Significance
Allosteric regulation is a crucial component in biochemical pathways, where ligand binding to an allosteric site modulates the enzymatic activity at a distant functional site. There is rapid growth in interest toward rational design of allosteric drugs. PDZ domain proteins (frequently encountered in signaling protein complexes) are classic examples of allostery without significant structural changes (dynamic allostery), where ligand binding leads to modulation in distal side-chain motions. So far dynamic allostery has been attributed to purely entropic effects. In contrast, here we show that a significant internal redistribution and rewiring of electrostatic interactions provide the basis of dynamic allostery in PDZ domain protein. A dramatic population shift in the pairwise hydrogen-bonded interactions involving side chains leads to the allosteric response.
Keywords: PDZ domain, dynamic allostery, population shift, molecular dynamics, electrostatic interactions
Abstract
Allosteric effect implies ligand binding at one site leading to structural and/or dynamical changes at a distant site. PDZ domains are classic examples of dynamic allostery without conformational changes, where distal side-chain dynamics is modulated on ligand binding and the origin has been attributed to entropic effects. In this work, we unearth the energetic basis of the observed dynamic allostery in a PDZ3 domain protein using molecular dynamics simulations. We demonstrate that electrostatic interaction provides a highly sensitive yardstick to probe the allosteric modulation in contrast to the traditionally used structure-based parameters. There is a significant population shift in the hydrogen-bonded network and salt bridges involving side chains on ligand binding. The ligand creates a local energetic perturbation that propagates in the form of dominolike changes in interresidue interaction pattern. There are significant changes in the nature of specific interactions (nonpolar/polar) between interresidue contacts and accompanied side-chain reorientations that drive the major redistribution of energy. Interestingly, this internal redistribution and rewiring of side-chain interactions led to large cancellations resulting in small change in the overall enthalpy of the protein, thus making it difficult to detect experimentally. In contrast to the prevailing focus on the entropic or dynamic effects, we show that the internal redistribution and population shift in specific electrostatic interactions drive the allosteric modulation in the PDZ3 domain protein.
Allosteric regulation of proteins plays a key role in physiological cell functions, biochemical and signal transduction pathways, and drug discovery (1–3). It has remained a challenge to understand how the thermodynamic perturbation caused by ligand binding at one site would propagate and modulate the structure and dynamics of distal regions of proteins. The prevailing models of structure-based allostery (4, 5) do not apply to the more recent examples of allostery without conformational change such as PDZ domain (6), CAP dimer (7), and met repressor (8). These examples have triggered the concept of dynamic allostery, where the side-chain dynamics is modulated on ligand binding and the origin has often been attributed to changes in the conformational entropy (9, 10). The modern view of allostery invokes a thermodynamic picture, where a population shift among preexisting conformational states occurs on binding the allosteric effector (11–13). It has also been suggested in the context of allostery without conformational change that “not observed does not imply that it is not there” (10) because crystallographic techniques may not resolve the relatively minor population shifts. An interesting idea has emerged that all proteins might be allosteric in nature (14).
PDZ domain has been a classic model system to study single domain allostery without major structural changes (9, 15, 16) (Fig. 1A). PDZ domains are evolutionary conserved protein–protein interaction modules associated with the cellular signaling and are implicated in localization of membrane receptors and ion channels (17, 18). They can dimerize with other modular protein domains (e.g., WW, SH2, SH3, PH, etc.) or can bind specific recognition sequences at the C terminus of proteins in a hydrophobic groove between β2 and α2 regions (17, 19). The signature of the allosteric effects observed in PDZ domains has been purely in terms of the dynamics of the side chains. Solution NMR studies (20, 21) as well as molecular dynamics (MD) simulations (22, 23) have confirmed that binding the effector ligand leads to substantial modulation of side-chain dynamics in the PDZ domain. In particular, Lee and coworkers have unearthed a hidden dynamic allostery in the PDZ3 domain, where deletion of a noncanonical distal α3 helix domain reduces the ligand binding affinity by 21 times (6). Their work has highlighted the role of differential side-chain motions toward the allosteric response and attributed the origin of dynamic allostery to purely entropic effects because the enthalpic contribution toward change in binding affinity on α3 helix deletion was minimal. Interestingly, they have also hinted toward a possibility that internal structural adjustments could lead to cancellations in individual changes of enthalpy, which will be demonstrated to be the case in our work.
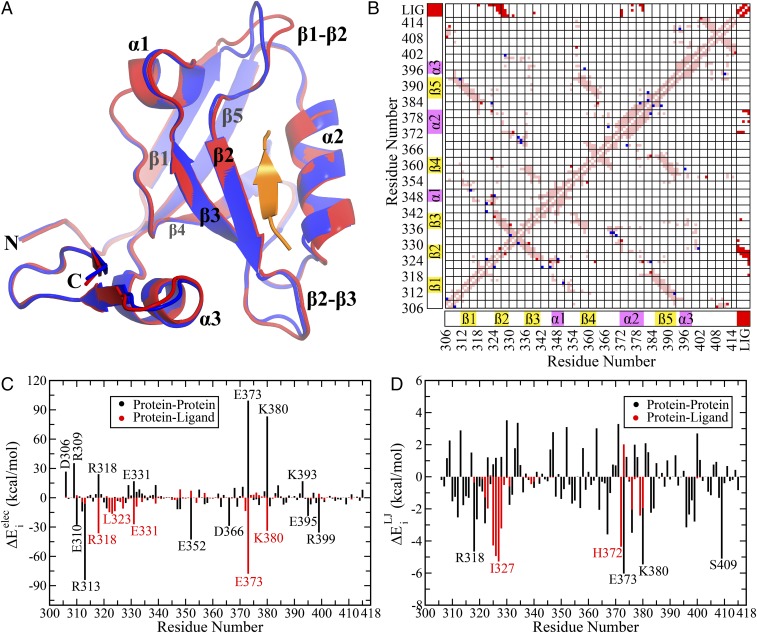
Fig. 1.
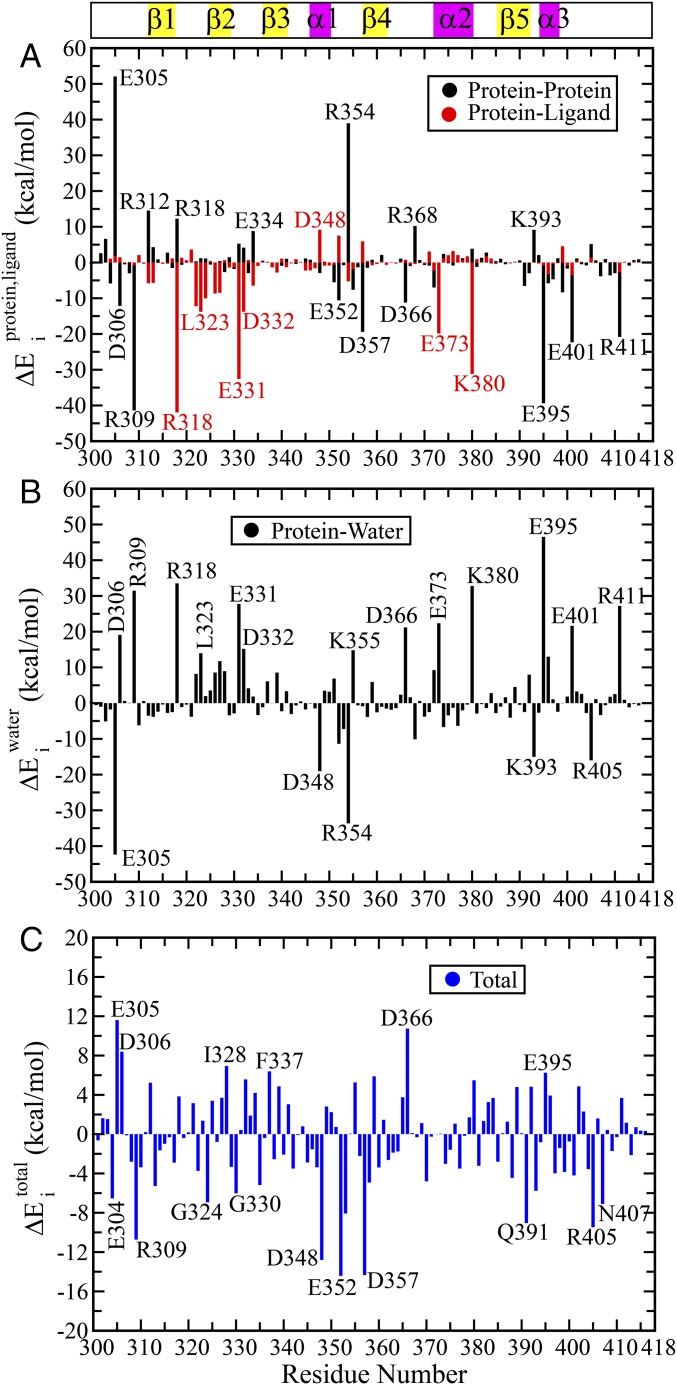
(A) Superposition of the structures after energy minimization of the respective crystal structures in the unbound and bound states (PDB IDs: 1BFE and 1BE9, respectively). Bound and unbound states are colored in red and blue, respectively. The peptide ligand is highlighted in orange. (B) Differential contact map between the bound and unbound states (energy minimized structures). Contacts unique in bound (including the contacts with the ligand) and unbound states are shown in red and blue, respectively. The common contacts are shown in light pink. The secondary structural elements have been highlighted along both axes as visual guides. (C) Electrostatic (Coulomb) and (D) van der Waals’ (Lennard-Jones) components of the residuewise change in total interaction energy between the bound and unbound states (). A few residues with significant change between the two states have been marked on each figure. The due to interaction with ligand () and other protein residues () have been marked in red and black, respectively. Numerical values are provided in Table S1.
Prior simulation studies have attempted to understand the allosteric communication pathways in terms of correlations in structural or energetic fluctuations in PDZ domain (24, 25). Karplus and coworkers have revealed two continuous correlation pathways in a PDZ2 domain and highlighted the existence of such pathways even in the absence of the ligand (24). A number of theoretical (23, 26, 27) and experimental (6, 28) studies propose multiple allosteric pathways for PDZ domains on the basis of evolutionary information (29, 30), local structural changes (22, 31, 32), heat-diffusion pathways (33, 34), and energy connectivity networks (35, 36). Most of the existing approaches look for correlated motions or energy fluctuations to characterize the allosteric effects. Although such correlations may demonstrate the effect of allostery, they do not explain the specific origin of such coupling. It has also been debated that the statistical analysis of evolutionarily coupled residues may not be true reporter of functional coupling because the evolutionary information does not include the molecular details of the interactions (31, 37). Thus, a molecular thermodynamic approach that uses the perturbations in the nonbonded interactions on ligand binding would provide a more direct view of the functional energetic coupling between the protein residues (38–40).
In this work, we argue that the protein dynamics is governed by the underlying energy landscape. Our objective is to understand the perturbation in the internal energy landscape of the allosteric protein due to ligand binding and how that manifests into functional changes in distal sites. To achieve this goal, we performed atomistic MD simulations both in the ligand bound and unbound states of a PDZ3 domain protein and compared the changes in nonbonded interactions energies for each residue as well as contributions from individual pairwise interactions. We track the large energetic perturbations on ligand binding as signature of allosteric effects. We show that despite the subtle structural changes, the protein–protein and protein–water electrostatic interactions undergo dramatic redistribution on ligand binding. We construct a residue-pairwise energetic perturbation network where the binding site and distal allosteric regions are connected by the noncanonical α3 helix, which has been suggested to play a significant role in the dynamic allostery in PDZ3 domain (6). Finally, we elucidate the molecular basis of this perturbation network to be a population shift between the pairwise hydrogen-bonded network involving the protein side chains. Based on our observations, we suggest a hidden energetic allostery driven by the population shift in specific electrostatic interactions and internal redistribution of nonbonded interactions to be the driving force behind the dynamic allostery in PDZ3 domain.
Results and Discussion
Side-Chain Rearrangement Leads to Dramatic Change in Energetics.
The basic premise of the dynamic allostery phenomenon has been the lack of structural changes between the ligand bound and unbound states as observed in the respective crystal structures. To be precise, the presence or absence of significant structural changes is evaluated based only on the backbone structure, and side chains are not invoked in this description in general. As a first step toward characterizing the structural features and differences (if any) between the available crystal structures for the PDZ3 domain with and without the ligand (PDB ID: 1BE9 and 1BFE, respectively), we performed energy minimization of the crystal structures while restraining the position of the backbone atoms. Only the side-chain atoms were allowed to move to investigate the significance of side-chain rearrangement on the interaction pattern. To maintain a consistent comparison, we used an identical length of the two protein structures (residues 306–415). Fig. 1A shows a superposition of the structures (bound and unbound) after energy minimization, which preserves the well-known characteristics of the crystal structures that there is almost no structural difference (in backbone) between the two states except minor rearrangement in the β1–β2 and β2–β3 loop regions.
Subsequently we turn our attention to the possible rearrangement of the side chains. Looking for differences in the residue pairwise contact map has been a popular choice for identifying the significant interaction pathways. After all, the nonbonded interactions are intimately coupled to the presence of contacts and their rearrangements. As a matter of fact, in the context of allosteric coupling it has been suggested that the residues that are in spatial proximity forming contacts are more likely to be coupled compared with the distal residues (37), and models have been proposed to construct a biophysical framework based on contacts acting as structural support for the propagation of information (41–43). We took a similar approach of identifying the possible differences in the side-chain contacts between the two end states by constructing the differential contact map (Materials and Methods) in Fig. 1B. The contacts present exclusively in the ligand bound and unbound states are marked in red and blue, respectively. This visual representation clearly indicates that majority of the pairwise contacts remain the same (light pink regions); whereas, there are a few interresidue contacts, which completely disappear or appear on ligand binding. Evidently the side-chain contact map reveals relatively minor structural rearrangement in the contact pattern.
