 values for each condition (color coded). (D) BtubC has an effect on BtubAB lifetimes. Increasing molecule ratios (BtubC to BtubAB) shifts the distribution to older filaments, by reducing the rate parameters at which the steps have to occur to induce a catastrophe. Values are again given above each data point, and error bars represent 95% confidence intervals.
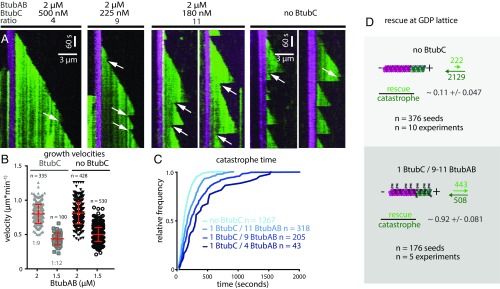
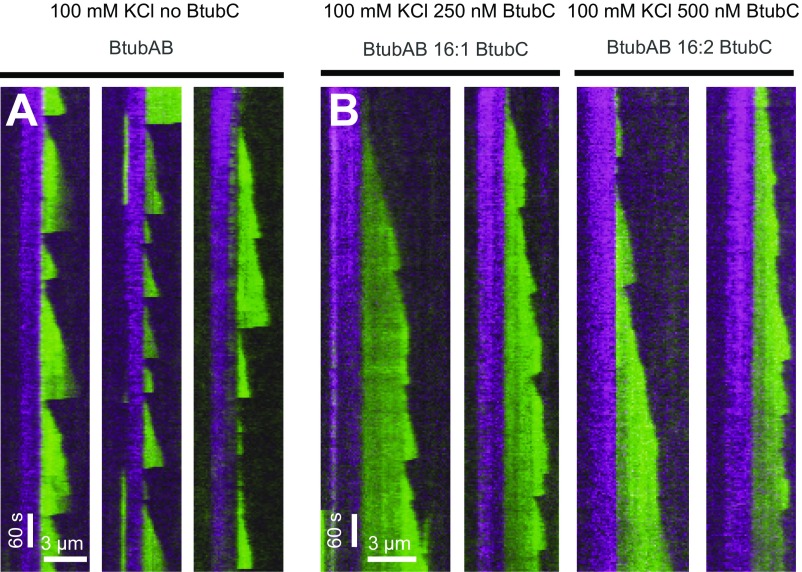
Finally, we asked what, if any, influence BtubC had on the dynamic properties of BtubAB filaments (Fig. 4A). Even at substoichiometric ratios, BtubAB filaments showed markedly reduced catastrophes but increased rescues from the GDP/GTP (green) ends of the filament with the result that close to stoichiometry, the filaments were fully stabilized and simply grew and were nondynamic (Fig. 4A, Left and Movie S9). Growth velocities were not significantly affected by the addition of BtubC (Fig. 4B). When using less BtubC, enough catastrophes could be observed to analyze rescue and catastrophe events along the green filament extensions (GDP/GTP ends) (Fig. 4A, Right and Movie S10). The analysis of catastrophe events with BtubC present revealed that the multistep nature of catastrophe stayed unchanged (Fig. S6 C and D). Catastrophe times (filament age when catastrophes occurred) were affected by BtubC, which increased because of the lower catastrophe rate parameters (Fig. 4C and Fig. S6 C and D). When using less BtubC, enough catastrophes could be observed to also analyze rescue frequency (Fig. 4D). Rescues increased roughly eightfold, meaning that BtubC may stabilize BtubAB filaments both through a reduction in catastrophe frequency but also through an increase in rescue frequency. As mentioned, in the BtubABC mini microtubules, BtubC binds mostly to BtubB but has also weaker interactions with at least two neighboring subunits along and across the protofilaments (Fig. 2A and Movie S3). We propose that cross-linking subunits in the mini microtubule lattice might provide the additional stabilization needed to prevent depolymerization but might also lead to more effective polymerization and even nucleation.
Fig. 4.
BtubAB dynamic instability is regulated by BtubC. (A) Kymographs of BtubAB filaments emanating from stabilized seeds (as in Fig. 3C) in the presence of varying amounts of BtubC. BtubC interferes with dynamic instability and inhibits it at stoichiometric amounts. Filaments grown in the presence of BtubC do not completely disassemble instead they start regrowth from the nonstabilized (green) GDP/GTP ends (arrows). (B) Scatter plots of BtubC-dependent growth velocities (the horizontal line above the scatter plots indicates the presence or absence of BtubC. BtubC was tested at different molecular ratios (1 BtubC to 11 BtubAB for 2 µM BtubAB and 1 BtubC to 9 BtubAB for 1.5 µM BtubAB). BtubC does not drastically disturb filament growth compared with BtubAB alone. Filaments grow slightly slower in the presence of BtubC (1:11) at 0.8 ± 0.14 µm⋅min−1, mean ± SD. compared with 0.81 ± 0.16 µm⋅min−1 mean ± SD without BtubAB at the same BtubAB concentration of 2 µM. Similarly, at the ratio BtubC to BtubAB (1:9), filaments grow slightly slower at 0.44 ± 0.08 µm⋅min−1, mean ± SD to 0.5 ± 0.1 µm⋅min−1 mean ± SD for a BtubAB concentration of 1.5 µM. (C) Cumulative catastrophe time distributions of filaments at different BtubC concentrations. Filament lifetimes are increased in the presence of BtubC, which is caused by a BtubC concentration dependent reduction in the rate parameter at which catastrophe occurs (Fig. S6 C and D). (D) Effect of BtubC on rescue/catastrophe ratios. In the presence of BtubC at ratios 1:9 and 1:11, 92 ± 8.1% of catastrophes are followed by rescue, whereas without BtubC, this occurs in approximately 11 ± 4.7% of observed events. In summary, BtubC stabilizes BtubAB filaments by increasing rescue events and reducing catastrophe frequency.
