Significance
Approximately a quarter of all land mammals are currently threatened, mostly by human activities including habitat loss and harvesting. Here, we provide the first biological map of priority areas that captures all three dimensions of mammalian biodiversity: taxonomic, phylogenetic, and traits. We find limited overlap in priority regions across the three dimensions and with currently protected areas, indicating that conservation planning should consider multiple dimensions of biodiversity to maximize biodiversity conservation. Our complementarity-based prioritization provides a conservation solution that can be incorporated in future conservation planning efforts aimed at helping protect not only species but also evolutionary potential and ecosystem function.
Keywords: complementarity, phylogenetic dimension, spatial conservation prioritization, taxonomic dimension, trait dimension
Abstract
Conservation priorities that are based on species distribution, endemism, and vulnerability may underrepresent biologically unique species as well as their functional roles and evolutionary histories. To ensure that priorities are biologically comprehensive, multiple dimensions of diversity must be considered. Further, understanding how the different dimensions relate to one another spatially is important for conservation prioritization, but the relationship remains poorly understood. Here, we use spatial conservation planning to (i) identify and compare priority regions for global mammal conservation across three key dimensions of biodiversity—taxonomic, phylogenetic, and traits—and (ii) determine the overlap of these regions with the locations of threatened species and existing protected areas. We show that priority areas for mammal conservation exhibit low overlap across the three dimensions, highlighting the need for an integrative approach for biodiversity conservation. Additionally, currently protected areas poorly represent the three dimensions of mammalian biodiversity. We identify areas of high conservation priority among and across the dimensions that should receive special attention for expanding the global protected area network. These high-priority areas, combined with areas of high priority for other taxonomic groups and with social, economic, and political considerations, provide a biological foundation for future conservation planning efforts.
Human activities are rapidly transforming the planet and are the primary causes of biodiversity loss (1–4). In response to growing concern about the future of biodiversity, the 10th Convention on Biological Diversity developed a strategic plan for 2011–2020 that introduced protection targets called the “Aichi Targets” to facilitate conservation action (5). One of the primary targets is to protect 17% of the global land surface (6), focusing on areas of particular importance for biodiversity and ecosystem services (7, 8). Traditionally, global priority areas have been identified based on richness, species endemism, and vulnerability (9, 10). Although these features are important in identifying key biodiversity regions, they focus on only one dimension of biodiversity—taxonomic diversity. Selecting regions based on species richness may not be the best conservation strategy, because richness does not reflect complementarity and thus could lead to priority areas with similar assemblage composition, at the cost of protecting unique assemblages (11, 12). Species are a product of ecological and evolutionary processes, and the species that we observe today represent only the tips of the tree of life. If we measure only species numbers, we might miss unique ecological and evolutionary information. Therefore the taxonomic dimension may not sufficiently capture other facets of diversity, such as evolutionary history and functional traits (13–15). Evolutionary history captures the uniqueness of linages through deep time (16) and can influence species’ susceptibility to extinction, because extinction risk is phylogenetically nonrandom (17–19). The trait dimension reflects ecological, morphological, and physiological strategies of species (20). Therefore losses in all three dimensions could have large consequences for biodiversity and ecosystem function (13).
There is clearly a need for spatial conservation priorities that account for multiple dimensions of biological diversity, including taxonomy, phylogeny, and traits (15, 21, 22). However, the relationships among these dimensions across the globe are poorly understood and vary depending on the metric used and the scale of the analysis (15, 23). Some studies have shown a high correlation among taxonomic, phylogenetic, and trait diversity of global mammals (23, 24), especially when using diversity indices affected by species richness, i.e., phylogenetic diversity (14) and functional diversity (20). For example, species richness was an effective surrogate for the functional and phylogenetic dimensions of local rodent assemblages in Manu (25). In contrast, Mazel et al. (15) identified strong geographical mismatches among global hotspots of taxonomic, phylogenetic, and functional diversity of mammals. Low geographical congruence was also found in prioritization analyses based on taxonomic, phylogenetic, and trait diversity of birds and mammals in Brazil (21). The problem with the lack of congruence among dimensions is that conservation planning based on only one dimension does not fully represent biodiversity (26, 27). Policy responses to biodiversity loss require knowledge of how these dimensions relate to each other and should use a more integrated approach by considering each dimension independently and all three jointly.