Finally, we turn our attention to the arguably most fundamental parameter: nonbonded interaction energy. Ligand binding is likely to cause significant local energetic perturbation in the binding site of the protein, which should propagate through the intraprotein interaction network to the allosteric site. Thus, we have investigated the changes in the residuewise nonbonded interaction energy () with the rest of the protein (and ligand). Figs. 1 C and D present the electrostatic () and van der Waals’ () components of the change in total interaction energy of the ith residue on ligand binding. Remarkably, we find that there is a wide range of variation in the change of electrostatic energy () (up to kcal/mol) for both protein-only and ligand-only interactions. On the other hand, the changes in van der Waals’ interaction energy () show relatively modest range of variation up to kcal/mol. Evidently, the nonbonded interactions, particularly the electrostatic interaction energy, provide a highly sensitive probe toward identifying the subtle structural changes. Thus, even though the backbone and side-chain structural parameters (e.g., RMSD, cutoff-based contact map, etc.) show a minor change, the interaction energies show a substantial change between the two states and in our subsequent analysis we shall further demonstrate that the electrostatic interaction energy indeed captures the nature of allosteric coupling in the PDZ3 domain. We further emphasize that our analysis proves that indeed there is structural change at the side-chain level (even in the crystal structure) that can be significant in terms of energetics (Fig. S1 for a structural view of side-chain rearrangement between the bound and unbound states).
Fig. S1.
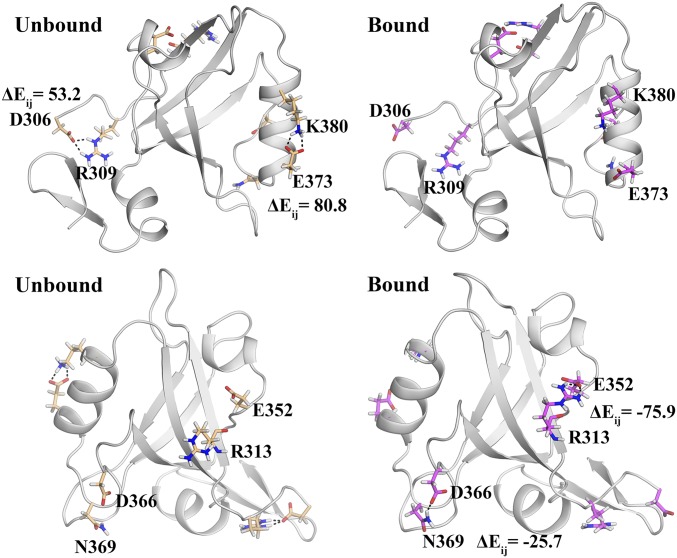
Energetic perturbation based on minimized crystal structures with identical backbone structure. The analysis shows large change in pairwise residue interaction energy () of certain residues on change in side-chain orientations. The figure highlights four such pairs of residues with large as a consequence of explicit hydrogen bond formation between the unbound and bound states.
The results presented in Fig. 1 confirm that the inherent energy landscapes of the bound and unbound states can be drastically different even when the backbone structures are almost identical. The subtle rearrangement and rewiring of the side-chain interactions can lead to dramatically different energetic coupling (Fig. S1 and Table S1). But the crystal structures may not be truly representative of respective structural ensemble in solution due to crystal packing and crystallization conditions. Moreover, a crystallographic view of single static structure often does not capture the biologically significant ensemble of conformations and dynamic fluctuations. Thus, to investigate the signatures of dynamic allostery in PDZ3 domain, we shall base our subsequent analyses on the MD simulation trajectories to capture the population of the significant interactions (between the side chains) that might lead to allosteric modulation.
Table S1.
The average interaction energy contribution due to various terms and comparison between the bound (B) and unbound (U) states using the energy minimized crystal structures for the unbound and bound state (PDB ID: 1BFE/1BE9)
| ResID | ||||||||||
| 306 | −95.0 | −68.3 | 26.7 | 1.1 | 27.9 | −5.6 | −5.8 | −0.2 | 0 | −0.2 |
| 307 | −2.9 | −4.1 | −1.2 | −0.2 | −1.4 | −10.6 | −11.2 | −0.6 | 0 | −0.6 |
| 308 | −13.8 | −13.9 | −0.1 | 0.1 | 0.0 | −6.8 | −5.6 | 1.2 | 0 | 1.2 |
| 309 | −140.4 | −105.0 | 35.4 | −0.6 | 34.8 | −7.9 | −5.6 | 2.3 | 0 | 2.3 |
| 310 | −9.8 | −37.5 | −27.7 | 2.6 | −25.0 | −6.6 | −8.1 | −1.5 | 0 | −1.5 |
| 311 | −6.7 | −5.3 | 1.4 | −0.1 | 1.3 | −8.8 | −10 | −1.2 | 0 | −1.2 |
| 312 | −169.9 | −183.9 | −14.0 | −4.6 | −18.6 | −11.5 | −14 | −2.5 | 0 | −2.5 |
| 313 | −71.5 | −155.7 | −84.2 | −6.3 | −90.5 | −10.1 | −7.2 | 2.9 | 0 | 2.9 |
| 314 | −23.1 | −21.6 | 1.4 | −0.4 | 1.1 | −18 | −19.8 | −1.8 | 0 | −1.8 |
| 315 | −11.9 | −9.0 | 2.9 | −0.4 | 2.5 | −10.6 | −11.8 | −1.2 | 0 | −1.2 |
| 316 | −11.7 | −15.6 | −3.9 | 0.0 | −3.9 | −17.8 | −19.1 | −1.3 | 0 | −1.3 |
| 317 | −14.8 | −11.1 | 3.7 | 0.2 | 3.9 | −10.4 | −8.9 | 1.5 | 0 | 1.5 |
| 318 | −115.4 | −91.2 | 24.2 | −36.4 | −12.2 | −15 | −19.7 | −4.7 | −0.4 | −5.1 |
| 319 | −10.3 | −6.6 | 3.7 | 0.5 | 4.2 | −0.8 | −3.4 | −2.7 | 0 | −2.7 |
| 320 | −3.1 | −7.9 | −4.8 | −0.1 | −4.9 | −1.3 | −1.9 | −0.6 | 0 | −0.6 |
| 321 | −6.5 | −17.5 | −10.9 | 1.6 | −9.3 | −3.6 | −4.5 | −0.9 | −0.1 | −1 |
| 322 | 1.0 | 1.0 | −0.1 | −14.8 | −14.9 | −0.9 | −3.8 | −2.9 | −1.3 | −4.2 |
| 323 | −4.1 | −6.9 | −2.8 | −16.4 | −19.1 | −18.1 | −16.9 | 1.2 | −2 | −0.8 |
| 324 | −3.9 | −3.1 | 0.8 | −13.8 | −13.1 | −3.9 | −4.4 | −0.5 | 0.1 | −0.4 |
| 325 | −5.6 | −8.9 | −3.3 | −1.7 | −5.0 | −19 | −18.2 | 0.8 | −4.3 | −3.5 |
| 326 | −15.5 | −18.5 | −3.0 | −4.4 | −7.4 | −10.9 | −9.7 | 1.2 | −4.9 | −3.7 |
| 327 | −6.1 | −5.0 | 1.1 | −11.1 | −10.0 | −15.8 | −16.6 | −0.8 | −5.3 | −6.1 |
| 328 | −10.8 | −16.2 | −5.4 | 0.0 | −5.3 | −14.5 | −13.3 | 1.2 | −3.2 | −2 |
| 329 | −28.2 | −15.3 | 12.9 | −1.4 | 11.6 | −7.9 | −8.1 | −0.1 | −0.5 | −0.6 |
| 330 | −12.4 | −10.1 | 2.3 | −0.7 | 1.6 | −9.1 | −5.6 | 3.5 | −0.1 | 3.4 |
| 331 | −65.3 | −48.4 | 16.9 | −27.2 | −10.3 | −5.5 | −7.2 | −1.7 | −0.6 | −2.3 |
| 332 | −16.2 | −10.2 | 6.0 | −9.1 | −3.1 | −1.6 | −2.7 | −1.1 | 0 | −1.1 |
| 333 | −10.1 | −1.5 | 8.6 | 0.5 | 9.1 | −1.7 | 0.1 | 1.8 | 0 | 1.8 |
| 334 | −68.7 | −64.4 | 4.3 | −4.4 | −0.1 | −5.4 | −2.1 | 3.4 | 0 | 3.4 |
| 335 | −9.0 | −6.8 | 2.2 | −0.6 | 1.6 | −8.8 | −8 | 0.7 | 0 | 0.7 |
| 336 | −12.2 | −14.8 | −2.6 | 0.5 | −2.1 | −18.4 | −19.4 | −1 | −0.1 | −1.1 |
| 337 | −19.9 | −17.8 | 2.2 | −0.2 | 2.0 | −25.1 | −24.9 | 0.2 | −0.1 | 0.1 |
| 338 | −18.6 | −15.8 | 2.8 | −1.3 | 1.5 | −21.7 | −21.9 | −0.2 | −0.1 | −0.3 |
| 339 | −23.9 | −10.9 | 13.0 | −6.0 | 6.9 | −9.9 | −10 | −0.2 | −0.5 | −0.7 |
| 340 | −6.1 | −6.4 | −0.4 | 1.1 | 0.7 | −13.6 | −14.3 | −0.7 | −0.3 | −1 |
| 341 | −5.6 | −4.0 | 1.6 | −1.3 | 0.3 | −17.9 | −17.6 | 0.3 | 0 | 0.3 |
| 342 | −5.4 | −6.2 | −0.8 | 0.2 | −0.6 | −9.2 | −9.6 | −0.4 | −0.1 | −0.5 |
| 343 | −11.2 | −11.3 | −0.1 | 0.6 | 0.5 | −4.3 | −4.1 | 0.2 | 0 | 0.2 |
| 344 | −14.5 | −14.0 | 0.5 | 0.2 | 0.7 | −2.1 | −2.3 | −0.2 | 0 | −0.2 |
| 345 | −8.5 | −7.8 | 0.6 | −2.2 | −1.6 | −3.5 | −3.8 | −0.3 | 0 | −0.3 |
| 346 | −3.1 | −3.8 | −0.7 | −2.2 | −3.0 | −14.9 | −13.2 | 1.7 | 0 | 1.7 |
| 347 | −6.2 | −17.9 | −11.8 | −1.9 | −13.7 | −11.8 | −9 | 2.8 | 0 | 2.8 |
| 348 | −44.0 | −55.7 | −11.7 | 8.6 | −3.1 | −7.5 | −8.8 | −1.2 | 0 | −1.2 |
| 349 | −6.2 | −5.9 | 0.3 | −0.8 | −0.5 | −7.7 | −7.5 | 0.2 | 0 | 0.2 |
| 350 | −19.0 | −19.8 | −0.8 | −0.9 | −1.7 | −2.9 | −4.3 | −1.4 | 0 | −1.4 |
| 351 | −22.7 | −22.5 | 0.2 | −0.4 | −0.3 | −1 | −1.8 | −0.8 | 0 | −0.8 |
| 352 | −94.7 | −137.5 | −42.8 | 7.2 | −35.6 | −4.9 | −1.8 | 3.1 | 0 | 3.1 |
| 353 | −3.8 | −4.0 | −0.2 | −0.3 | −0.5 | −18.7 | −19.2 | −0.5 | 0 | −0.5 |
| 354 | −128.3 | −129.1 | −0.8 | −5.3 | −6.1 | −14.5 | −16.2 | −1.7 | 0 | −1.7 |
| 355 | −171.0 | −162.7 | 8.3 | 0.4 | 8.7 | −10 | −11.4 | −1.5 | 0 | −1.5 |
| 356 | −13.9 | −13.7 | 0.3 | −0.2 | 0.1 | −8.7 | −8.7 | 0 | 0 | 0 |
| 357 | −143.1 | −149.7 | −6.6 | 4.9 | −1.7 | −11.4 | −13.3 | −2 | 0 | −2 |
| 358 | −45.7 | −51.0 | −5.3 | 0.7 | −4.6 | −19.6 | −17.8 | 1.8 | 0 | 1.8 |
| 359 | −5.8 | −6.2 | −0.4 | −0.7 | −1.1 | −19.6 | −20.5 | −0.8 | 0 | −0.8 |