In summary, we think based on our results it is justified to call the filaments formed by BtubAB mini microtubules (Figs. S7–S9). Many striking similarities exist: The structure of the BtubAB heterodimer is closely related to αβ-tubulin (7), with the difference that both sites are exchangeable, capable of hydrolyzing GTP. Protofilaments contain alternating A and B subunits, as was suggested (7, 20). Mini microtubules have overall polarity, with all subunits pointing in the same direction and M loops making the only lateral contacts linking protofilaments laterally to each other. They also have a seam, being mostly B-lattice with one row of A-lattice. Mini microtubules display dynamic instability with a complex aging cap, a hallmark of eukaryotic microtubules. Mini microtubule dynamics are regulated by an associated protein, BtubC, leading to stable, cytoskeletal rather than cytomotive filaments. The regulation of mini microtubules by BtubC is reminiscent of what certain microtubule associated proteins (MAPs), such as DCX, Clasp, and Mal3, do to microtubules (21–23).
Fig. S7.
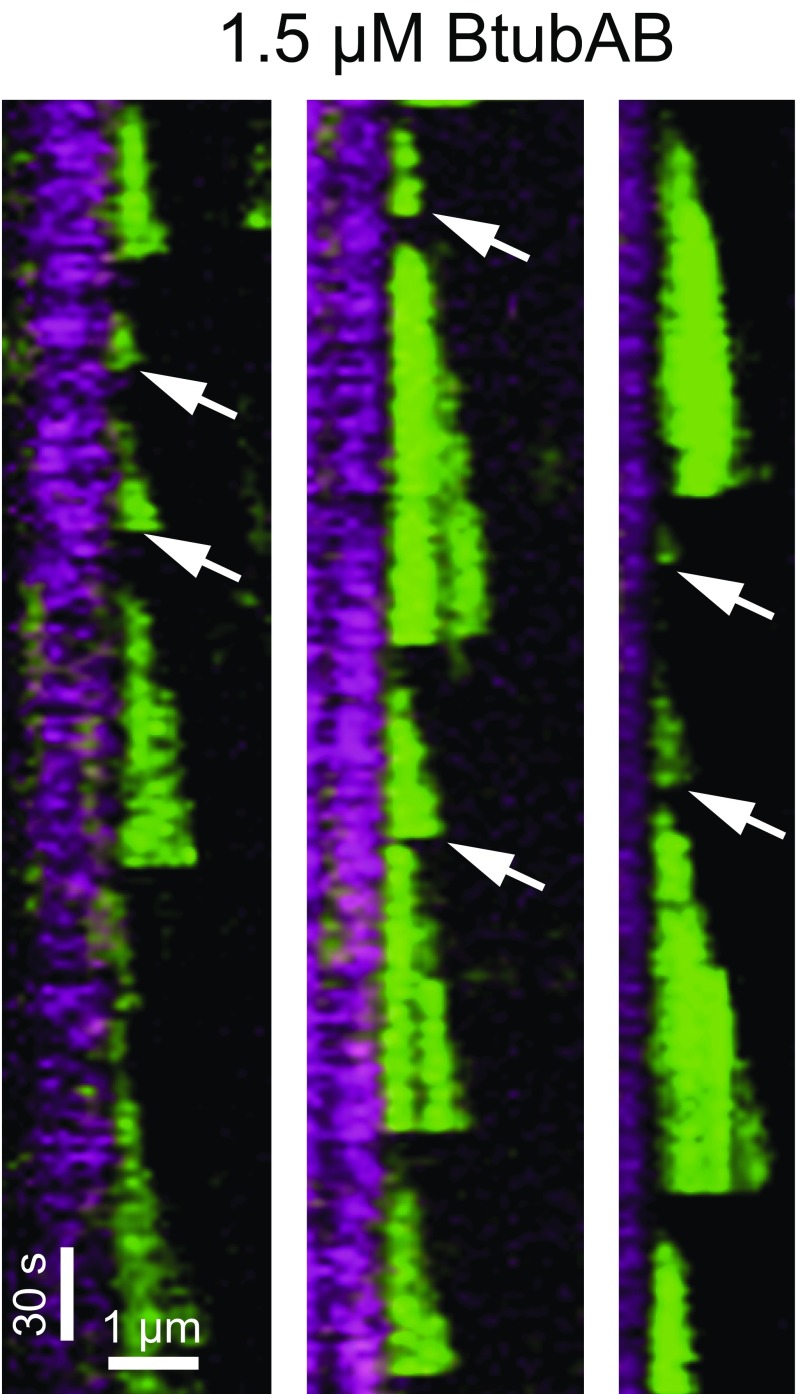
BtubAB dynamic instability at 1.5 µM. GMPCPP-stabilized seeds are shown in magenta with green filament elongations at 1.5 µM free BtubAB. Note that extensions over short periods and lengths (arrows) could be detected to reliably obtain data of catastrophe times and filament lengths.
Fig. S9.