Global priority regions for mammal conservation have been identified (9, 10, 28, 29), but few have included different [i.e., taxonomic, phylogenetic, and functional (15, 21)] dimensions of mammal biodiversity simultaneously, and few have used a complementarity-based prioritization framework globally (30–33). Further, all prioritization analyses for mammals incorporate taxonomic, phylogenetic, and functional information by using alpha diversity indexes (15, 21, 23). Selecting priority areas for conservation based on high alpha diversity could omit some species/lineages/traits, because alpha diversity indices do not take into account the differences in composition among sites (12). Complementarity is derived from beta diversity and is considered a core principle of systematic conservation planning to ensure that conservation actions are directed to all species, not just to those occurring in species-rich hotspots (7, 34). Complementarity-based selection of priority areas allows the representation of all biological features without much duplication of sites with similar species, resulting in a more cost-effective conservation solution (12). For example, Strecker et al. (22) performed the first complementarity-based prioritization, considering separately taxonomic, phylogenetic, and trait diversity in freshwater fish, highlighting the use of this approach for the optimal allocation of limited conservation resources that incorporates multiple dimensions of biodiversity.
Here we present a spatial conservation prioritization for terrestrial mammals using taxonomic, phylogenetic, and trait information and a complementarity-based approach to site selection. Our aims were (i) to identify and compare priority regions for global terrestrial mammal conservation, based on taxonomic, phylogenetic, and trait dimensions and (ii) to evaluate how those priority areas relate to threatened species’ distribution and existing protected areas. Our work on the global prioritization of mammalian biodiversity across the dimensions can be used as a biological layer from which priority areas can be identified and subsequently combined with political, social, and economic considerations (e.g., ref. 31).
Results and Discussion
Priority Areas for Global Mammal Conservation Across the Three Dimensions of Biodiversity.
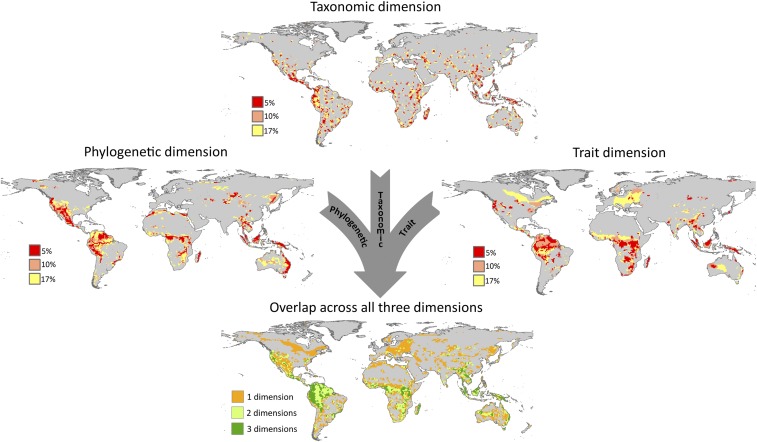
We found that important areas for mammal conservation had strikingly different spatial patterns for the three dimensions of biodiversity (Figs. 1 and 2). For the top 17% of sites with the highest conservation values, priority areas based on taxonomic dimension were spatially scattered, whereas the phylogeny- and trait-based priority areas were more contiguous (Fig. 1; for maps with all values see Fig. S1). Similarly, Carwardine et al. (35) and Dobrovolski et al. (32) found a scattered pattern of global priority areas for mammals, and Albuquerque et al. (36) also found the same diffused priority areas for mammals, birds, and amphibians. The areas we identified by using the taxonomic dimension were likely spatially scattered because of the prioritization algorithm we used. The core-area algorithm reduces biological loss by retaining the core of each species’ geographic range, even when they occur in species-poor regions, and it favors species with restricted ranges in the final solution.
Fig. 1.
The top 17% of cells selected according to zonation prioritization based on the taxonomic dimension, phylogenetic dimension, trait dimension and on the overlap across the three dimensions. In the overlap map, cells in which all three dimensions overlap are shown in dark green, those in which two dimensions overlap are shown in light green, and those selected only by one dimension are shown in orange.