| 360 | −7.6 | −8.4 | −0.8 | −0.4 | −1.2 | −16.9 | −16.6 | 0.3 | 0 | 0.3 |
| 361 | −16.9 | −16.6 | 0.3 | 0.6 | 0.8 | −11.7 | −12.2 | −0.5 | 0 | −0.5 |
| 362 | −13.3 | −9.0 | 4.3 | −0.7 | 3.5 | −14.8 | −15 | −0.2 | 0 | −0.2 |
| 363 | −18.6 | −28.1 | −9.5 | −0.7 | −10.2 | −7.7 | −4.6 | 3 | 0 | 3 |
| 364 | −2.4 | 0.5 | 2.8 | −0.3 | 2.5 | 0.4 | −1.7 | −2.1 | 0 | −2.1 |
| 365 | −12.5 | −12.6 | −0.1 | 0.3 | 0.2 | −6.4 | −6.8 | −0.3 | 0 | −0.3 |
| 366 | −81.6 | −110.3 | −28.8 | 0.3 | −28.5 | −5.2 | −5.6 | −0.4 | 0 | −0.4 |
| 367 | −7.1 | −6.7 | 0.4 | −0.7 | −0.3 | −15.5 | −19.1 | −3.6 | 0 | −3.6 |
| 368 | −153.3 | −143.3 | 10.1 | −0.5 | 9.5 | −6 | −7 | −1 | 0 | −1 |
| 369 | −11.8 | −21.0 | −9.2 | −0.6 | −9.8 | −3.9 | −3.1 | 0.8 | 0 | 0.8 |
| 370 | −0.7 | 1.0 | 1.7 | −0.3 | 1.4 | −8.3 | −7.6 | 0.7 | 0 | 0.7 |
| 371 | −36.3 | −25.1 | 11.2 | 1.5 | 12.6 | −8.9 | −5.6 | 3.3 | 0 | 3.3 |
| 372 | −40.9 | −45.3 | −4.4 | −13.6 | −18.0 | −10.9 | −14.1 | −3.2 | −4.4 | −7.6 |
| 373 | −129.6 | −30.2 | 99.4 | −77.8 | 21.6 | −0.5 | −6.5 | −6 | 2 | −4 |
| 374 | −27.2 | −24.3 | 2.8 | 2.7 | 5.5 | −9.6 | −8.4 | 1.2 | 0 | 1.2 |
| 375 | −22.9 | −25.3 | −2.3 | 3.1 | 0.7 | −10.1 | −10.4 | −0.3 | −0.2 | −0.5 |
| 376 | −24.5 | −26.1 | −1.6 | 5.3 | 3.6 | −6.2 | −9.7 | −3.5 | −2 | −5.5 |
| 377 | −24.0 | −31.5 | −7.5 | 2.7 | −4.7 | −11.3 | −9.3 | 2 | −0.2 | 1.8 |
| 378 | −19.8 | −19.5 | 0.3 | 1.3 | 1.6 | −8.8 | −7.6 | 1.2 | −0.1 | 1.1 |
| 379 | −32.5 | −26.0 | 6.6 | 1.8 | 8.4 | −16 | −18.2 | −2.2 | −2.4 | −4.6 |
| 380 | −164.6 | −81.4 | 83.2 | −33.8 | 49.5 | −3.8 | −9.3 | −5.5 | −2 | −7.5 |
| 381 | −13.9 | −22.7 | −8.7 | 0.1 | −8.7 | −5 | −2.9 | 2.1 | 0 | 2.1 |
| 382 | −5.0 | −5.6 | −0.6 | −1.0 | −1.6 | −7.8 | −6.3 | 1.5 | 0 | 1.5 |
| 383 | −25.1 | −20.4 | 4.7 | 1.9 | 6.6 | 0.5 | −1.8 | −2.2 | 0 | −2.2 |
| 384 | −30.0 | −17.3 | 12.7 | 0.1 | 12.8 | −7.1 | −9.1 | −2 | 0 | −2 |
| 385 | −15.6 | −14.2 | 1.4 | −0.4 | 1.0 | −9.7 | −8.9 | 0.9 | 0 | 0.9 |
| 386 | −4.9 | −11.9 | −7.0 | −0.2 | −7.2 | −17.5 | −17.6 | −0.1 | 0 | −0.1 |
| 387 | −19.4 | −24.5 | −5.1 | 0.3 | −4.8 | −9.9 | −9.6 | 0.2 | 0 | 0.2 |
| 388 | −16.8 | −14.5 | 2.3 | −0.1 | 2.2 | −18.9 | −19.6 | −0.6 | 0 | −0.6 |
| 389 | −14.0 | −14.2 | −0.2 | 0.1 | −0.1 | −15.2 | −14.2 | 1 | 0 | 1 |
| 390 | −14.6 | −12.9 | 1.7 | 0.1 | 1.8 | −12.4 | −12.6 | −0.2 | 0 | −0.2 |
| 391 | −25.0 | −27.4 | −2.3 | 0.0 | −2.3 | −16.8 | −16.8 | 0 | 0 | 0 |
| 392 | −22.5 | −18.1 | 4.5 | −0.1 | 4.4 | −26.9 | −25.6 | 1.3 | 0 | 1.3 |
| 393 | −190.0 | −173.1 | 16.9 | −0.1 | 16.8 | −12 | −12.6 | −0.5 | 0 | −0.5 |
| 394 | −4.5 | −5.6 | −1.1 | −0.3 | −1.4 | −17.3 | −17.4 | 0 | 0 | 0 |
| 395 | −136.5 | −155.0 | −18.5 | −0.5 | −19.0 | −5.7 | −5.3 | 0.4 | 0 | 0.4 |
| 396 | −117.9 | −124.7 | −6.8 | −1.2 | −8.0 | −6.7 | −9.9 | −3.2 | 0 | −3.2 |
| 397 | −32.3 | −27.2 | 5.1 | −0.7 | 4.4 | −20.2 | −22.6 | −2.4 | −0.1 | −2.5 |
| 398 | −20.6 | −26.0 | −5.5 | −0.4 | −5.9 | −8.5 | −10 | −1.5 | 0 | −1.5 |
| 399 | −174.6 | −210.2 | −35.6 | 4.1 | −31.5 | −3.3 | −6.1 | −2.8 | 0 | −2.8 |
| 400 | −9.5 | −9.0 | 0.5 | −1.1 | −0.6 | −15.5 | −12.8 | 2.7 | −0.1 | 2.6 |
| 401 | −148.5 | −152.9 | −4.4 | −2.6 | −7.0 | −1.7 | −0.6 | 1.1 | 0 | 1.1 |
| 402 | −7.7 | −7.8 | 0.0 | 0.3 | 0.3 | −3.2 | −3.5 | −0.3 | 0 | −0.3 |
| 403 | −23.1 | −21.3 | 1.9 | 0.3 | 2.1 | −4.6 | −4.7 | 0 | 0 | 0 |
| 404 | −15.1 | −15.9 | −0.9 | 0.1 | −0.7 | −9.4 | −10.3 | −0.9 | 0 | −0.9 |
| 405 | −68.4 | −71.7 | −3.2 | 0.9 | −2.3 | −12.1 | −13.2 | −1.2 | 0 | −1.2 |
| 406 | −1.6 | −3.1 | −1.6 | −0.1 | −1.7 | −14.7 | −13.9 | 0.8 | 0 | 0.8 |
| 407 | −14.5 | −16.6 | −2.1 | 0.0 | −2.1 | −9.6 | −8.6 | 0.9 | 0 | 0.9 |
| 408 | 0.2 | 0.3 | 0.1 | 0.0 | 0.2 | −1.3 | −2.6 | −1.3 | 0 | −1.3 |
| 409 | −20.4 | −27.1 | −6.8 | 0.1 | −6.7 | 0.2 | −4.9 | −5.1 | 0 | −5.1 |
| 410 | −13.3 | −12.8 | 0.5 | 0.1 | 0.5 | −3.5 | −3.9 | −0.4 | 0 | −0.4 |
| 411 | −88.1 | −84.6 | 3.5 | −1.7 | 1.8 | −13.5 | −12.8 | 0.7 | 0 | 0.7 |
| 412 | −5.1 | −5.1 | 0.1 | 0.2 | 0.3 | −17.5 | −17.8 | −0.3 | 0 | −0.3 |
| 413 | −13.8 | −12.7 | 1.1 | −0.1 | 1.0 | −11.5 | −11.7 | −0.2 | 0 | −0.2 |
| 414 | −23.9 | −27.0 | −3.1 | 0.4 | −2.7 | −8.1 | −7.6 | 0.4 | 0 | 0.4 |
| 415 | −45.9 | −37.8 | 8.2 | 0.0 | 8.2 | −3.6 | −4.4 | −0.8 | 0 | −0.8 |
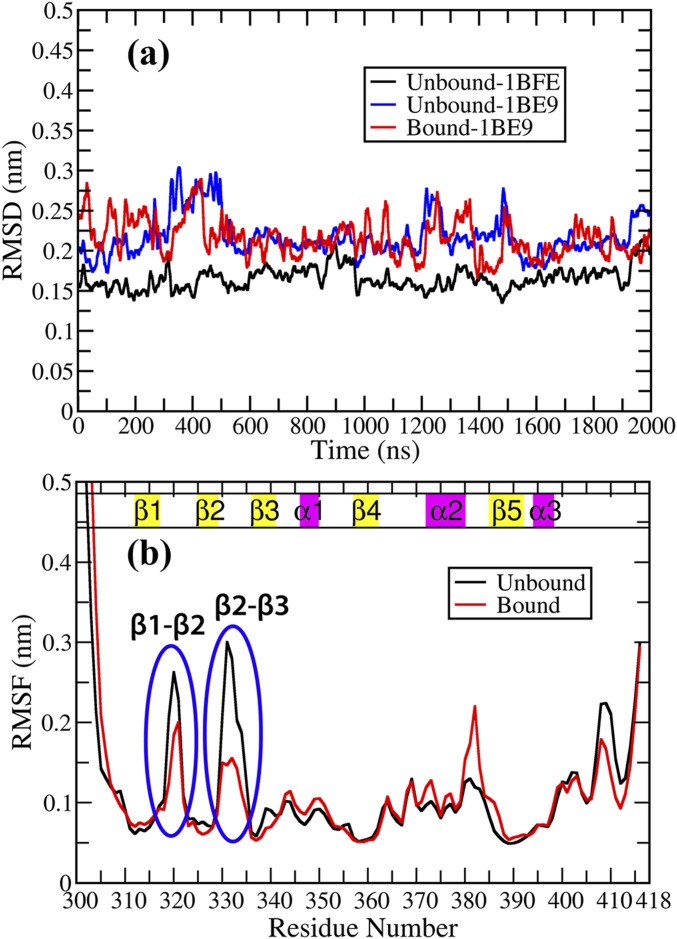
The local dynamical variations in the PDZ3 domain has been captured in the residuewise root mean square fluctuations (RMSF) with respect to the average structure for both the ligand bound and unbound states (Fig. S2B). Overall the fluctuations decrease in the presence of the ligand in the β1–β2 and β2–β3 loop regions and toward the C-terminal region. In contrast, the fluctuation increases in the presence of ligand around residues 381–385 (α2–β5 loop). This emphasizes greater flexibility and plasticity in the unbound state compared with the bound state as already discussed in the literature (25). Although it has been well established that the overall dynamics of the PDZ3 domain is modulated on ligand binding (hence the term dynamic allostery), but the reason behind it is not completely clear. We shall argue that dynamics is governed by the underlying energy landscape. Thus, understanding the perturbation or modulation in the intraprotein interaction network is likely to provide the answer.
Fig. S2.
RMSD and RMSF for the MD trajectories (2 μs) (A) Backbone RMSD on comparison with the crystal structures of PDZ3 in unbound and bound state [PDB IDs: 1BFE (unbound) and 1BE9 (bound)]. The system is found to be stable and well equilibrated across all of the production runs. (B) Residue based RMSF profile for PDZ3 domain in the unbound state (black) and bound state (red). There is overall decrease in fluctuations in the ligand-bound state compared with the unbound state exhibiting flexibility in the domain in the absence of ligand and subsequently ligand-binding activity quenches fluctuations in the loop regions prominently.
Contact Maps Do Not Capture the Specific Nature of Contacts (Ionic/Polar/Nonpolar).
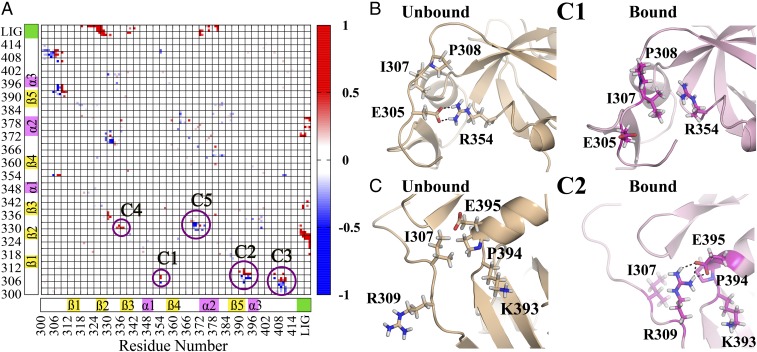
We have shown in Fig. 1B that the side-chain contact maps undergo only a minor rearrangement. Of course, proteins in solution are flexible entities and thus contacts may break/form with certain population. To understand the differences in the contact pattern in the dynamic system, we have computed the differential contact frequency map (Cij; See Materials and Methods) between the trajectories for bound and unbound states (Fig. 2A). Five major regions (C1 to C5) are marked on Fig. 2A corresponding to the significant changes in the intraprotein contact map (involving both backbone and side-chain atoms). The numerical values for residue pairs with |Cij| > 0.5 are shown in Table S2. Red and blue regions signify exclusive contacts present in bound and unbound states, respectively.
Fig. 2.
(A) Differential contact frequency map between the bound and unbound states as obtained from the MD simulation trajectories. The regions with high differential contact frequency () are highlighted as clusters (C1 to C5). Positive values (red regions) and negative values (blue regions) indicate exclusive contacts present in the bound and unbound states, respectively. The values of differential contact frequency of these residue pairs have been shown in Table S2. (B) The representative snapshots showing changes in contact pattern between unbound and bound states for cluster C1 (ionic: R354–E305 to nonpolar: R354–I307) and (C) C2 (nonpolar: E395–I307 to ionic: E395–R309).
Table S2.