Schematic comparison of BtubAB mini microtubules and eukaryotic 13-protofilament microtubules. Apart from previously known similarity of the subunits themselves and the heterodimer (7), the two structures conserve overall polarity (chemically different ends because all subunits point in the same direction), the seam where the nature of lateral contacts between protofilaments change and alternating AB/αβ protofilaments. The M loop connecting protofilaments is also conserved. In contrast to microtubules, both BtubA and BtubB contain GDP in the polymerized and matured filament and, of course, the protofilament number is vastly different, leading to much thinner mini microtubules (and a small helical twist that is not depicted here).
The most obvious structural difference between microtubules and BtubAB mini microtubules is the greatly reduced number of protofilaments, from 13 to 4, or possibly 5 if there exists a difference between tubes polymerized in vitro and in vivo (8). We suggest that after the likely horizontal gene transfer of the btubABC genes from a eukaryotic organism (7, 16) (or, alternatively, its evolution from a primitive tubulin precursor) (24), the size of the microtubules was reduced during subsequent evolution, possibly to fit better into the much smaller bacterial cells where they acquired a yet to be determined function. It will be especially interesting to see whether mini microtubules have a role in stalk formation in Prosthecobacter because they tend to be located there (8) and how regulation of filament dynamics by BtubC is used in the cellular context, especially because it was reported recently that BtubC might link mini microtubules to biological membranes (10). In the meantime, BtubABC will provide an exciting biochemical tool for the detailed atomistic investigation of filament dynamics, including treadmilling and dynamic instability, and will also provide an exciting building block for synthetic biology approaches to molecular machines.
Materials and Methods
Plasmid Construction.
To obtain a plasmid that codes for untagged proteins BtubA and BtubB from P. dejongeii DSM 12251 (PdBtubAB), a stop codon (TAA) was introduced before the C-terminal hexahistidine tag in the BtubAB bacterial T7 expression plasmid used (7) by Q5 mutagenesis (New England Biolabs). The plasmid maintains the intergenic region between BtubA and BtubB as present in the source genome. The gene btubC (previously known as the bacterial kinesin light chain BKLC) from P. dejongeii DSM 12251 was amplified from genomic DNA and was cloned into vector pET15b by using Gibson Assembly (New England Biolabs). The resulting construct encoded an N-terminal hexahistidine tag followed by PdBtubC (MGSSHHHHHHSSGLLPRGSH-1-257). The synthesized gene (Integrated DNA Technologies, IDT) btubC (previously known as the bacterial kinesin light chain BKLC) from P. vanneervenii [National Center for Biotechnology Information (NCBI) nucleotide database ID code AM041148.3] was cloned into plasmid pHis17 by using Gibson Assembly (New England Biolabs), yielding the construct encoding P. vanneervenii BtubC (PvBtubC) followed by a C-terminal hexahistidine tag (1-256-KLHHHHHH).
Protein Expression and Purification.
Untagged proteins BtubA and BtubB from P. dejongeii DSM 12251 were coexpressed and copurified by using a protocol adapted from described methods (7). The plasmid encoding untagged BtubAB was used to transform C41(DE3) E. coli cells (Lucigen) by electroporation. Sixty milliliters of 2xTY media supplemented with 100 μg/mL ampicillin were inoculated with a single colony from the plate, and were grown at 200 rpm, 37 °C overnight. The culture was then used to inoculate 6 L 2xTY media with 100 μg/mL ampicillin. After reaching an OD600 of 0.6–1.0 at 200 rpm, 37 °C, the expression was induced with 1 mM isopropyl β-d-thiogalactoside for 3 h at the same temperature, and the cells were harvested by centrifugation. The whole pellet was resuspended in 300 mL of buffer A (20 mM Tris/HCl, 1 mM sodium azide, 5 mM magnesium acetate, pH 8.5) containing DNase I, RNase A (Sigma), and EDTA-free protease inhibitor tablets (Roche). Cells were disrupted at 25 kPSI in a cell disruptor (Constant Systems), and the lysate was cleared by centrifugation at 35,000 rpm in a 45 Ti rotor (Beckman) for 25 min at 4 °C. The cleared lysate was loaded onto a Q Sepharose HiLoad HP column (GE Healthcare), which was then washed with stepwise increases of NaCl in buffer A: 0, 100, 150, 250, 300, and 500 mM. The eluates were analyzed by SDS/PAGE, and the fractions containing BtubAB (mainly at 100 and 150 mM NaCl) were pooled and concentrated in Centriprep concentrators [30 kDa molecular weight cutoff (MWCO), Millipore]. The concentrated proteins were loaded onto a Sephacryl S300 26/60 column (GE Healthcare) equilibrated with buffer B (20 mM Tris/HCl, 1 mM sodium azide, 1 mM EDTA, pH 7.5). Fractions containing pure BtubAB as determined by SDS/PAGE were concentrated in Centriprep concentrators (30 kDa MWCO, Millipore) to 20 mg/mL and flash frozen in liquid nitrogen.
P. dejongeii BtubC (PdBtubC, MGSSHHHHHHSSGLLPRGSH-1-257) was expressed in C41(DE3) cells the same way as described above for BtubAB. For purification, the entire pellet was resuspended in 250 mL of buffer C [50 mM Tris/HCl, 200 mM NaCl, 20 mM imidazole, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 1 mM sodium azide, pH 8.0] supplemented with DNase I, RNase A (Sigma), and EDTA-free protease inhibitor tablets (Roche). Cells were disrupted at 25 kPSI in a cell disruptor (Constant Systems), and the lysate was cleared by centrifugation at 35,000 rpm in a 45 Ti rotor (Beckman) for 25 min at 4 °C. The cleared lysate was loaded onto a 5-mL HisTrap HP column (GE Healthcare) and was washed with stepwise increases of imidazole in buffer C: 0, 50, 100, 200, 500, and 1,000 mM. The elution was analyzed by SDS/PAGE, and the fractions containing PdBtubC (mostly at 100 mM imidazole) were pooled and concentrated in Centriprep concentrators (30 kDa MWCO, Millipore). The concentrated proteins were loaded onto a Sephacryl S200 26/60 column (GE Healthcare) equilibrated with buffer D (50 mM Tris/HCl, 200 mM NaCl, 1 mM TCEP, 1 mM sodium azide, 1 mM EDTA, pH 8.0). Fractions containing pure PdBtubC as determined by SDS/PAGE were concentrated in Centriprep concentrators (30 kDa MWCO, Millipore) to 10 mg/mL and flash frozen in liquid nitrogen.