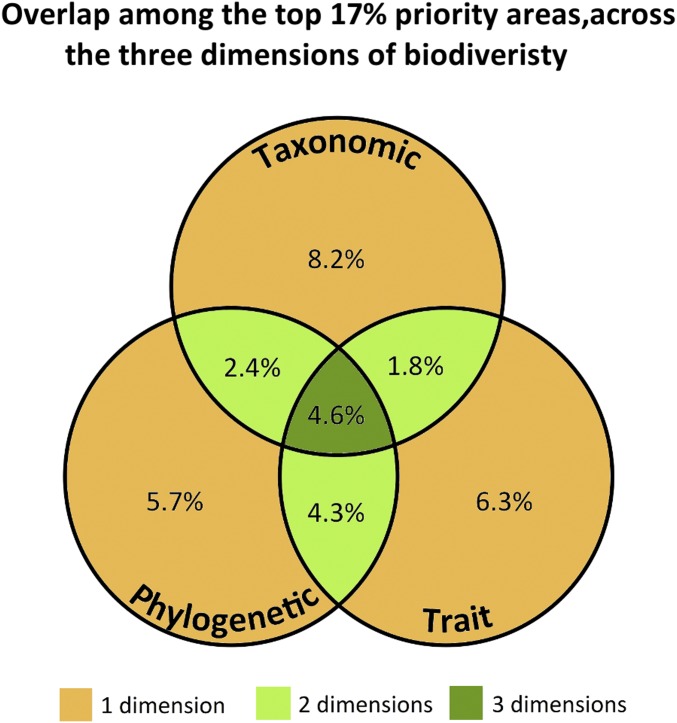
Fig. 2.
Venn diagram showing the proportion of the land surface where we can observe the overlap between the top 17% priority areas across the taxonomic, phylogenetic, and trait dimensions of biodiversity, referent to the overlap map presented in Fig. 1. For example, only 1.8% of the land was selected as an area of priority for both the taxonomic and trait dimensions. The color scheme is that used in the overlap map in Fig. 1.
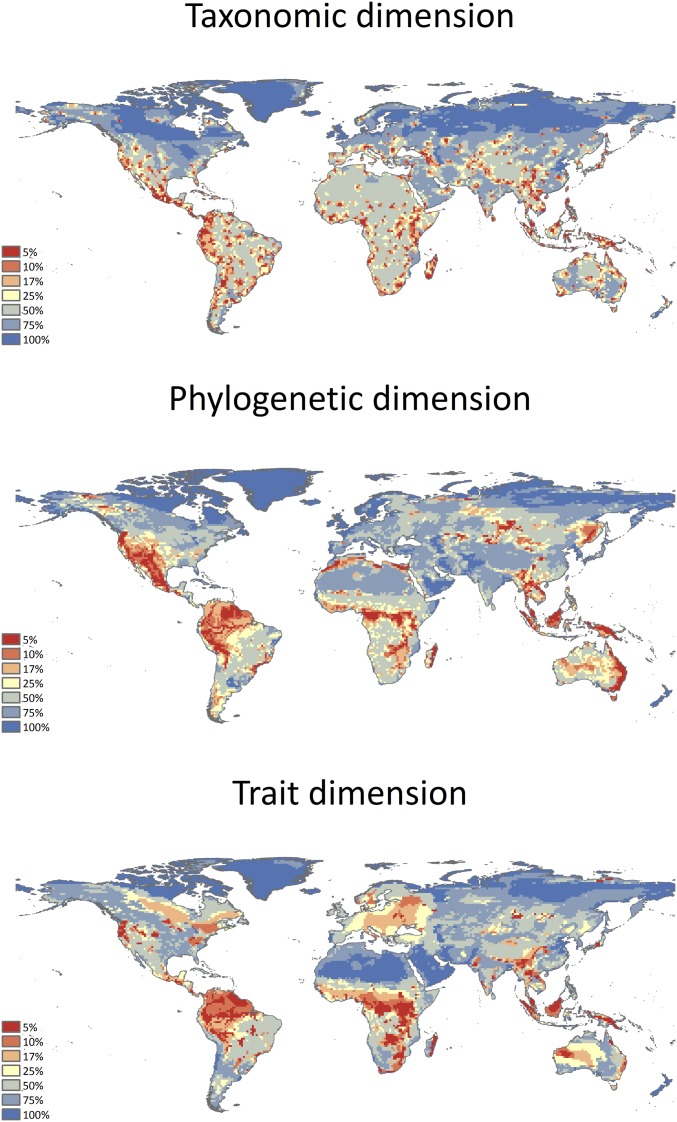
Fig. S1.
Priority map for conservation of terrestrial mammals according to Zonation analysis, using CAZ. (Top) The map based on the taxonomic dimension. (Middle) The map based on the phylogenetic dimension. (Bottom) The map based on the traits dimension.