List of residues with net contact frequency (), positive value would indicate a contact between residue pairs i and j to be present exclusively in bound state and negative value states contact is predominantly formed in unbound state
| ResID | Residues list with net contact frequency |
| G303 | S409 (−0.6) |
| E304 | S408 (−0.5) |
| E305 | R354 (−0.8), Y392 (−0.8), G410 (−0.7) |
| D306 | S409 (0.7), R411 (0.6) |
| I307 | P394 (−0.9), K393 (−0.9), V406 (−0.8), S409 (0.7), E395 (−0.6), R411 (0.9), R354 (0.9) |
| P308 | R354 (0.6), Q391 (−0.9) |
| R309 | Y392 (0.9), K393 (0.8), P394 (0.9), E395 (0.7), V406 (0.8), G410 (0.8) |
| E310 | Y392 (0.9) |
| E331 | G335 (0.7), A370 (−0.5) |
| D332 | N369 (−0.6) |
| G335 | E331 (0.7) |
| R354 | E305 (−0.8), I307 (0.9), P308 (0.6) |
| N369 | D332 (−0.6) |
| A370 | E331 (−0.5) |
| Q391 | P308 (−0.9) |
| Y392 | E305 (−0.8), R309 (0.9), E310 (0.9) |
| K393 | I307 (−0.9), R309 (0.8) |
| P394 | I307 (−0.9), R309 (0.9) |
| E395 | I307 (−0.6), R309 (0.7) |
| V406 | I307 (−0.8), R309 (0.8) |
| S408 | E304 (−0.5) |
| S409 | G303 (−0.6), D306 (0.7) I307 (0.7) |
| G410 | E305 (−0.7), R309 (0.8) |
| R411 | D306 (0.6), I307 (0.9) |
| K5 | I328 (0.8), G329 (0.7), H372 (0.8) |
| Q6 | N326 (1.0), I327 (0.9), I328 (0.9), S339 (0.8), F340 (0.7), H372 (0.7) |
| T7 | F325 (0.7), N326 (1.0), I327 (1.0), I328 (0.9), H372 (1.0), A376 (1.0) |
| S8 | F325 (1.0), N326 (1.0), I327 (1.0), L342 (0.7), K380 (0.6) |
| V9 | R318 (0.8), T321 (0.7), G322 (1.0), L323 (1.0), G324 (1.0), F325 (1.0), N326 (1.0), I327 (1.0), A376 (0.9), L379 (1.0), K380 (0.7) |
We notice that the N terminus region (residues 305–309) is involved in contact(s) with the α1–β4 (C1), β5–α3 (C2), and C terminus (C3) loop/coil regions. For all these clusters there is an upward movement (N to C terminus) on going from unbound to bound state. This minor change in contact pattern seems insignificant because the contact map is agnostic to the specific nature of the contacts (ionic/polar/nonpolar). Let us focus on two specific cases for the clusters C1 (Fig. 2B) and C2 (Fig. 2C) to understand the nature of interactions between these contacts. For cluster C1, there exists a salt bridge (ionic interaction) between the oppositely charged species E305 (negative)–R354 (positive) in the unbound state, whereas in the bound state this converts to nonpolar contacts between hydrophobic I307/P308 and charged R354 (positive). Similarly, for the cluster C2, there is nonpolar contact between I307 (hydrophobic) and E395 (negative) in the unbound state, and this converts to a salt bridge between R309 (positive) and E395 (negative) in the bound state.
Note that in both the cases, the contact region shifts by only two residues in the N-terminal region, whereas the specific nature of the interaction dramatically changes between nonpolar to ionic and vice versa. Thus, we argue that looking at purely structural parameters (distance/position based) is not enough to understand the allosteric modulation unless the chemical identity (charge distribution) is being considered. Thus, in subsequent sections we shall exclusively focus on the energetics as the yardstick to identify the interaction network that connects the binding site to the allosteric site. Here the nonbonded interaction energy captures both the structural parameters (through distance dependence) and chemical identities (through partial charges for electrostatic and Lennard-Jones parameters for van der Waals’ interactions) of the molecular systems.
Electrostatic Energy Is the Key Determinant of Allosteric Modulation in PDZ3 Domain.
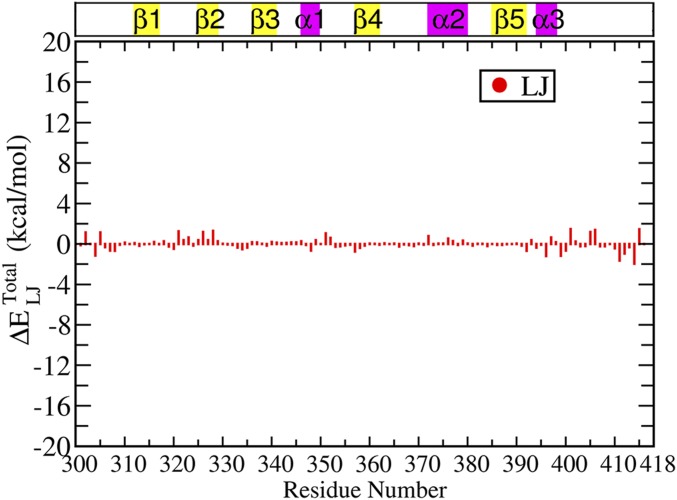
So far we have established that nonbonded interactions provide the most sensitive yardstick to capture the structural rearrangement compared with any purely position/distance-based parameters. Now we are going to demonstrate the perturbation in the intraprotein interaction network caused by the ligand and how this perturbation might propagate through pairwise interactions between protein residues. For this purpose, we have evaluated the average interaction energy of each residue with the various components of its environment, namely protein, ligand, and water. The changes in each of these energy terms were calculated between the ligand bound and unbound states (Materials and Methods for details). All of these results are summarized in Fig. 3A: protein-only and ligand-only, Fig. 3B: water-only and Fig. 3C: total (protein + ligand + water). Here we focus on only the electrostatic component of the respective terms in Fig. 3, where < > indicates the ensemble average over respective bound and unbound trajectories. Corresponding van der Waals’ (Lennard-Jones) terms are much smaller in magnitude (<3 kcal mol) as shown in Fig. S3 and Table S3. The numerical values of for all residues are provided in Table S3. The corresponding error bars for the average interaction energy for the unbound and bound states are provided in Table S4.
Fig. 3.
Residuewise changes in average electrostatic energy between bound and unbound states () as obtained from MD simulation trajectories: (A) The contributions due to ligand and rest of the protein have been shown separately as red and black lines, respectively. (B) Contribution due to water only, where average interaction energy between the ith residue and water molecules with 2-nm cutoff radius has been used to compute . (C) The due to the full environment comprising of ligand, protein, and water molecules within 2 nm, i.e., . The numerical values of all terms are provided in Table S3.
Fig. S3.
Van der Waals’ (Lennard-Jones) component of the residuewise change in total interaction energy between the bound and unbound states . The change in interaction energy is less than 3 kcal/mol, which is relatively very low compared with the difference in the electrostatic interaction energy between the bound and unbound state. This highlights the major contribution of electrostatic effect in the allosteric phenomenon on ligand binding.
Table S3.
The average interaction energy contributions due to various terms and comparison between the bound (B) and unbound (U) states averaged over the respective molecular dynamics trajectories where
| ResID | ||||||||||
| 301 | −31.4 | −31.5 | −0.1 | 0.0 | −0.1 | −19.4 | −20.0 | −0.6 | −0.7 | −0.1 |
| 302 | −33.3 | −30.7 | 2.6 | 0.0 | 2.6 | −16.7 | −17.7 | −1.0 | 1.6 | 1.2 |
| 303 | −44.5 | −37.9 | 6.6 | 0.0 | 6.6 | −10.8 | −15.9 | −5.1 | 1.5 | 0 |
| 304 | −10.7 | −16.5 | −5.8 | 1.1 | −4.7 | −189.8 | −191.6 | −1.8 | −6.5 | −1.2 |
| 305 | −66.5 | −14.4 | 52.1 | 1.9 | 54.0 | −165.2 | −207.6 | −42.4 | 11.6 | 1.2 |
| 306 | 4.9 | −7.2 | −12.1 | 1.5 | −10.6 | −209.0 | −189.9 | 19.1 | 8.5 | −0.3 |
| 307 | −28.5 | −29.0 | −0.5 | −0.2 | −0.7 | −12.8 | −12.2 | 0.6 | −0.1 | −0.7 |
| 308 | −30.1 | −33.1 | −3.0 | 0.1 | −2.9 | −13.7 | −13.7 | 0.0 | −2.9 | −0.7 |
| 309 | −164.8 | −206.1 | −41.3 | −0.8 | −42.1 | −26.2 | 5.3 | 31.5 | −10.6 | −0.1 |
| 310 | −34.9 | −34.2 | 0.7 | 2.1 | 2.8 | −167.7 | −173.9 | −6.2 | −3.4 | 0.1 |
| 311 | −23.0 | −23.3 | −0.3 | −0.1 | −0.4 | −19.8 | −19.2 | 0.6 | 0.2 | 0 |
| 312 | −206.6 | −192.1 | 14.5 | −5.8 | 8.7 | 20.2 | 16.7 | −3.5 | 5.2 | 0.1 |
| 313 | −101.4 | −97.2 | 4.2 | −5.7 | −1.5 | −98.4 | −102.2 | −3.8 | −5.3 | −0.2 |
| 314 | −53.6 | −52.8 | 0.8 | −0.1 | 0.7 | 3.4 | 1.1 | −2.3 | −1.6 | 0 |
| 315 | −30.4 | −30.6 | −0.2 | −0.4 | −0.6 | −17.2 | −17.6 | −0.4 | −1.0 | 0 |
| 316 | −45.1 | −42.3 | 2.8 | −0.3 | 2.5 | −5.6 | −8.4 | −2.8 | −0.3 | 0.2 |
| 317 | −34.4 | −35.9 | −1.5 | 1.2 | −0.3 | −20.2 | −22.7 | −2.5 | −2.8 | 0 |
| 318 | −127.6 | −115.3 | 12.3 | −41.9 | −29.6 | −66.4 | −32.9 | 33.5 | 3.9 | 0.3 |