PvBtubC (1-256-KLHHHHHH) was also expressed in C41(DE3) cells as described above for BtubAB. The entire pellet was resuspended in 200 mL of buffer E [50 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES)/NaOH, 500 mM NaCl, 20 mM Imidazole, 1 mM TCEP, 1 mM sodium azide, 5% (vol/vol) glycerol, pH 9.0] supplemented with DNase I, RNase A (Sigma), and EDTA-free protease inhibitor tablets (Roche). Cells were disrupted at 25 kpsi in a cell disruptor (Constant Systems), and the lysate was cleared by centrifugation at 35,000 rpm in a 45 Ti rotor (Beckman) for 25 min at 4 °C. The cleared lysate was loaded onto a 5-mL HisTrap HP column (GE Healthcare) and was washed with stepwise increases of imidazole in buffer E: 0, 30, 100, 200, 500, and 1,000 mM. Eluates were analyzed by SDS/PAGE, and the fractions containing BtubC (mostly at 100 and 200 mM imidazole) were pooled and concentrated in Centriprep concentrators (30 kDa MWCO, Millipore). The concentrated proteins were loaded onto a Superdex 75 16/60 (GE Healthcare) equilibrated with buffer F [50 mM CHES/NaOH, 500 mM NaCl, 1 mM TCEP, 1 mM EDTA, 5% (vol/vol) glycerol, pH 9.0]. Fractions containing pure BtubC as determined by SDS/PAGE were concentrated in Centriprep concentrators (30 kDa MWCO, Millipore) to 20 mg/mL and flash frozen in liquid nitrogen.
Pelleting Assays.
Each reaction contained 15 μM PdBtubAB (15 µM heterodimer: 15 μM BtubA and 15 μM BtubB coexpressed and copurified); 0, 3, 15, or 60 μM PdBtubC in buffer containing 50 mM Hepes/NaOH (pH 7.5); 10 mM magnesium acetate; 100 mM potassium acetate; and 1 mM EGTA in a total volume of 120 μL. At the beginning, a 10-μL sample was taken from each reaction. For prespinning, all of the reactions were centrifuged at 50,000 rpm for 10 min at 10 °C in a TLA100 rotor (Beckman), and a 10-μL sample was taken from the supernatant of each reaction. The supernatants after the prespin were used in subsequent pelleting assays, induced by the addition of 1 mM GTP, and after 10 min of incubation, at room temperature. The reactions were then centrifuged at 50,000 rpm for 20 min at 10 °C in a TLA100 rotor (Beckman). The supernatant from each reaction was removed, and a sample of 10 μL was taken. Each pellet was resuspended in 100 μL of 0.1% (wt/vol) SDS solution, and a sample of 10 μL was taken. All of the 10-μL samples were analyzed by SDS/PAGE after adding the same amount of loading buffer.
CryoET Sample Preparation and Imaging.
PdBtubAB was polymerized before being deposited on EM grids. The polymerization reaction contained 40 μM BtubAB (heterodimer: 40 μM BtubA and 40 μM BtubB coexpressed and copurified) in buffer containing 50 mM Hepes/NaOH (pH 7.7), 5 mM magnesium acetate, 100 mM potassium acetate, and 1 mM EGTA in a total volume of 50 μL. The reaction was induced by the addition of 1 mM GTP and was left for 10 min to incubate at room temperature. Protein A conjugated with 10-nm colloidal gold (Cell Microscopy Centre, Utrecht University) was added to the polymerization reaction at a ratio of 3:7 (gold:reaction, vol/vol) before 2.5 μL of the reaction was pipetted onto freshly glow-discharged Cu/Rh Quantifoil R2/2 200 mesh holey carbon grids (Quantifoil). The grids were blotted for 1.5 s with a blotting force of −15, a drain time of 0.5 s, and were flash frozen in liquid nitrogen-cooled liquid ethane by using an FEI Vitrobot Mark IV. The Vitrobot chamber was set to 10 °C and 100% relative humidity. Electron cryotomographic data were collected on an FEI Krios microscope operated at 300 kV, and equipped with a Quantum energy filter (Gatan), which was set to 20 eV slit width around the zero loss peak. Data were acquired by using SerialEM software on a K2 direct electron detector in counting mode at a calibrated pixel size of 2.17 Å. Tilt series were typically collected ±60° with 3° tilt increment at 2.5- to 3-μm underfocus with a combined dose of approximately 80 e/Å2 over the entire tilt series.
CryoET Data Analysis and Subtomogram Averaging.