In our study, priority areas based on phylogeny and traits were generally similar, but there were important differences as well. For example, parts of Australia were more valuable in the phylogeny-based solution than in the trait-based solution. Almost half of the native land mammals from Australia are either monotremes or marsupials, lineages that diverged early in mammalian evolution and have a unique evolutionary history, making them phylogenetically distinct. However, the ecological traits of Australian marsupials are functionally similar to those of placentals and are classic examples of ecological convergence (37). The preponderance of these distinct lineages may explain why Australia had more areas of importance when we considered priorities for the phylogenetic dimension rather than trait dimension, and the results suggest we are successfully capturing different facets of biodiversity in our prioritization analyses.
The overlap of important areas across the three dimensions of biodiversity was low, only 4.6% of the global land area (Figs. 1 and 2). Many of the overlapping regions have been identified previously as important areas for conservation because of their high species richness and number of threatened and restricted-range species, including mammals and other vertebrates, invertebrates, and trees (10, 31–33, 36, 38). These areas also were included in the global priority map for the expansion of protected areas to achieve the goals of 17% of global protected land and triple the average protection of vertebrate species’ ranges (39). Further, some of the areas that we identify here as important for all dimensions of mammal biodiversity are already recognized as Biodiversity Hotspots (40) and High Biodiversity Wilderness Areas (41). For example, the Tropical Andes in the Neotropics, Madagascar, Sundaland and Indo-Burma regions in Indo-Malay realm, and the forest of eastern Australia are biodiversity hotspots because of their high vulnerability and irreplaceability (40). Likewise, the Amazon and New Guinea are identified by Conservation International as High-Biodiversity Wilderness Areas because of their mostly intact original vegetation cover and high species richness and endemism (41). We found that these areas are important for all three dimensions of biodiversity and that they harbor not only high species richness and endemism but also unique evolutionary lineages and distinct ecological traits, underscoring the importance of protecting these regions.
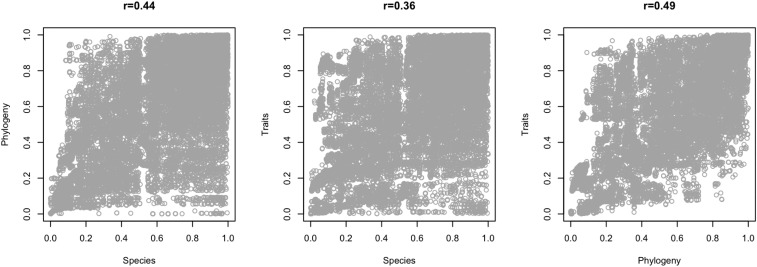
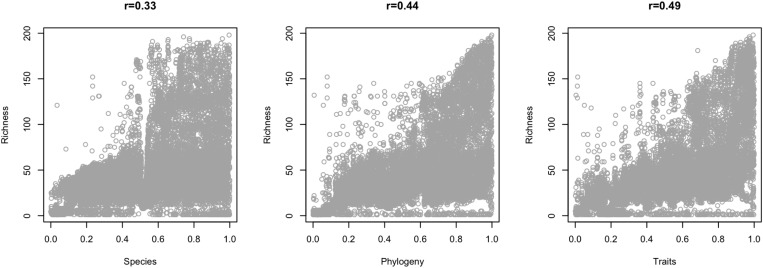
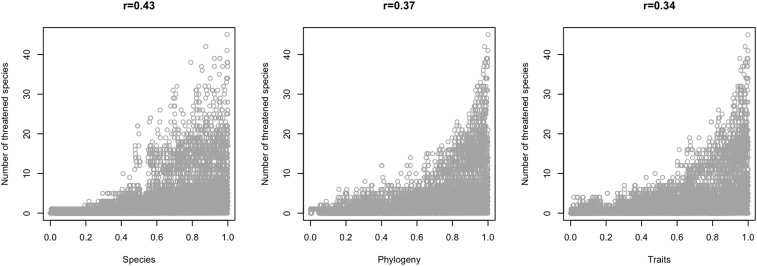
The conservation values according to our prioritization analysis for each of the different dimensions were correlated (Table 1 and Fig. S2), but correlation values were small, and the correlation plots did not show clear trends, again indicating the low congruence of conservation priorities among the three dimensions of mammal biodiversity. The conservation values were also positively correlated with species richness and with the number of threatened species, showing that places with higher conservation values for all dimensions usually had higher species richness (Table 1 and Fig. S3), and consistently harbored a greater number of threatened species (Table 1 and Fig. S4). The spatial mismatch among the taxonomic, phylogenetic, and functional components of biodiversity has been observed globally for birds (26), freshwater fishes (22), and mammals (15). Such lack of congruence triggers a conservation conflict, because conservation strategies based only on taxonomic diversity do not include important phylogenetic and functional hotspots (15, 26).