| 319 | −41.9 | −42.5 | −0.6 | 1.3 | 0.7 | −13.2 | −14.3 | −1.1 | −0.4 | −0.3 |
| 320 | −31.8 | −31.9 | −0.1 | 0.7 | 0.6 | −25.2 | −25.6 | −0.4 | 0.2 | −0.5 |
| 321 | −37.8 | −34.5 | 3.3 | 3.6 | 6.9 | −21.3 | −25.1 | −3.8 | 3.1 | 1.3 |
| 322 | −33.6 | −33.3 | 0.3 | −12.2 | −11.9 | −11.4 | −3.2 | 8.2 | −3.7 | 0.4 |
| 323 | −30.9 | −29.7 | 1.2 | −13.8 | −12.6 | −10.2 | 3.8 | 14.0 | 1.4 | 0.6 |
| 324 | −33.1 | −31.9 | 1.2 | −10.0 | −8.8 | −9.4 | −7.4 | 2.0 | −6.8 | −0.2 |
| 325 | −39.6 | −40.2 | −0.6 | 0.4 | −0.2 | −12.3 | −8.7 | 3.6 | 3.4 | 0.4 |
| 326 | −46.1 | −46.8 | −0.7 | −8.7 | −9.4 | −20.6 | −12.0 | 8.6 | −0.8 | 1.2 |
| 327 | −37.9 | −37.5 | 0.4 | −8.5 | −8.1 | −15.6 | −3.9 | 11.7 | 3.6 | 0.4 |
| 328 | −42.0 | −44.7 | −2.7 | 0.7 | −2.0 | −10.5 | −1.6 | 8.9 | 6.9 | 1.3 |
| 329 | −44.8 | −43.3 | 1.5 | −1.4 | 0.1 | −2.2 | −5.6 | −3.4 | −3.3 | 0.3 |
| 330 | −42.6 | −44.4 | −1.8 | −1.3 | −3.1 | −11.3 | −14.2 | −2.9 | −6.0 | 0 |
| 331 | 28.2 | 33.5 | 5.3 | −32.6 | −27.3 | −254.3 | −226.5 | 27.8 | 0.5 | −0.1 |
| 332 | 47.3 | 51.4 | 4.1 | −13.8 | −9.7 | −265.4 | −250.2 | 15.2 | 5.5 | −0.1 |
| 333 | −37.4 | −40.3 | −2.9 | 0.7 | −2.2 | −14.3 | −10.1 | 4.2 | 2.0 | −0.4 |
| 334 | −8.9 | −0.1 | 8.8 | −6.5 | 2.3 | −212.3 | −210.5 | 1.8 | 4.1 | −0.5 |
| 335 | −31.5 | −32.4 | −0.9 | −0.9 | −1.8 | −10.5 | −13.9 | −3.4 | −5.2 | −0.4 |
| 336 | −43.3 | −42.8 | 0.5 | 0.3 | 0.8 | −5.1 | −6.2 | −1.1 | −0.3 | 0.2 |
| 337 | −49.1 | −48.9 | 0.2 | 0.1 | 0.3 | −6.1 | 0.0 | 6.1 | 6.4 | 0.2 |
| 338 | −42.7 | −43.9 | −1.2 | −1.3 | −2.5 | −0.5 | −0.5 | 0.0 | −2.5 | 0 |
| 339 | −43.7 | −44.6 | −0.9 | −2.8 | −3.7 | −11.4 | −2.9 | 8.5 | 4.8 | −0.2 |
| 340 | −38.0 | −38.9 | −0.9 | 1.1 | 0.2 | −7.5 | −9.8 | −2.3 | −2.1 | 0.2 |
| 341 | −37.3 | −36.2 | 1.1 | −1.3 | −0.2 | −11.7 | −8.4 | 3.3 | 3.1 | 0.1 |
| 342 | −35.6 | −35.9 | −0.3 | −0.2 | −0.5 | −13.4 | −16.5 | −3.1 | −3.6 | 0.1 |
| 343 | −40.6 | −41.0 | −0.4 | 0.9 | 0.5 | −8.9 | −9.5 | −0.6 | −0.1 | 0.1 |
| 344 | −44.3 | −44.1 | 0.2 | 0.2 | 0.4 | −5.6 | −5.1 | 0.5 | 0.9 | 0.2 |
| 345 | −33.0 | −31.9 | 1.1 | −2.3 | −1.2 | −11.1 | −12.9 | −1.8 | −3.0 | 0.2 |
| 346 | −30.4 | −29.6 | 0.8 | −2.2 | −1.4 | −11.1 | −11.2 | −0.1 | −1.5 | 0.3 |
| 347 | −35.3 | −35.6 | −0.3 | −1.6 | −1.9 | −17.0 | −18.5 | −1.5 | −3.4 | −0.1 |
| 348 | −33.1 | −36.1 | −3.0 | 9.2 | 6.2 | −162.4 | −181.5 | −19.1 | −12.9 | −0.7 |
| 349 | −28.8 | −28.5 | 0.3 | −0.9 | −0.6 | −22.7 | −19.2 | 3.5 | 2.9 | 0.4 |
| 350 | −33.7 | −33.9 | −0.2 | −0.8 | −1.0 | −16.9 | −13.7 | 3.2 | 2.2 | 0 |
| 351 | −40.6 | −46.2 | −5.6 | −0.6 | −6.2 | −11.7 | −4.8 | 6.9 | 0.7 | 1.1 |
| 352 | −39.5 | −50.1 | −10.6 | 7.6 | −3.0 | −156.1 | −167.4 | −11.3 | −14.3 | 0.6 |
| 353 | −29.0 | −29.8 | −0.8 | −0.1 | −0.9 | −4.4 | −11.7 | −7.3 | −8.2 | −0.3 |
| 354 | −203.1 | −164.1 | 39.0 | −5.3 | 33.7 | 39.9 | 6.2 | −33.7 | 0.0 | −0.2 |
| 355 | −163.2 | −170.8 | −7.6 | −1.9 | −9.5 | −43.2 | −28.4 | 14.8 | 5.3 | −0.1 |
| 356 | −50.3 | −51.6 | −1.3 | −0.3 | −1.6 | 2.6 | 2.0 | −0.6 | −2.2 | −0.1 |
| 357 | −126.9 | −146.2 | −19.3 | 5.9 | −13.4 | −82.6 | −83.5 | −0.9 | −14.3 | −0.8 |
| 358 | −69.2 | −70.7 | −1.5 | 0.4 | −1.1 | −6.8 | −10.7 | −3.9 | −5.0 | −0.4 |
| 359 | −35.0 | −34.2 | 0.8 | −0.8 | 0.0 | −10.3 | −4.4 | 5.9 | 5.9 | −0.2 |
| 360 | −35.7 | −36.1 | −0.4 | −0.4 | −0.8 | −10.7 | −13.2 | −2.5 | −3.3 | 0.1 |
| 361 | −48.8 | −46.7 | 2.1 | 0.4 | 2.5 | −10.7 | −11.8 | −1.1 | 1.4 | 0 |
| 362 | −35.4 | −35.9 | −0.5 | −0.7 | −1.2 | −4.5 | −6.0 | −1.5 | −2.7 | −0.1 |
| 363 | −36.5 | −36.5 | 0.0 | −0.1 | −0.1 | −38.6 | −40.5 | −1.9 | −2.0 | 0.1 |
| 364 | −30.1 | −30.1 | 0.0 | −0.3 | −0.3 | −13.5 | −14.9 | −1.4 | −1.7 | 0 |
| 365 | −52.1 | −51.0 | 1.1 | 0.3 | 1.4 | −11.5 | −9.1 | 2.4 | 3.8 | 0.1 |
| 366 | −72.2 | −83.4 | −11.2 | 0.8 | −10.4 | −125.7 | −104.6 | 21.1 | 10.7 | −0.3 |
| 367 | −29.8 | −30.8 | −1.0 | −0.6 | −1.6 | −11.3 | −9.7 | 1.6 | 0.0 | 0 |
| 368 | −176.5 | −166.3 | 10.2 | −0.4 | 9.8 | −13.6 | −23.7 | −10.1 | −0.3 | −0.1 |
| 369 | −45.4 | −44.7 | 0.7 | −0.2 | 0.5 | −31.7 | −31.0 | 0.7 | 1.2 | −0.2 |
| 370 | −35.1 | −35.8 | −0.7 | −0.4 | −1.1 | −10.5 | −14.3 | −3.8 | −4.9 | 0.1 |
| 371 | −54.7 | −55.6 | −0.9 | 3.1 | 2.2 | −2.2 | −4.7 | −2.5 | −0.3 | −0.1 |
| 372 | −51.4 | −58.4 | −7.0 | −2.3 | −9.3 | −17.3 | −8.1 | 9.2 | −0.1 | 0.8 |
| 373 | −18.1 | −20.6 | −2.5 | −19.9 | −22.4 | −201.8 | −179.4 | 22.4 | 0.0 | −0.1 |
| 374 | −45.1 | −42.7 | 2.4 | 1.3 | 3.7 | −21.8 | −28.4 | −6.6 | −2.9 | 0.1 |
| 375 | −45.4 | −45.6 | −0.2 | 2.1 | 1.9 | 8.2 | 4.7 | −3.5 | −1.6 | 0.1 |
| 376 | −46.5 | −47.3 | −0.8 | 3.2 | 2.4 | 3.8 | 2.5 | −1.3 | 1.1 | 0.5 |
| 377 | −47.2 | −46.4 | 0.8 | 2.1 | 2.9 | 5.9 | −0.4 | −6.3 | −3.4 | 0.3 |
| 378 | −41.3 | −40.1 | 1.2 | 0.6 | 1.8 | −1.3 | −3.4 | −2.1 | −0.3 | −0.1 |
| 379 | −44.2 | −43.8 | 0.4 | 1.7 | 2.1 | 4.8 | 4.4 | −0.4 | 1.7 | 0.3 |
| 380 | −97.1 | −93.3 | 3.8 | −31.2 | −27.4 | −111.5 | −78.7 | 32.8 | 5.4 | 0 |
| 381 | −45.1 | −46.4 | −1.3 | 0.9 | −0.4 | −30.9 | −33.9 | −3.0 | −3.4 | −0.2 |
| 382 | −36.6 | −35.3 | 1.3 | 0.3 | 1.6 | −13.1 | −13.3 | −0.2 | 1.4 | 0 |
| 383 | −40.4 | −37.7 | 2.7 | 1.9 | 4.6 | −16.1 | −17.5 | −1.4 | 3.2 | 0 |
| 384 | −39.1 | −39.4 | −0.3 | 1.1 | 0.8 | −29.4 | −26.6 | 2.8 | 3.6 | −0.2 |
| 385 | −40.5 | −40.1 | 0.4 | −0.4 | 0.0 | −21.3 | −24.0 | −2.7 | −2.7 | 0 |
| 386 | −40.8 | −39.7 | 1.1 | 0.0 | 1.1 | −3.1 | −4.1 | −1.0 | 0.1 | −0.1 |
| 387 | −44.5 | −45.0 | −0.5 | 0.1 | −0.4 | −15.7 | −14.0 | 1.7 | 1.3 | −0.1 |
| 388 | −47.1 | −47.4 | −0.3 | −0.2 | −0.5 | 1.4 | −2.7 | −4.1 | −4.6 | 0 |
| 389 | −44.6 | −44.5 | 0.1 | 0.2 | 0.3 | −5.2 | −0.6 | 4.6 | 4.9 | 0 |
| 390 | −47.0 | −46.4 | 0.6 | 0.0 | 0.6 | 18.1 | 17.6 | −0.5 | 0.1 | 0 |
| 391 | −51.5 | −58.1 | −6.6 | 0.0 | −6.6 | −21.6 | −24.0 | −2.4 | −9.0 | −0.2 |
| 392 | −45.0 | −48.0 | −3.0 | −0.1 | −3.1 | −11.6 | −3.6 | 8.0 | 4.9 | −0.7 |
| 393 | −182.1 | −173.0 | 9.1 | 0.1 | 9.2 | 13.1 | −1.9 | −15.0 | −5.8 | 0.4 |
| 394 | −46.2 | −44.1 | 2.1 | −0.3 | 1.8 | 0.4 | −2.3 | −2.7 | −0.9 | −0.4 |
| 395 | −28.1 | −67.5 | −39.4 | −0.9 | −40.3 | −170.1 | −123.6 | 46.5 | 6.2 | −0.1 |
| 396 | −38.8 | −44.7 | −5.9 | −3.3 | −9.2 | −161.3 | −148.3 | 13.0 | 3.8 | −1.2 |
| 397 | −41.7 | −46.3 | −4.6 | −0.4 | −5.0 | −4.0 | −3.0 | 1.0 | −4.0 | 0.6 |
| 398 | −44.3 | −43.2 | 1.1 | −0.2 | 0.9 | −6.3 | −8.7 | −2.4 | −1.5 | 0.2 |
| 399 | −250.6 | −258.9 | −8.3 | 4.5 | −3.8 | 53.2 | 53.2 | 0.0 | −3.8 | −1.2 |
| 400 | −43.4 | −45.1 | −1.7 | −0.9 | −2.6 | −16.8 | −15.0 | 1.8 | −0.8 | −0.7 |
| 401 | −46.1 | −68.4 | −22.3 | −3.5 | −25.8 | −145.3 | −123.7 | 21.6 | −4.2 | 1.5 |
| 402 | −32.9 | −31.8 | 1.1 | 0.5 | 1.6 | −18.3 | −15.0 | 3.3 | 4.9 | 0.2 |
| 403 | −40.9 | −41.4 | −0.5 | 0.2 | −0.3 | −29.1 | −26.5 | 2.6 | 2.3 | −0.2 |
| 404 | −39.8 | −40.7 | −0.9 | 0.0 | −0.9 | −15.0 | −17.7 | −2.7 | −3.6 | −0.2 |
| 405 | −133.7 | −128.6 | 5.1 | 1.4 | 6.5 | −59.6 | −75.6 | −16.0 | −9.5 | 1.2 |
| 406 | −43.9 | −43.3 | 0.6 | −0.2 | 0.4 | −8.2 | −7.0 | 1.2 | 1.6 | 1.4 |
| 407 | −41.8 | −45.6 | −3.8 | 0.1 | −3.7 | −33.8 | −37.2 | −3.4 | −7.1 | −0.2 |
| 408 | −29.0 | −28.0 | 1.0 | 0.0 | 1.0 | −28.9 | −29.5 | −0.6 | 0.4 | −0.2 |
| 409 | −40.1 | −43.7 | −3.6 | 0.1 | −3.5 | −18.4 | −16.6 | 1.8 | −1.7 | 0 |
| 410 | −39.9 | −42.9 | −3.0 | 0.2 | −2.8 | −6.9 | −4.4 | 2.5 | −0.3 | −0.4 |
| 411 | −165.2 | −186.1 | −20.9 | −2.7 | −23.6 | −3.4 | 23.8 | 27.2 | 3.6 | −1.6 |
| 412 | −43.5 | −43.5 | 0.0 | 0.2 | 0.2 | −12.6 | −11.7 | 0.9 | 1.1 | −0.9 |
| 413 | −39.5 | −40.4 | −0.9 | −0.1 | −1.0 | −7.1 | −8.3 | −1.2 | −2.2 | −0.3 |
| 414 | −44.9 | −44.2 | 0.7 | 0.3 | 1.0 | −20.7 | −21.0 | −0.3 | 0.7 | −2 |
| 415 | −43.6 | −42.7 | 0.9 | 0.1 | 1.0 | −31.7 | −32.4 | −0.7 | 0.3 | 1.5 |
Table S4.