Tilt series were aligned and reconstructed in three dimensions by using the IMOD package. Subtomograms along BtubAB filaments were selected by using described methods (25). Each subtomogram was extracted as a 3D volume by using RELION (11), with a box size of 160 × 160 × 160 voxels. CTF estimation for each subtomogram was conducted with CTFFIND (26). Finally, 3,338 particles (volumes) were used for RELION subtomogram averaging refinement (27). The resulting model was used to determine the helical parameters of BtubAB filaments by visualization in UCSF CHIMERA (28), and the refined model was also used as the initial reference for the high-resolution BtubAB filament helical reconstruction.
CryoEM Sample Preparation and Imaging.
BtubAB was polymerized and grids for cryoEM with helical reconstruction were prepared in the same way as for cryoET (above), except that fiducial gold was omitted. For BtubABC cryoEM sample preparation, 40 μM PdBtubAB (40 µM heterodimer, 40 μM BtubA, and 40 μM BtubB coexpressed and copurified, untagged) and 45 μM PdBtubC (tagged) were polymerized in 50 μL of buffer containing 50 mM Hepes/NaOH (pH 8.0), 5 mM magnesium acetate, 100 mM potassium acetate, 200 mM NaCl, 1 mM EGTA, and 1 mM GTP. After 10 min incubation at room temperature, the polymerization reaction was gently mixed, and 2 μL of the reaction was pipetted onto freshly glow-discharged Quantifoil Au R2/2 holey carbon 200 mesh grids (Quantifoil). The grids were blotted for 3.5 s with a blotting force of −15, a drain time of 0.5 s, and were flash frozen in liquid nitrogen-cooled liquid ethane by using an FEI Vitrobot Mark IV. The Vitrobot chamber was set to 10 °C and 100% humidity. CryoEM data collection for both BtubAB and BtubABC filaments followed the same protocol. Micrographs of BtubAB or BtubABC filaments were collected on a FEI Tecnai G2 Polara microscope operating at 300 kV. Data were acquired on a Falcon III direct electron detector prototype in integrating mode at 30 frames per s and at a calibrated pixel size of 1.34 Å and a total dose of approximately 40 e/Å2 by using the automated acquisition software EPU (FEI). Images were collected at 1.5–3.0 μm underfocus over 1.5 s.
Helical Reconstruction of BtubAB and BtubABC Filaments, Model Building, and Refinement.
For BtubAB filament reconstruction, 1,394 movies were used from two separate 24-h microscope sessions. Movie frames were combined into images after initial whole-image motion correction with MOTIONCORR (29), and CTF parameters were estimated by using GCTF (30). All further processing was performed with RELION (31). A few hundred helical segments were picked manually in boxes of 280 pixels and 2D classified. A few good classes were selected for automatic template-based autopicking in RELION with the helical option enabled so that the filaments were picked reliably. Overlapping segments 40 Å apart were picked. The autopicking algorithm also tracked which filaments individual particles were derived from, to be able to create two half sets for FSC determination that separate only complete filaments. The autopicking process yielded ∼300,000 particles, which were assessed by one round of 2D classification, yielding 263,972 particles. Using rough pseudohelical parameters of the BtubAB filaments as derived manually from the cryoET reconstruction (averaging BtubA and BtubB and ignoring the seam) of twist = −90° and rise = 10 Å, a 3D refinement with helical averaging was performed. The cryoET reconstruction was used as the reference structure, filtered to 30 Å. The helical parameters were refined in RELION to twist = −90.7° and rise = 10.4 Å. No mask was used during 3D refinement. In postprocessing, the resulting two half maps were used to assess final resolution by gold standard FSC, combined, masked, and a sharpening B factors was applied, yielding a final map at 4.2 Å resolution (FSC 0.143 criterion) (Fig. S1).
For BtubABC filament reconstruction, 6,105 movies were used from four separate 24-h microscope sessions. As above, whole-image motion correction and CTF estimation were performed and initial classes for helical autopicking in RELION were produced. The same box size of 280 pixels was used. Autopicking yielded 345,777 particles, this time picked 80 Å apart. Two-dimensional classification was used to select the best particles from these autopicked particles, yielding 257,656 particles that were used in 3D refinement. The initial helical parameters were twist = −5.6 ° (−0.7° × 4 × 2) and rise = 80.0 Å (∼40 Å × 2) that describe symmetry along one protofilament only, not averaging BtubA and BtubB and preserving the seam, but not using all symmetry available in the structure because the four protofilaments were not averaged. Helical parameters refined to twist = −5.54° and rise = 79.31 Å. Subsequent movie refinement in RELION and particle polishing yielded particles with higher signal-to-noise ratios because the final reconstruction and postprocessing (masking central 30% of the map for determination of overall parameters) produced the highest resolution map at 3.6 Å, as assessed by the gold standard FSC procedure implemented in RELION (0.143 FSC criterion) (Fig. S3). For atomic fitting and refinement, a central portion of the map, covering BtubAB (not BtubC because of significantly weaker density) of the cryoEM filament density was cut out by using REFMAC and fitted with previous crystal structures of BtubA and BtubB [Protein Data Bank (PDB) ID code 2BTQ]. Assignment of BtubAB to their corresponding parts of the density map was done through manual inspection of side chain densities and was clear (Figs. S3 and S4). The atomic model was manually adjusted with MAIN (32), and subsequent refinement against the cryoEM density was performed with REFMAC (33) in reciprocal space after back-transforming the cutout density into structure factors (REFMAC SFCALC mode) (34). Model quality was assessed by using the standard R factor and further in MOLPROBITY (35) for stereochemical plausibility. Statistics are summarized in Table S1. To produce an atomic model of the entire BtubABC filament, four BtubAB heterodimers and the PvBtubC crystal structure (below) were fitted into the cryoEM densities of the four protofilaments by using CHIMERA (28) and then expanding helical symmetry (twist = −5.54° and rise = 79.31 Å).