Table 1.
Kendall rank correlation coefficients (τ) between the conservation priority values given by the spatial prioritization analysis for each pair of dimension of biodiversity (species, phylogeny, and traits) and with species richness and the number of threatened mammal species
| Conservation priority | |||||
| Taxonomic dimension | Phylogenetic dimension | Trait dimension | Species richness | Number of threatened species | |
| Conservation priority for taxonomic dimension | 1.00 | 0.30 | 0.25 | 0.34 | 0.44 |
| Conservation priority for phylogenetic dimension | 0.30 | 1.00 | 0.43 | 0.46 | 0.37 |
| Conservation priority for trait dimension | 0.25 | 0.43 | 1.00 | 0.50 | 0.34 |
All correlation coefficients were significant for P < 0.001.
Fig. S2.
Correlation plots of the conservation value of each cell resulting from Zonation analysis based on species occurrences (Species), phylogenetic groups (Phylogeny), and trait values (Traits). The Kendall correlation (τ) coefficient is indicated above the plots.
Fig. S3.
Plots of the correlation between mammal species richness per cell and the conservation rank of each cell resulting from Zonation analysis based on species occurrences (Species), phylogenetic groups (Phylogeny), and trait values (Traits). The Kendall correlation (τ) coefficient is indicated above the plots.
Fig. S4.
Plots of the correlation between the number of threatened mammal species per cell and the conservation rank of each cell resulting from Zonation analysis based on species occurrences (Species), phylogenetic groups (Phylogeny), and trait values (Traits). The Kendall correlation (τ) coefficient is indicated above the plots.
Priority Areas and Currently Protected Areas.
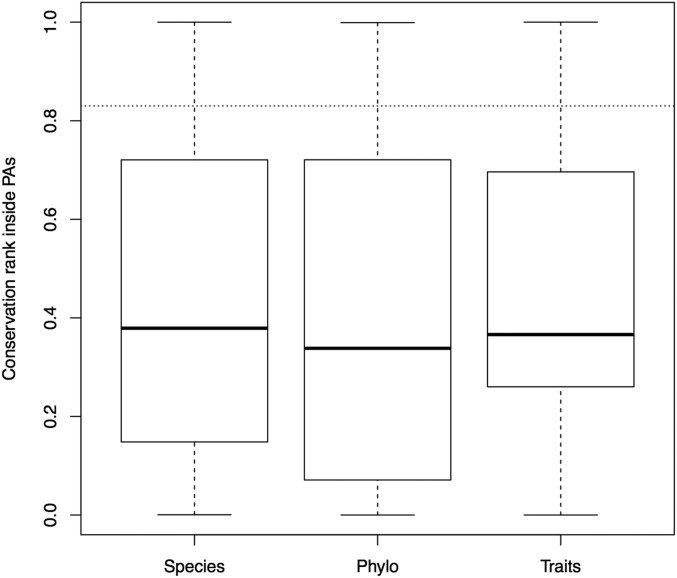
Currently protected areas performed poorly in terms of protecting the species, phylogenetic diversity, trait dimensions of mammal biodiversity and also in protecting threatened species (Table 2). The average conservation values of cells currently under protection were significantly lower than would be expected by randomly selecting areas around the globe, and this finding was consistent for each of the three dimensions and for all three dimensions jointly (Fig. S5). Similarly, Guilhaumon et al. (27) found that the Mediterranean marine protected areas system did not harbor more taxonomic diversity, phylogenetic diversity, or functional diversity of coastal fish than would be expected by chance. Jenkins et al. (38) showed that location of protected areas in the United States contrasts with regions where high numbers of endemic and threatened species are located, leaving unique species unprotected. Some of the priority areas highlighted by Jenkins et al. (38) concur with those that we identify as important across the three dimensions of mammal biodiversity, especially in the western United States. Several studies (e.g., refs. 42–44) have shown that the global network of protected areas is ineffective and is biased toward residual locations; these places are considered cheap to protect because they are remote or of low economic value, making it easier to set them aside for protection (7, 44, 45). Our findings combined with these other studies at local and regional scales demonstrate that currently protected areas are not maximizing the coverage of biodiversity.