Error bars for the calculated values for bound state (protein contribution only) and unbound state
| ResID | ||||
| 301 | −31.4 | 0.3 | −31.5 | 0.5 |
| 302 | −33.3 | 0.6 | −30.7 | 0.6 |
| 303 | −44.5 | 1 | −37.9 | 0.5 |
| 304 | −10.7 | 1.6 | −16.5 | 6.5 |
| 305 | −66.5 | 6.6 | −14.4 | 3.7 |
| 306 | 4.9 | 3.5 | −7.2 | 3.6 |
| 307 | −28.5 | 0.1 | −29 | 0.9 |
| 308 | −30.1 | 0.2 | −33.1 | 0.5 |
| 309 | −164.7 | 1.9 | −206.1 | 3.1 |
| 310 | −34.9 | 1.1 | −34.2 | 1.5 |
| 311 | −23 | 0.1 | −23.3 | 0.2 |
| 312 | −206.6 | 1.1 | −192.1 | 4 |
| 313 | −101.4 | 0.7 | −97.2 | 1 |
| 314 | −53.6 | 0.1 | −52.8 | 0.4 |
| 315 | −30.4 | 0.1 | −30.6 | 0.3 |
| 316 | −45.1 | 0.2 | −42.3 | 4.9 |
| 317 | −34.4 | 0.6 | −35.9 | 0.1 |
| 318 | −127.6 | 4.8 | −115.3 | 17.1 |
| 319 | −41.9 | 0.3 | −42.5 | 0.5 |
| 320 | −31.8 | 0.3 | −31.9 | 0.1 |
| 321 | −37.8 | 1.6 | −34.5 | 0.1 |
| 322 | −33.6 | 0.9 | −33.3 | 0.3 |
| 323 | −30.8 | 0.3 | −29.7 | 0.4 |
| 324 | −33.1 | 0.3 | −31.9 | 0.4 |
| 325 | −39.6 | 0.1 | −40.2 | 0.1 |
| 326 | −46.1 | 0.1 | −46.7 | 0.3 |
| 327 | −37.9 | 0.1 | −37.5 | 0.2 |
| 328 | −42 | 0.5 | −44.7 | 0.3 |
| 329 | −44.8 | 1.3 | −43.3 | 3.9 |
| 330 | −42.6 | 1.2 | −44.4 | 2.1 |
| 331 | 28.2 | 1 | 33.5 | 1 |
| 332 | 47.3 | 1.2 | 51.4 | 1 |
| 333 | −37.4 | 1.1 | −40.3 | 0.5 |
| 334 | −8.9 | 4.8 | −0.1 | 3.2 |
| 335 | −31.4 | 0.4 | −32.4 | 0.3 |
| 336 | −43.3 | 0.4 | −42.8 | 0.3 |
| 337 | −49.1 | 0.2 | −48.9 | 0.3 |
| 338 | −42.7 | 1.7 | −43.9 | 0.2 |
| 339 | −43.7 | 0.3 | −44.6 | 0.4 |
| 340 | −38 | 0.2 | −38.9 | 0.4 |
| 341 | −37.2 | 0.2 | −36.2 | 0.2 |
| 342 | −35.6 | 0.1 | −35.9 | 0 |
| 343 | −40.6 | 0.1 | −41 | 0 |
| 344 | −44.3 | 0.1 | −44.1 | 0.1 |
| 345 | −33 | 0.2 | −31.9 | 0.2 |
| 346 | −30.4 | 0.5 | −29.6 | 0.4 |
| 347 | −35.3 | 0.2 | −35.6 | 0.2 |
| 348 | −33.1 | 0.6 | −36.1 | 0.9 |
| 349 | −28.8 | 0.1 | −28.5 | 0.1 |
| 350 | −33.7 | 0.1 | −33.9 | 0.1 |
| 351 | −40.6 | 0.2 | −46.2 | 1.4 |
| 352 | −39.5 | 1 | −50.1 | 1.5 |
| 353 | −29 | 0.1 | −29.7 | 0.5 |
| 354 | −203.1 | 5 | −164.1 | 1.9 |
| 355 | −163.2 | 3 | −170.8 | 1.7 |
| 356 | −50.3 | 0.8 | −51.6 | 0.1 |
| 357 | −126.9 | 2.3 | −146.2 | 9.3 |
| 358 | −69.2 | 0.3 | −70.7 | 0.4 |
| 359 | −35 | 0.6 | −34.2 | 0.2 |
| 360 | −35.7 | 0.2 | −36.1 | 0.2 |
| 361 | −48.8 | 0.3 | −46.7 | 0.2 |
| 362 | −35.4 | 0.1 | −35.9 | 0.3 |
| 363 | −36.5 | 0.4 | −36.5 | 2.5 |
| 364 | −30.1 | 0.2 | −30.1 | 0.1 |
| 365 | −52.1 | 0.1 | −51 | 0.2 |
| 366 | −72.2 | 3.5 | −83.4 | 2.5 |
| 367 | −29.8 | 0.4 | −30.8 | 0.5 |
| 368 | −176.5 | 2.8 | −166.3 | 2.2 |
| 369 | −45.4 | 1 | −44.7 | 1.8 |
| 370 | −35.1 | 0.3 | −35.8 | 0.6 |
| 371 | −54.7 | 0.7 | −55.6 | 0.5 |
| 372 | −51.4 | 1.1 | −58.4 | 3.2 |
| 373 | −18.1 | 2.4 | −20.6 | 1.7 |
| 374 | −45.1 | 0.5 | −42.7 | 0.3 |
| 375 | −45.4 | 0.2 | −45.6 | 0.4 |
| 376 | −46.5 | 0.2 | −47.3 | 0.3 |
| 377 | −47.2 | 0.2 | −46.4 | 4 |
| 378 | −41.3 | 0.1 | −40.1 | 0.2 |
| 379 | −44.1 | 0.8 | −43.8 | 4.7 |
| 380 | −97.1 | 0.8 | −93.3 | 3.2 |
| 381 | −45.1 | 0.2 | −46.3 | 5.2 |
| 382 | −36.6 | 0.4 | −35.3 | 3.4 |
| 383 | −40.4 | 0.7 | −37.7 | 1.7 |
| 384 | −39.1 | 0.5 | −39.4 | 0.3 |
| 385 | −40.5 | 0.3 | −40.1 | 2.5 |
| 386 | −40.7 | 0.2 | −39.7 | 4 |
| 387 | −44.5 | 0.1 | −45 | 0.2 |
| 388 | −47.1 | 0.1 | −47.4 | 0.1 |
| 389 | −44.6 | 0.1 | −44.5 | 0.1 |
| 390 | −47 | 0 | −46.4 | 0.1 |
| 391 | −51.5 | 0.2 | −58.1 | 0.6 |
| 392 | −45 | 1 | −48 | 0.3 |
| 393 | −182.1 | 7.3 | −173 | 2.6 |
| 394 | −46.2 | 0.2 | −44.1 | 0.4 |
| 395 | −28.1 | 2 | −67.5 | 3.4 |
| 396 | −38.8 | 1.1 | −44.7 | 1.2 |
| 397 | −41.7 | 1 | −46.3 | 0.5 |
| 398 | −44.3 | 0.5 | −43.1 | 0.6 |
| 399 | −250.6 | 9.6 | −258.9 | 7.2 |
| 400 | −43.4 | 0.3 | −45.1 | 0.3 |
| 401 | −46.1 | 7.3 | −68.4 | 2.2 |
| 402 | −32.9 | 0.4 | −31.7 | 0.2 |
| 403 | −40.9 | 0.3 | −41.4 | 0.3 |
| 404 | −39.8 | 0.3 | −40.6 | 0.3 |
| 405 | −133.7 | 1.2 | −128.6 | 0.8 |
| 406 | −43.9 | 0.2 | −43.3 | 0.1 |
| 407 | −41.7 | 1.1 | −45.6 | 0.3 |
| 408 | −29 | 2.2 | −28 | 0.3 |
| 409 | −40.1 | 5.9 | −43.7 | 0.7 |
| 410 | −39.9 | 0.7 | −42.9 | 0.2 |
| 411 | −165.2 | 6.2 | −186.1 | 3.4 |
| 412 | −43.5 | 0.2 | −43.5 | 0.1 |
| 413 | −39.5 | 0.1 | −40.4 | 0.1 |
| 414 | −44.9 | 0.4 | −44.2 | 0.5 |
| 415 | −43.6 | 0.4 | −42.7 | 0.4 |
All of the values are in kcal/mol. represents the average error for the 2-µs trajectory.
Our results unequivocally prove that the electrostatic interaction energy provides a sensitive yardstick toward capturing the allosteric effects in this system. Although the van der Waals’ interactions are crucial toward attaining the functional structure of the protein, the allosteric modulation due to ligand binding seems to be strongly associated with the electrostatic interactions. Our findings provide further support to the already established view regarding the significant role of electrostatic interactions in biomolecular functions including allostery (38, 44–48).
Evidently, a negative signifies an interaction more favorable in the bound state compared with the unbound state and vice versa. In Fig. 3A we can easily recognize the residues interacting strongly and favorably with the ligand (red lines), for example, R318, E331, E373, and K380 have contributions more negative than −30 kcal/mol. On the other hand, there are residues (black lines) that do not interact with the ligand at all or spatially far away from the ligand, but still exhibit very large magnitude of , for example, E305, R309, R354, E395, and E401, and so forth. These residues clearly demonstrate the effect of allosteric modulation in terms of their energetics. Thus, interactions with the ligand are leading to internal structural rearrangements (or population shift) in the protein in such a way that the intraprotein interaction network is significantly perturbed.
Another interesting observation to make here is the contribution from the ligand and protein-only interactions toward are often in the reverse direction, for example, R312, R318, E331, E334, and E352, and so forth. The favorable interaction with the ligand () forces the side chains of these residues to attain certain orientations that lead to unfavorable interactions (or break previously favorable interactions) with the other residues of the protein (). Such locally unfavorable protein–protein interaction is likely to initiate further downstream rearrangement to release the energetic stress, much like a domino effect. We must also note that there are positive values for a few residues, for example, E305, R312, R318, E334, R354, R368, and K393, and so forth. This signifies that there exist certain favorable interactions for these residues in the unbound state, which were broken in the process of ligand binding and subsequent allosteric modulation. We shall further dissect the molecular basis of these long-range perturbations in the subsequent sections.
Prior studies on allostery have primarily focused on the structure and dynamics of the protein itself; whereas, a few studies highlight the effect of solvation forces and water-mediated interactions toward allosteric modulation (49, 50). Fig. 3B demonstrates the modulation in the residuewise solvation energy (to be precise, the average electrostatic interaction energy with water) on ligand binding. Here the electrostatic interaction energy for each residue was calculated with respect to the water molecules within 2-nm cutoff. As expected the solvent exerts a dielectric screening effect that goes in the reverse direction of the electrostatic contributions due to protein and ligand only. The residuewise changes in the total interaction energy including the solvation energy () is shown in Fig. 3C. Evidently, the contributions due to the charged residues still remain the most significant. In addition, Fig. 3C highlights the relatively large values and oscillatory patterns for the residues in and around the binding site, for example, G324, I328, G330, and F337, and so forth. Most of these residues have favorable interaction with the ligand, but an unfavorable desolvation penalty associated with ligand binding. The local solvation environment may have substantially altered for certain residues; for example, for residues D306, D348, E352, D366, and R405 we observe that . This implies that the solvation energy may overcompensate the changes due to protein–protein interactions in these cases due to redistribution in the protein–protein versus protein–water interaction pattern. Future research could explore the more specific roles of water-mediated interactions toward allosteric regulation.
An interesting feature of the total reported in Fig. 3C is that there is an oscillatory pattern of residues with large positive and large negative values. This indicates that the ligand binding leads to a massive internal rearrangement or redistribution of the nonbonded interactions. There is a clear separation of the residues in terms of favorable and unfavorable interactions. Due to these cancellation effects, the total nonbonded interaction energy of the protein is perturbed to a much lower extent compared with the local perturbation or energy redistribution at the residue level. Thus, it is not that enthalpy does not play a role in the dynamic allostery observed in PDZ3 domain; rather it plays a significant role in terms of the local rearrangement and rewiring of the specific interactions.
Energetic Perturbation Network Connects the Binding Site and Allosteric Site.
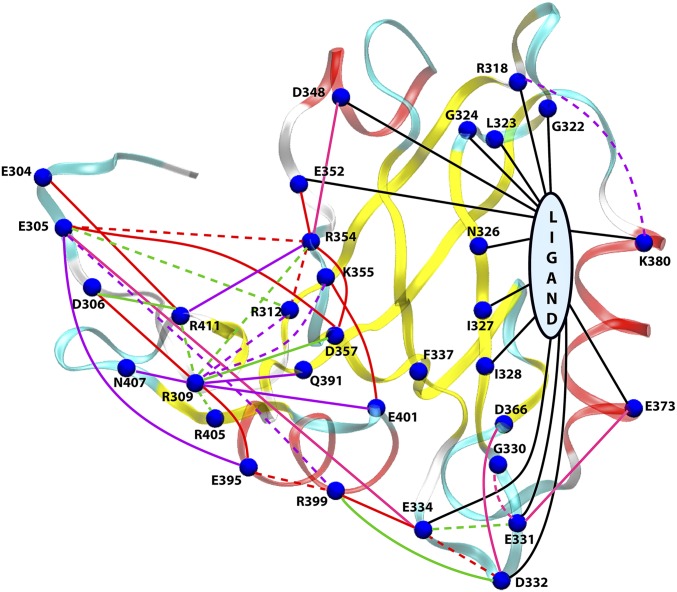
Now that we have identified the protein residues that undergo a magnificent energetic perturbation between the bound and unbound states, we attempt to dissect the residue-pairwise contributions toward this change so that we can build a connectivity network of the energetic perturbation. To achieve this, we have dissected the (shown in Fig. 3A) into all residue-pairwise contribution terms: , where . Note that involves the electrostatic component of the interaction only. The numerical values of for residues with kcal/mol have been shown in Table S5. The values are summarized and visualized in Fig. 4, which now leads to an energetic perturbation network that dramatically connects the ligand-binding site to the allosteric side (distal regions) of PDZ3 domain. The caption of Fig. 4 provides a detailed description of the visualization scheme used to build the network.
Table S5.
Breakdown of perturbation in pairwise interactions, where || > 2 kcal/mol
| ResID | Residue-pairwise contribution: (kcal/mol) |
| E304 | R411 (−10.8) |
| E305 | R354 (57.8), D357 (−11.4), R312 (8.8), K393 (6.4), E396 (−5.0), E395 (−4.9), R399 (4.7), E334 (−3.4), K355 (3.2), R405 (3.1), E401 (−3.1), E352 (−2.6), E304 (2.3), R368 (2.1), N407 (−2.1) |
| D306 | R309 (−12.1), R411 (−9.5), E395 (3.7), R354 (2.8) |
| R309 | E395 (−41.3), D306 (−12.1), R411 (9.9), D357 (−7.3), R405 (6.8), R354 (6.1), K355 (5.3), Q391 (−5.1), E401 (−4.9), R312 (4.9), E310 (4.7), N407 (−4.3), P394 (3.6), K393 (3.6), D348 (−2.9), E396 (−2.7), I307 (−2.6), E304 (−2.4), E352 (−2.4), R399 (2.3) |
| R312 | R354 (13.1), E305 (8.8), E310 (−8.4), P308 (−5.2), R309 (4.9) |
| R318 | K380 (4.9), N381 (−4.5), T321 (3.9), G383 (3.8) |
| E331 | E334 (8.3), E373 (−3.6), G330 (3.2), E401 (2.9), R399 (−2.9), E396 (2.6), K355 (−2.4), H372 (−2.2), A370 (2.2), Q374 (2.2), G333 (−2.0) |
| D332 | E334 (15.6), R399 (−8.4), E396 (4.2), D366 (−3.5), R368 (2.9), G333 (−2.7) |
| E334 | D332 (15.6), R399 (−13.3), E331 (8.3), K393 (4.7), E305 (−3.4) |
| E352 | R354 (−11.1) |
| R354 | E305 (57.8), R312 (13.1), D357 (−11.3), E352 (−11.1), R309 (6.1), E310 (−6.0), G351 (−5.0), P308 (−4.3), R411 (−4.0), D348 (−3.9), D306 (2.8), R313 (2.1), R405 (−2.1) |
| K355 | E401 (−13.2), R309 (5.3), E305 (3.2), Y397 (2.8), E304 (−2.4), E331 (−2.4) |
| D357 | E305 (−11.4), R354 (−11.3), R309 (−7.3), E310 (4.7), P308 (4.3), E395 (3.0), R411 (−2.7) |
| D366 | D332 (−3.5) |
| R368 | E334 (3.0), D332 (2.9), K393 (2.3), E305 (2.1) |
| H372 | E331 (−2.2) |
| E373 | E331 (-3.6) |
| K380 | R318 (4.9) |
| Q391 | R309 (−5.1), K393 (−3.5) |
| K393 | E305 (6.4), E395 (−4.7), E334 (4.7), R309 (3.6), Q391 (−3.5), R368 (2.3), P308 (2.0) |
| E395 | R309 (−41.3), R399 (11.0), E305 (−4.9), K393 (−4.7), E396 (−4.5), D306 (3.7), E310 (3.3), D357 (3.0), P394 (−2.8), R411 (−2.6), R312 (−2.5), I307 (2.3) |
| R399 | E334 (−13.3), E395 (11.0), D332 (−8.4), E305 (4.7), E331 (−2.8), R309 (2.3), E405 (−2.0) |
| E401 | K355 (−13.2), Y397 (−6.8), R309 (−4.9), E305 (−3.1), E331 (2.9) |
| R405 | R309 (6.8), E305 (3.1) |
| N407 | R309 (−4.3) |
| R411 | E304 (−10.8), R309 (9.9), D306 (−9.5), E310 (−4.1), R354 (−4.0) |
Fig. 4.