BtubC Crystallization, Data Collection, and Structure Determination.
Initial crystallization conditions for PvBtubC were found by using our in-house nanolitre crystallization facility (36) using 20 mg/mL BtubC in 50 mM CHES/NaOH, 500 mM NaCl, 1 mM TCEP, 5% glycerol, pH 9.0 and 100 + 100-nL sitting drops in MRC crystallization plates at room temperature. After optimization, the best crystals were obtained in drops containing 5–8.1% wt/vol PEG 8000, 5–8.1% wt/vol PEG 1000, 200 mM lithium sulfate, 100 mM Tris/acetic acid at pH 8.5. Crystals were cryoprotected by using 30% vol/vol glycerol and flash frozen in liquid nitrogen for data collection. A native dataset to 2.5 Å was collected on beamline ID29 at the European Synchrotron Radiation Facility (ESRF) synchrotron (Table S1). For phasing, a mercury derivative was prepared by soaking BtubC crystals in 5% wt/vol PEG 8000, 5% wt/vol PEG 1000, 420 mM lithium sulfate, 100 mM Tris/acetic acid, 600 mM NaCl, pH 8.5 supplemented with 0.5 mM thiomersal (EMTS) for 2 h with subsequent cryoprotection (30% vol/vol glycerol) and freezing in liquid nitrogen for data collection. A derivative dataset was collected to 2.8 Å resolution on beamline ID30B at the ESRF synchrotron (Table S1). The structure was solved by SAD with CRANK2 (37), leading to a rough model describing two BtubC chains in the asymmetric unit of the crystals. The initial model was refined against the native data at 2.5 Å resolution in REFMAC (33), and the resulting phases were used for further rounds of automated model building with BUCCANEER (38) and REFMAC refinement until a complete model was obtained. The model was adjusted by using MAIN (32) and refined with REFMAC and PHENIX (39) in several rounds.
Coordinate and Map Depositions.
Coordinates and structure factors of the P. vanneervenii BtubC crystal structure were deposited with PDB ID code 5O01. The BtubABC filament structure was deposited with PDB ID code 5O09, and the corresponding 3.6 Å cryoEM 3D map was deposited in the EM Data Bank with accession no. EMD-3726.
Polymerization and Labeling of PdBtubAB for TIRF Microscopy.
Surface exposed lysine residues were used to label BtubAB filaments by ATTO dyes (Jena Bioscience and Pierce). Filament labeling with fluorescent dyes was performed by adopting procedures for tubulin (40). After affinity purification or gel filtration, BtubAB was cycled through polymerization and depolymerization. Filaments were polymerized by the addition of 2 mM GTP and 5 mM MgCl2 for 15 min. The reaction was centrifuged at 100,000 × g in a TLA 100 rotor (Beckman Coulter) for 10 min at 20 °C. The supernatant was discarded, and the pellet resuspended and allowed to depolymerize in buffer (80 mM Pipes/KOH, pH 6.9, 1 mM EGTA, 0.5 mM EDTA) at 37 °C or room temperature containing no nucleotide until the solution turned clear. Then, at a ratio of 1:2 (protein:dye), cycled BtubAB was incubated with dye in buffer L (80 mM Pipes/KOH, 75 mM potassium acetate, pH 7.5, 1 mM EGTA, 0.5 mM EDTA, 0.2 mM TCEP) in the presence of 5 mM MgCl2 and 2 mM GTP to induce polymerization (30 min to 1 h). The reaction was quenched by using an equal volume of buffer L, supplemented with 75 mM potassium glutamate, and again 2 mM GTP was added. After 1 min, BtubAB filaments were separated from nonconjugated dye and monomeric BtubAB by centrifugation at 100,000 × g for 20 min over a 20% glycerol cushion (buffer L with 75 mM potassium glutamate) by using a Beckman TLA 100 rotor. After aspirating the supernatant, the pellet was washed carefully with and resuspended in buffer R (80 mM Pipes/KOH, pH 6.9, 1 mM EGTA, 1 mM EDTA, 0.2 mM TCEP) by using a cut 200-µL pipette tip. Subsequently filaments were allowed to disassemble, and protein aggregates were removed by an additional spin at 200,000 × g for 10 min. After depolymerization, the protein to label ratio was determined by using a NanoDrop ND-1000 Spectrophotometer. Protein concentrations were determined by using the calculated extinction coefficients.
In Vitro Single Filament Polymerization Assays.
Microscope chambers were constructed from two silanised coverslips that were sandwiched and separated by spacers of Parafilm as described (41, 42). Channels were incubated with anti-biotin antibody diluted 1:100 (Sigma) in PBS for 3 min for assays by using stabilized BtubAB seeds followed by two sequential washes with 40 µL of PBS. Generally, surfaces were blocked for 30 min up to 1 h with 1% Pluronic F127 (Sigma) in PBS after functionalization with antibodies. Then channels were rinsed with 100 µL of motility buffer MB (BRB80, 80 mM Pipes/KOH, pH 6.9, 5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 0.2 mM TCEP, 0.01% Tween 20, 2 mg⋅mL−1 BSA without and supplemented with 75 mM potassium acetate unless stated differently). Before imaging, channels were equilibrated in 20 µL of motility buffer (buffer MB), containing an oxygen scavenging reaction system composed of 40 mM glucose, 40 µg⋅mL−1 glucose oxidase, 16 µg⋅mL−1 catalase, 1 mM DTT, 1 mM GTP. Labeled BtubAB alone and with BtubC in buffer MBG (buffer MB plus anti-fade, 2 mM GTP and 0.2% methylcellulose, 400 centipoise) was added in two subsequent steps of 15 µL by using Atto488 or Atto655 labeled BtubAB at different concentrations and labeling ratios (1:14 and 1:3 labeled:unlabeled). All quantifications were made from experiments that used low BtubAB labeling ratios. High labeling ratios affected growth and nucleation because filaments were observed only at slightly higher concentrations of 3–4 µM instead of 2–3 µM. Also, note the slower elongation from seeds in experiments with 2 µM BtubAB (Figs. S7 and S8) in comparison with Figs. 3D and 4A.