A comprehensive network view of the perturbation in pairwise electrostatic interaction energies (). Visualization scheme: (i) The blue spheres indicate residues with or kcal/mol. A few residues with large , but kcal/mol have been highlighted as blue spheres as well, e.g., R312, R354, K355, R399, E401, and R411. (ii) Connections with negative and positive values are indicated with solid and dashed lines, respectively, i.e., a solid (or dashed) line indicates a contact more (or less) favorable in bound (or unbound) state. (iii) The connections are colored on the basis of magnitude of kcal/mol (red), > 6 kcal/mol (green), > 4 kcal/mol (purple), and > 3 kcal/mol (pink). The black lines represent connections between peptide ligand and residues with > 6 kcal/mol. The connections based on kcal/mol were considered only for residues that are directly perturbed on ligand binding. The values for all significant pairs have been reported in Table S5.
A detailed analysis of the network presented in Fig. 4 gives us a multitude of significant insights into the nature of the communication pathways between the binding site and allosteric site: (i) Direct perturbation by ligand: The solid black lines indicates that the ligand perturbs most of the binding site residues [in β1–β2 loop, β2 sheet, and α2 helix (K380, E373)], but also induces changes in energetics of the β2–β3 loop and distant residues (D348, E352) through long-range electrostatic interactions. Our analysis successfully captures the effect on residues already known for their involvement in ligand binding, for example, R318, G322, N326, I327, I328, D332, and K380 (27, 29, 31, 33, 35). (ii) Perturbation in intraprotein interactions: Interestingly, we can observe an extensive energetic redistribution at the distal side (possibly, the allosteric site) comprised of the N and C termini, α3 helix, and α1–β4 unstructured regions. The solid lines indicate the interactions that have become more favorable in the ligand-bound state compared with unbound state, and the dashed lines indicate more unfavorable ones. The presence of comparable number of solid and dashed lines in the allosteric site indicates that an extensive rearrangement and rewiring of interactions have taken place in this region. In particular, a few pairs connected by red lines ( kcal/mol) indicate that D332–E334, E395–R399, and E305–R354 interactions are more favorable in the unbound state; whereas, E334–R399, E395–R309, R309–D306, E305–D357, E304–R411, E352–R354, R354–D357, and K355–E401 become more favorable in the bound state. As we can speculate many of these pairwise interactions between charged residues would involve strong electrostatic interaction (salt bridges) or hydrogen-bonded interactions as we shall show in the next section.
Interestingly, the binding site and allosteric site are connected through the α3 helix mediated by a salt bridge interaction between E334 and R399. Thus, our energy perturbation network reestablishes the experimental results of Lee and coworkers regarding the significant role of α3 helix toward hidden dynamic allostery in PDZ3 domain (6) and subsequent simulation studies showing the hydrogen-bonded interactions between the α3 helix and β2–β3 loop region that controls this connection (51).
As we have already seen before, both and values show large variation with opposite signs that leads to cancellations resulting in small change in the total energy/enthalpy of the protein. Similarly, a few residues have large values of with kcal/mol, for example, R312, R354, K355, R399, E401, and R411 (Table S5 for a detailed breakdown). This observation again highlights the phenomena of massive internal rearrangement without changing the total interaction energy due to cancellation between pairwise interactions in reverse directions. The residues with large number of connections as shown in Fig. 4 are likely to play the role of intermediate hubs in the network of energetic propagation of allosteric modulation.
Population Shift of Hydrogen-Bonded Network Leads to Allosteric Modulation.
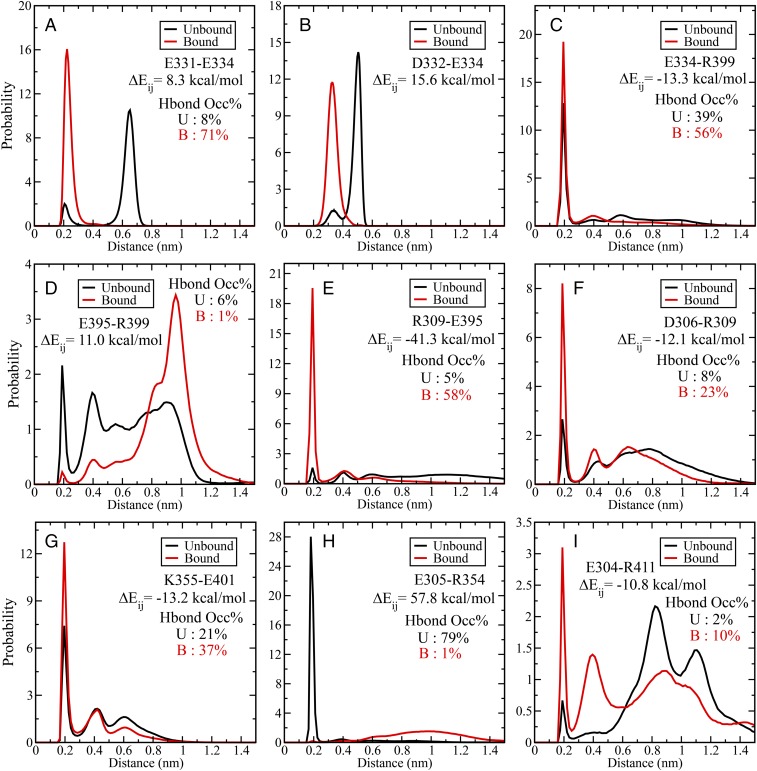
So far we have unraveled that ligand binding leads to significant perturbation in the intraprotein electrostatic interaction pattern. Let us now investigate the nature of these specific interactions that rearrange and rewire on ligand binding. Toward this goal we have shortlisted the residue pairs with kcal/mol and found that most of these pairs are capable of forming hydrogen bonds (H bonds) either through side chains or backbones. Fig. 5 shows representative snapshots from the unbound and bound trajectories that highlight the possible differences in the H-bonding pattern between these selected residue pairs. Interestingly, these H bonds are not exclusive in nature, that is, a certain pair that forms H bond in unbound state may be present in the bound state as well, but with a different population.
Fig. 5.
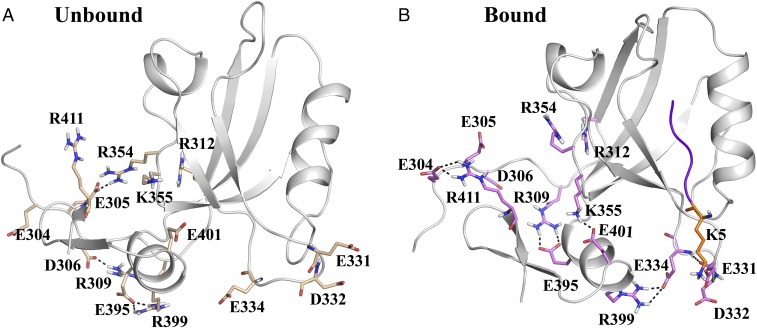
Rearrangement and rewiring of side-chain interaction network between (A) unbound and (B) bound states. These pairs have been chosen based on large (Fig. 6 for values). Note that these specific interactions can be transient and not all hydrogen bonds are present at all frames due to inherent dynamical fluctuations (Fig. 6 for population distribution of these interactions). We have shown selected frames that highlight the nature of the hydrogen-bonded network leading to the electrostatic coupling between the binding site and allosteric site. Polar contacts (e.g., hydrogen bonds or salt bridges) formed between the side chains are shown using dashed lines.
This aspect of population shift of the pairwise interactions has been further elucidated in Fig. 6. Here we show the population distribution of the minimum approach distance between a set of representative residue pairs with large values. The minimum distance has been computed between all possible pairs of atoms (including hydrogen) between the residues. For most of the cases we observe that there are at least two peaks where one corresponds to strong interaction at short distance (around 0.2 nm) and broken/weaker interactions at larger distances for both the unbound and bound states. The strong peaks observed around 0.2 nm signify existence of specific polar interactions (e.g., hydrogen bond or salt bridge) between these pairs. For example, Fig. 6A shows the interaction between E331 and E334, where the peak around 0.2 nm signifies the H-bonded interaction between the backbone atoms of these residues (Fig. 5B for a representative structure). The population of this state is significantly higher in the bound state compared with unbound state, but it is still present in the unbound state. Similar observations can be made for other pairs as well.
Fig. 6.
(A–I) Probability distribution of pairwise minimum distance between residue pairs with kcal/mol for unbound (black line) and bound (red line) states. The values have been indicated for all pairs. The strong peak around 0.2 nm would signify presence of polar contact (e.g., hydrogen bond or salt bridge). The hydrogen bond occupancy (percentage) based on standard geometric criteria has been marked for all of the pairs (except D332–E334).
There are examples where this population shift in pairwise interaction is very subtle, for example, E334–R399 (Fig. 6C) and K355–E401 (Fig. 6G). Here the peak for the H-bonded species gets stronger (increased population) without changing the overall shape of the distribution function. However, these pairs exhibit large values (around −13 kcal/mol). There are only two examples, where the distribution function changes in an almost exclusive nonoverlapping manner and very large values for , for example, E305–R354 (Fig. 6H; kcal/mol) and R309–E395 (Fig. 6e; kcal/mol). As indicated by the signs of the values and distance distribution, the E305–R354 salt bridge interaction exists almost exclusively in the unbound state. On the other hand, the R309–E395 interaction becomes highly favorable in the ligand-bound state. A structural implication of this observation is that in the unbound state the disordered N terminus region interacts strongly with the α1–β4 coil region (involving R354), whereas on ligand binding the interactions shift to α3 helix region (R309–E395) implicated as mediator in dynamic allostery, and also the C terminus coil region (e.g., E304–R411). Fig. 5 provides a structural view of these specific H-bonded interactions that undergoes subtle to dramatic population shift on ligand binding.
The above analysis also provides a clear mechanistic picture into the initiation and propagation of the energetic perturbation through the population shift in the H-bonded network much like trapeze artists changing their partners. As shown in Fig. 5, the positively charged lysine (K5) of ligand interacts strongly with the E331 and E332 residues in the β2–β3 loop, which in turn makes the backbone H bonding between E331 and E334 stronger (Fig. 6A). This controls the orientation of E334 side chain to increase the population of the H bond (salt bridge) formed with R399 in the α3 helix (Fig. 6C). The role of the interaction between β2–β3 loop and α3 helix has been already shown in an earlier study (51). Interestingly, there exists a significant population (6%) for the intrahelical i − i + 4 salt bridge formation between the side chains of E395–R399 in the unbound state (Fig. 6D). This interaction gets weaker (1%) in the ligand-bound state as E395 starts interacting with R309, and R399 interacting with E334 (Figs. 5 and 6 E and C). Thus, the combination of Figs. 5 and 6 paints a detailed mechanistic picture of the rearrangement of H-bonded interactions induced by ligand and resultant changes in pairwise electrostatic interaction energies. Also, we have proven the molecular basis of the strong influence of the extradomain α3 helix on the ligand binding and the conformational preference of side-chain rotamers. These dynamic interactions and energetic coupling between the side-chain orientations/interactions provide the key toward the dynamic allosteric modulation in PDZ3 domain.
We must note that for most of the residue pairs dissected in Fig. 6, there is a significant tightening of the polar contacts (H bonds) on ligand binding. Hence, the underlying free energy landscape of these pairwise interactions become narrower, thus providing a direct connection with the entropic view of dynamic allostery, where the tightening of the H-bonding interactions leads to reduced conformational entropy for these residues.
Conclusions
The basic premise of this work is to demonstrate that the dynamic allostery phenomena in the PDZ3 domain protein originates from the modulation of underlying energy landscape dictated by ligand binding. Although the prevailing view considers dynamic allostery to be purely entropy driven, we argue and demonstrate that there exists significant energetic redistribution in terms of the specific electrostatic interactions between the protein residues. First, we demonstrate that the structure/position-based parameters like RMSD and contact map, and so forth, are not able to capture the extent of modulation in specific interactions that undergo on ligand binding. It is crucial to understand the nature of the rearranging contacts, namely hydrogen bonding, salt bridge, nonpolar, and so forth. The nonbonded interaction energy provides the most fundamental and robust yardstick for capturing the subtle changes in the side-chain orientation.
We have shown that the electrostatic interaction energy becomes the key determinant in distinguishing the structural ensemble between the ligand bound and unbound states, and elucidating the allosteric modulation. There exist extensive competing interactions and cancellation effects due to interactions between the protein residues, protein–protein, protein–ligand, and protein–water interactions. Such cancellations lead to relatively minor changes in the total enthalpy of the protein, whereas there exists substantial rearrangement or redistribution of the interactions at a local level. We have identified the allosteric network by decomposing the average pairwise interaction energies for all PDZ3 domain residues and their perturbation on ligand binding. This network clearly connects the ligand-binding site with the distal allosteric site through the α3 helix domain, which has been implicated to play a crucial role in mediating the dynamic allostery in this system.
Our detailed analysis has identified an extended network of hydrogen-bonded pairs that control the interaction network. The population distribution of these pairwise interactions indicate that a population shift mechanism prevails, where the preexisting conformational distribution gets modulated on ligand binding with the tightening of most of the H-bonding interactions on ligand binding (related to the previous reports of reduction in conformational entropy). Our study identifies the role of specific electrostatic interactions and their population shift toward the allosteric modulation in PDZ3 domain. Moreover, we elucidate the molecular basis of the allosteric modulation in the N- and C-terminal regions through major rearrangement in the interaction pattern, which would provide valuable mechanistic insights into the role of PDZ3 domains in cell signaling. Because the PDZ domains are usually chained together in sequence, it is interesting that the allosteric effects propagate to the termini regions, possibly leading to global response in terms of spatial arrangement of these domains on binding with the effector ligands/proteins.
We must note that the substantial rearrangement of electrostatic interactions on ligand binding and associated structural population shift might alter the pKa values of the titratable residues. That would modulate the population of the protonation states of these residues with further changes in the electrostatic interaction network. Similar effects have been reported earlier for other enzymatic systems (52), and future research could explore the intricacies of such altered population of protonation states with possible consequence of pH-dependent allostery.
Our approach of energetic perturbation map would be useful in identification of putative target sites for allosteric drugs. There is a rapidly growing interest in allosteric drugs in contrast to the competitive orthosteric inhibitors due to their selectivity and ability to both enhance and inhibit the activity in a controllable manner. Prior studies have suggested that allosteric drugs work through “anchor” and “driver” atoms, where the anchor atom attaches itself to the binding pocket without causing any conformational change and the driver atom exerts a “push” or “pull” action to modulate the conformational ensemble toward the active or inactive states (53). A rigorous analysis of the energetic balance of such interactions (electrostatic, van der Waals’, and water mediated) along with their coupling with the inherent energy flow network of the enzyme as elucidated in the current work would be essential for rational design of allosteric drugs.
Materials and Methods
Molecular Dynamics Simulation.
The peptide bound and unbound structures for molecular dynamics (MD) simulations were obtained from the crystal structure of PDZ3 domain bound with peptide CRIPT (PDB ID: 1BE9). The unbound structure was obtained by removing the peptide CRIPT (KQTSV) and equilibrating for 100 ns. The C terminus of the protein and N termini of both protein and ligand were capped by N-methyl amide and acetyl groups, respectively. All MD simulations were performed using GROMACS 5.0.7 software (54) with Amber99SB-Ildn force field (55) and TIP4P-Ew water model (56). The resultant solvated boxes contained around 8,800 water molecules for all of the systems. The protonation states for the titratable residues were determined using MCCE (multi-conformation continuum electrostatics) method (57) as follows: all Asp, Glu, Arg, and Lys residues were charged and His residues were neutral. Although the MCCE protocol identified a minor population of the protonated charged species for the two histidine residues, we have decided to use the neutral species with major population for both bound and unbound states. The systems were neutralized by adding appropriate number of Na+ ions. The structures were energy minimized followed by two-step equilibration, namely NVT equilibration followed by NPT equilibration. Temperature was controlled through velocity rescaling (58) at 300 K with a time constant of 0.1 ps, and pressure was controlled using Parrinello–Rahman barostat (59) at 1 bar. The particle mesh Ewald algorithm (60) was applied to calculate long-range electrostatic interactions. The cutoff for short-range electrostatics and van der Waals’ interaction was 1.0 nm. Four independent MD simulations of 500 ns each were performed (total 2.0 μs) for the PDZ3 domain in the bound and unbound states with LINCS (linear constraint solver) constraints for all bonds (61) and frames were recorded at every 2 ps. The stability and convergence of the simulated trajectories were ensured based on backbone RMSD computed over the production run for both the states (Fig. S2A).
Differential Contact Map.
Two residues were defined to be in contact if the distance between any two atoms of these residues was less than 4.5 Å (62). Because a particular contact may form and break during the course of a dynamic trajectory, we defined a contact frequency map by , where is the number of frames where the residues i and j were in contact and N is the total number of frames. Thus, for a contact that is present in all of the frames. The differential contact map () is the difference between the contact frequency map () obtained from simulations of the bound state and the unbound state: , where the values of would lie between −1 and +1, and and would indicate a contact between residue pairs i and j to be present exclusively in unbound and bound states exclusively, respectively.
Perturbation in Nonbonded Interaction Energies.
The average nonbonded interaction energy was computed for each residue () as well as all residue pairs () and compared between the ligand bound and unbound states in the following manner. The change in average nonbonded energy of ith residue is given by: , where the notation indicates an ensemble average over the trajectory for that particular state (bound/unbound). Note that this difference in average energy can be further broken down in terms of the contributions from ligand, protein, and water in the following manner:
where the terms , , and denote the change in the average nonbonded interaction energy between the bound (b) and unbound (u) state due to the interactions between the ith residue and ligand/protein/water, respectively. Here the interaction energies were calculated for all atoms of protein and ligand, whereas for computing the interaction energy with water molecules a large cutoff of 2 nm has been used. We have separately computed the contributions due to Lennard-Jones (LJ) and electrostatic (Coulomb) nonbonded interactions to , but the LJ terms were generally found to be numerically much smaller than the respective electrostatic terms, so we have primarily focused on the electrostatic interactions while dissecting the perturbation in pairwise interactions . This has helped us to discover the significant role of specific electrostatic interactions toward observed allosteric response.
Acknowledgments
A.K. thanks Department of Biotechnology, India, for Senior Research Fellowship and PhD program of AcSIR. S.C. thanks the Department of Science and Technology, India, for the Ramanujan Fellowship (SR/S2/RJN-84/2012) and Council of Scientific and Industrial Research (CSIR), India, for funding from XIIth 5-year plan on Multiscale Modelling (CSC0129). We thank CSIR 4-PI supercomputing facility for the computational resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7480.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705311114/-/DCSupplemental.
References
- 1.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 2.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szilágyi A, Nussinov R, Csermely P. Allo-network drugs: Extension of the allosteric drug concept to protein-protein interaction and signaling networks. Curr Top Med Chem. 2013;13:64–77. doi: 10.2174/1568026611313010007. [DOI] [PubMed] [Google Scholar]
- 4.Koshland DE, Jr, Némethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 5.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 6.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafferty JB, Somers WS, Saint-Girons I, Phillips SEV. Three-dimensional crystal structures of Escherichia coli met repressor with and without corepressor. Nature. 1989;341:705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]
- 9.Cooper A, Dryden DTF. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 10.Nussinov R, Tsai CJ. Allostery without a conformational change? Revisiting the paradigm. Curr Opin Struct Biol. 2015;30:17–24. doi: 10.1016/j.sbi.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Tsai CJ, Nussinov R. A unified view of “how allostery works”. PLOS Comput Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussinov R, Tsai CJ, Liu J. Principles of allosteric interactions in cell signaling. J Am Chem Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 14.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 15.Feher VA, Durrant JD, Van Wart AT, Amaro RE. Computational approaches to mapping allosteric pathways. Curr Opin Struct Biol. 2014;25:98–103. doi: 10.1016/j.sbi.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 17.Lee HJ, Zheng JJ. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 19.Doyle DA, et al. Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 20.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J Mol Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, et al. Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E. Biochemistry. 2010;49:9280–9291. doi: 10.1021/bi101131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhulesia A, Gsponer J, Vendruscolo M. Mapping of two networks of residues that exhibit structural and dynamical changes upon binding in a PDZ domain protein. J Am Chem Soc. 2008;130:8931–8939. doi: 10.1021/ja0752080. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, Knecht V, Stock G. Long-range conformational response of a PDZ domain to ligand binding and release: A molecular dynamics study. J Chem Theory Comput. 2016;12:870–878. doi: 10.1021/acs.jctc.5b01009. [DOI] [PubMed] [Google Scholar]
- 24.Kong Y, Karplus M. Signaling pathways of PDZ2 domain: A molecular dynamics interaction correlation analysis. Proteins. 2009;74:145–154. doi: 10.1002/prot.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morra G, Genoni A, Colombo G. Mechanisms of differential allosteric modulation in homologous proteins: Insights from the analysis of internal dynamics and energetics of PDZ domains. J Chem Theory Comput. 2014;10:5677–5689. doi: 10.1021/ct500326g. [DOI] [PubMed] [Google Scholar]
- 26.Kalescky R, Liu J, Tao P. Identifying key residues for protein allostery through rigid residue scan. J Phys Chem A. 2015;119:1689–1700. doi: 10.1021/jp5083455. [DOI] [PubMed] [Google Scholar]
- 27.Sharp K, Skinner JJ. Pump-probe molecular dynamics as a tool for studying protein motion and long range coupling. Proteins. 2006;65:347–361. doi: 10.1002/prot.21146. [DOI] [PubMed] [Google Scholar]
- 28.Fuentes EJ, Gilmore SA, Mauldin RV, Lee AL. Evaluation of energetic and dynamic coupling networks in a PDZ domain protein. J Mol Biol. 2006;364:337–351. doi: 10.1016/j.jmb.2006.08.076. [DOI] [PubMed] [Google Scholar]
- 29.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 30.Dima RI, Thirumalai D. Determination of network of residues that regulate allostery in protein families using sequence analysis. Protein Sci. 2006;15:258–268. doi: 10.1110/ps.051767306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: Analysis through perturbation response scanning. PLOS Comput Biol. 2011;7:e1002154. doi: 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raimondi F, Felline A, Seeber M, Mariani S, Fanelli F. A mixed protein structure network and elastic network model approach to predict the structural communication in biomolecular systems: The PDZ2 domain from tyrosine phosphatase 1E as a case study. J Chem Theory Comput. 2013;9:2504–2518. doi: 10.1021/ct400096f. [DOI] [PubMed] [Google Scholar]
- 33.Ota N, Agard DA. Intramolecular signaling pathways revealed by modeling anisotropic thermal diffusion. J Mol Biol. 2005;351:345–354. doi: 10.1016/j.jmb.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 34.Miño-Galaz GA. Allosteric communication pathways and thermal rectification in PDZ-2 protein: A computational study. J Phys Chem B. 2015;119:6179–6189. doi: 10.1021/acs.jpcb.5b02228. [DOI] [PubMed] [Google Scholar]
- 35.Ho BK, Agard DA. Conserved tertiary couplings stabilize elements in the PDZ fold, leading to characteristic patterns of domain conformational flexibility. Protein Sci. 2010;19:398–411. doi: 10.1002/pro.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikura T, Iwata Y, Hatano T, Yamato T. Energy exchange network of inter-residue interactions within a thermally fluctuating protein molecule: A computational study. J Comput Chem. 2015;36:1709–1718. doi: 10.1002/jcc.23989. [DOI] [PubMed] [Google Scholar]
- 37.Chi CN, et al. Reassessing a sparse energetic network within a single protein domain. Proc Natl Acad Sci USA. 2008;105:4679–4684. doi: 10.1073/pnas.0711732105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Oelschlaeger P, Florián J, Goodman MF, Warshel A. Simulating the effect of DNA polymerase mutations on transition-state energetics and fidelity: Evaluating amino acid group contribution and allosteric coupling for ionized residues in human pol beta. Biochemistry. 2006;45:7036–7048. doi: 10.1021/bi060147o. [DOI] [PubMed] [Google Scholar]
- 39.Vijayabaskar MS, Vishveshwara S. Interaction energy based protein structure networks. Biophys J. 2010;99:3704–3715. doi: 10.1016/j.bpj.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharyya M, Vishveshwara S. Probing the allosteric mechanism in pyrrolysyl-tRNA synthetase using energy-weighted network formalism. Biochemistry. 2011;50:6225–6236. doi: 10.1021/bi200306u. [DOI] [PubMed] [Google Scholar]
- 41.Di Paola L, Giuliani A. Protein contact network topology: A natural language for allostery. Curr Opin Struct Biol. 2015;31:43–48. doi: 10.1016/j.sbi.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Johnson QR, Lindsay RJ, Nellas RB, Fernandez EJ, Shen T. Mapping allostery through computational glycine scanning and correlation analysis of residue-residue contacts. Biochemistry. 2015;54:1534–1541. doi: 10.1021/bi501152d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Bedem H, Bhabha G, Yang K, Wright PE, Fraser JS. Automated identification of functional dynamic contact networks from X-ray crystallography. Nat Methods. 2013;10:896–902. doi: 10.1038/nmeth.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warshel A. Electrostatic basis of structure-function correlation in proteins. Acc Chem Res. 1981;14:284–290. [Google Scholar]
- 45.Warshel A, Aqvist J. Electrostatic energy and macromolecular function. Annu Rev Biophys Biophys Chem. 1991;20:267–298. doi: 10.1146/annurev.bb.20.060191.001411. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura H. Roles of electrostatic interaction in proteins. Q Rev Biophys. 1996;29:1–90. doi: 10.1017/s0033583500005746. [DOI] [PubMed] [Google Scholar]
- 47.Sharp KA, Honig B. Electrostatic interactions in macromolecules: Theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. ChemBioChem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 49.Prakash P, Sayyed-Ahmad A, Gorfe AA. The role of conserved waters in conformational transitions of Q61H K-ras. PLOS Comput Biol. 2012;8:e1002394. doi: 10.1371/journal.pcbi.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buchli B, et al. Kinetic response of a photoperturbed allosteric protein. Proc Natl Acad Sci USA. 2013;110:11725–11730. doi: 10.1073/pnas.1306323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mostarda S, Gfeller D, Rao F. Beyond the binding site: the role of the β2-β3 loop and extra-domain structures in PDZ domains. PLOS Comput Biol. 2012;8:e1002429. doi: 10.1371/journal.pcbi.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexov E. Calculating proton uptake/release and binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association: Application to plasmepsin, cathepsin D and endothiapepsin-pepstatin complexes. Proteins. 2004;56:572–584. doi: 10.1002/prot.20107. [DOI] [PubMed] [Google Scholar]
- 53.Nussinov R, Tsai C-J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 55.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horn HW, et al. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 57.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 59.Parrinello M, Rahman A. Polymorphic transitions in single-crystals - A new molecular-dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 60.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 61.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 62.Yuan C, Chen H, Kihara D. Effective inter-residue contact definitions for accurate protein fold recognition. BMC Bioinformatics. 2012;13:292. doi: 10.1186/1471-2105-13-292. [DOI] [PMC free article] [PubMed] [Google Scholar]