Fig. S8.
Strongly fluorescently labeled BtubAB dynamics and BtubC-dependent stabilization in 100 mM KCl. Dual-color kymographs of filaments emanating from stabilized seeds grown alone (A) and in the presence of BtubC (B). BtubAB concentration is the same as used for quantification and shown in Fig. 3. Here, BtubAB is labeled 1:3, and experiments were performed under different salt concentration (100 mM KCl). Still, stabilization occurs. Note that growth seems to be affected as indicated by the shorter extensions formed from seed compared with Fig. 3 and generally slower growth velocities. Images were acquired by using the same settings as described in Materials and Methods.
Seeded polymerization.
In general, Atto488-labeled BtubAB filaments were grown from Atto655-marked BtubAB seeds in buffer MBG. In experiments with seeded polymerization, to demonstrate treadmilling and polarity, Atto655-marked BtubAB filaments were preassembled within the reaction chamber while imaging. After sufficient formation of at least 20 filaments per field of view, a careful wash containing Atto488-labeled BtubAB was done while imaging. The wash replaced nonpolymerized Atto655-labeled BtubAB but also some filaments because they were not immobilized.
Immobilized and stable BtubAB filament seeds.
Buffer MBG contained only 0.05% methylcellulose to keep extensions with their tips close to the field of view and to help with washing and buffer exchanges. First, BtubAB filaments were assembled from Atto655-labeled BtubAB and biotinylated BtubAB (labeled with biotin) at a ratio of 20 nonbiotinylated Atto655-labeled BtubAB molecules to 1 biotinylated one (40 µM final) in buffer MB containing 1 mM GMPCPP, a slowly hydrolysable GTP analog, for approximately 1 h. After functionalizing the reaction chamber with antibodies against biotin and blocking as above, 20 µL of biotinylated, Atto655-labeled seeds (diluted at least 200×) in buffer MBG (containing 2 mM GTP instead of 0.2 mM GMPCPP) were allowed to bind for a maximum of 2 min. Incubation times depended on the seed dilution, e.g., at 200× diluted seeds a short flush for approximately 30 s was sufficient to obtain good fields. Afterward unbound seeds and remaining GMPCPP were removed by additional washes with 100 µL of buffer MBG. Seed binding to the surface was checked under the microscope to ensure sufficient coverage. Finally, polymerization buffer MBG containing monomeric Atto488-labeled BtubAB was added. Imaging commenced immediately after that. All microscope-based assays were conducted at 24 ± 1 °C.
TIRF Imaging.
Images and time series of dynamic BtubAB filaments were recorded on a back illuminated EMCCD camera (iXonEM+ DU‐897E, Andor Technology) by using an inverted fluorescence microscope (Nikon) equipped with a 100× Nikon, 1.49 N.A. oil, APO TIRF objective and controlled by μManager (https://micro-manager.org/wiki/Micro-Manager). For excitation of filaments in TIRF, 488-nm and 561-nm lasers were used (both 150 mW, Coherent Sapphire, Coherent Cube). Images were acquired sequentially in multicolor experiments, by switching emission filters between GFP and Cy3 (Chroma Technology). Acquisition intervals varied from 3 to 5 s between images with exposures of 50–100 ms.
TIRF Image Analysis.
Measurement of velocities.
For all quantifications, the first 30 min of experiments were analyzed. The ImageJ plugin “Kymograph” by Jens Rietdorf and Arne Seitz was used to determine growth velocity. A line of three-pixel width was drawn manually originating from individual seeds and covering the corresponding filament extension and subsequently used to generate the kymograph. The elongation over time yields a slope revealing the average growth velocity of an individual plus end. Growth velocities for BtubAB alone and in presence of unlabeled BtubC were obtained from Atto488 extensions for different BtuBAB and BtubC concentrations as indicated in the experiments. Only free ends were considered for the analysis.
Quantification of catastrophe times and length.
Filaments were counted when they started growing as a point-like signal from an immobilized seed elongating into a filament. Catastrophe time and length were obtained from filament extensions containing a defined growing end. The time a filament spends growing until its sudden disappearance is the catastrophe time or lifetime, whereas the length is the distance the filament grew. Both were measured manually in kymographs by drawing a line along the x axis to determine the final extension before shrinkage (that is disappearance of the signal representing the dynamic filament extension) and along the y axis to obtain the time required to form the extension (with a cut off of three frames for 5-s intervals and five frames in case of 3-s intervals). After each catastrophe, a filament was considered to restart a growth shrinkage cycle de novo. The new growth shrinkage cycle happened at the end of stabilized seeds but also after a rescue on nonstabilized extensions (note this regrowth is especially the case in the presence of BtubC). A new event was defined through a phase of rapid shortening (when shortened by a minimum of 5–10 pixels in two of the subsequent images for long filaments in the presence of BtubC) followed by filament growth as indicated through a clear slope from the plus end over five subsequent images. Thus, fluctuations caused by thermal motion of the growing filament tips in and out of the field of view could be avoided. An example that we could detect short extensions is shown in Fig. S7. All data were plotted and analyzed by using Prism (GraphPad). Analysis of filament lifetimes by using catastrophe time was conducted in analogy to Gardner et al. (19). According to Odde et al. (43, 44), they can be described by using a gamma distribution where the predicted number of catastrophes at a given time t are
Here, n is the number of steps required to induce catastrophe, and r is the rate at which such a step occurs. Cumulative distributions were generated by using the tabulate exact cumulative frequency option in Prism (GraphPad Software). Fits of the gamma distribution to the experimental data were performed in Prism by using the regular fit option with least squared analysis. The cumulative density distribution of the gamma function was applied by using the implemented incomplete and complete gamma function (Y = 1 − igamma(n,k × x)/gamma(n)) keeping n either constant n = 1 or variable starting at an initial value of 3. When n = 1, the gamma function becomes exponential (exponential decay), such as for single-step processes where catastrophes are equally likely to occur. Obtained parameters and goodness of fits are shown in Fig. S6.
Supplementary Material
Acknowledgments
We thank Minmin Yu (Medical Research Council Laboratory of Molecular Biology) for help with synchrotron data collection and Marija Zanic (Vanderbilt University) for helpful discussions regarding catastrophe analysis. This work was funded by Medical Research Council Grant U105184326 (to J.L.) and Wellcome Trust Grant 095514/Z/11/Z (to J.L.). X.D. is a recipient of a Boehringer Ingelheim Fonds PhD Fellowship, G.F. was recipient of German Academy of Sciences Leopoldina Fellowship LPDS 2013-06, and T.A.M.B. is recipient of European Molecular Biology Organization Advanced Fellowship aALTF 778-2015.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5O01 and 5O09), and Electron Microscopy Data Bank, emdatabank.org (EMDB accession code EMD-3726).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705062114/-/DCSupplemental.
References
- 1.Amos L, Klug A. Arrangement of subunits in flagellar microtubules. J Cell Sci. 1974;14:523–549. doi: 10.1242/jcs.14.3.523. [DOI] [PubMed] [Google Scholar]
- 2.Brinkley W. Microtubules: A brief historical perspective. J Struct Biol. 1997;118:84–86. doi: 10.1006/jsbi.1997.3854. [DOI] [PubMed] [Google Scholar]
- 3.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins C, et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci USA. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlieper D, Oliva MA, Andreu JM, Löwe J. Structure of bacterial tubulin BtubA/B: Evidence for horizontal gene transfer. Proc Natl Acad Sci USA. 2005;102:9170–9175. doi: 10.1073/pnas.0502859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pilhofer M, Jensen GJ. The bacterial cytoskeleton: More than twisted filaments. Curr Opin Cell Biol. 2013;25:125–133. doi: 10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akendengue L, et al. Bacterial kinesin light chain (Bklc) links the Btub cytoskeleton to membranes. Sci Rep. 2017;7:45668. doi: 10.1038/srep45668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Song YH, Mandelkow E. Recombinant kinesin motor domain binds to beta-tubulin and decorates microtubules with a B surface lattice. Proc Natl Acad Sci USA. 1993;90:1671–1675. doi: 10.1073/pnas.90.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sosa H, Milligan RA. Three-dimensional structure of ncd-decorated microtubules obtained by a back-projection method. J Mol Biol. 1996;260:743–755. doi: 10.1006/jmbi.1996.0434. [DOI] [PubMed] [Google Scholar]
- 16.Pilhofer M, et al. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel Two-Step Gene Walking method. Nucleic Acids Res. 2007;35:e135. doi: 10.1093/nar/gkm836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: Rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Sontag CA, Staley JT, Erickson HP. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J Cell Biol. 2005;169:233–238. doi: 10.1083/jcb.200410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Bassam J, et al. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fourniol FJ, et al. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Galiano AJ, et al. Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J Biol Chem. 2011;286:19789–19803. doi: 10.1074/jbc.M111.230094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 27.Bharat TA, Russo CJ, Löwe J, Passmore LA, Scheres SH. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69:1342–1357. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88:311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Pannu NS, et al. Recent advances in the CRANK software suite for experimental phasing. Acta Crystallogr D Biol Crystallogr. 2011;67:331–337. doi: 10.1107/S0907444910052224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 40.Hyman AA. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J Cell Sci Suppl. 1991;14:125–127. doi: 10.1242/jcs.1991.supplement_14.25. [DOI] [PubMed] [Google Scholar]
- 41.Fink G, et al. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 42.Gell C, et al. Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 43.Odde DJ, Cassimeris L, Buettner HM. Kinetics of microtubule catastrophe assessed by probabilistic analysis. Biophys J. 1995;69:796–802. doi: 10.1016/S0006-3495(95)79953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odde DJ, Buettner HM. Autocorrelation function and power spectrum of two-state random processes used in neurite guidance. Biophys J. 1998;75:1189–1196. doi: 10.1016/S0006-3495(98)74038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeytuni N, et al. MamA as a model protein for structure-based insight into the evolutionary origins of magnetotactic bacteria. PLoS One. 2015;10:e0130394. doi: 10.1371/journal.pone.0130394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pernigo S, Lamprecht A, Steiner RA, Dodding MP. Structural basis for kinesin-1:cargo recognition. Science. 2013;340:356–359. doi: 10.1126/science.1234264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.