Table 2.
Percentage of global total of species richness, phylogenetic diversity (14), trait diversity (20), and number of threatened species that are retained within the top 17% of the important regions for each dimension (taxonomic, phylogenetic, and functional) and across all three dimensions (overlap priorities) from the zonation prioritization analyses and in currently protected areas
| % of the global total | Zonation prioritization | ||||
| Taxonomic dimension, % | Phylogenic dimension, % | Trait dimension, % | Overlap priorities | Currently protected areas, % | |
| Species richness | 99 | 83 | 71 | 70 | 62 |
| Phylogenetic diversity | 99 | 90 | 87 | 80 | 74 |
| Trait diversity | 99 | 91 | 80 | 80 | 78 |
| Threatened species | 99 | 72 | 56 | 57 | 37 |
Fig. S5.
Conservation values inside currently protected areas (PAs) from the conservation solutions based on the three dimensions of biodiversity (species, phylogeny, and traits). The dotted line indicates the 17% target for conservation, showing that species, phylogeny, and traits that are rare are poorly represented within protected areas.
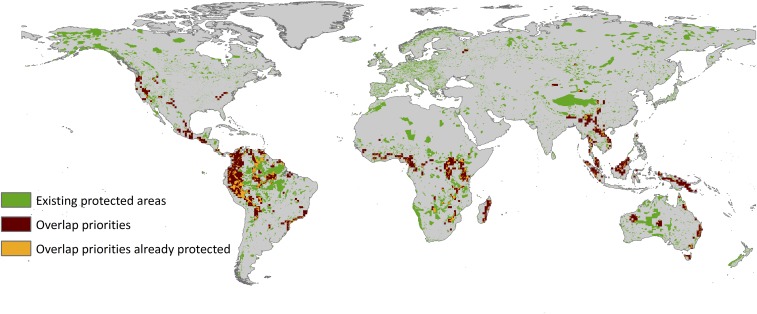
Of the 4.6% of the area that we indicated as having priority for conservation across all dimensions of mammal biodiversity (consensus map, Fig. 3), only 1% is currently protected, leaving vulnerable 3.6% of the most important areas for all three dimensions of mammal diversity conservation. Considering that ∼14% of the Earth’s land area is already protected (46), the 3.6% of unprotected land that we identified here as key areas for mammal conservation could inform efforts to expand the system of currently protected areas to achieve the Aichi Target 11. However, setting aside areas for conservation incurs socioeconomic and political costs, such as land acquisition value, social conflicts, and political willingness (7, 35). Therefore, the priority areas provide a key biodiversity layer for future conservation planning that should be considered along with the priority areas of other taxonomic groups and economic, social, and political considerations (7, 12, 35). They also highlight biologically important, unprotected regions of the globe that can be used to guide future conservation planning at both regional and global scales.
Fig. 3.
The overlap across areas important for the taxonomic, phylogenetic, and trait dimensions (brown), the current network of protected areas (green), and areas of overlap that are already protected (orange).
Conclusion
We show that priority areas for taxonomic, phylogenetic, and functional dimensions of biodiversity are strongly spatially mismatched, suggesting that conservation planning based in only one dimension of biodiversity could undermine the conservation of the other two. Effective conservation planning should maintain taxonomic, phylogenetic, and functional diversity to ensure biodiversity persistence in a changing world (47). Here, we identify a set of areas that are high conservation priorities across all three dimensions; this information can be used to inform conservation actions to expand the current network of protected areas to achieve the 17% target proposed by the Convention on Biological Diversity strategic plan for 2011–2020. The current system of protected areas poorly protects species richness, phylogenetic diversity, functional diversity, and threatened species; the important areas that we identified could perform better for all these aspects of biodiversity. The small fraction of areas (4.6%) where there was overlap across the dimensions should receive special conservation attention. Those areas retain many restricted-range species that have distinct evolutionary history and unique traits. Our work presents a global effort to identify important areas for terrestrial mammal conservation across the taxonomic, phylogenetic, and functional dimensions of biodiversity using a complementarity-based analysis. The key areas identified here can be used as a biological foundation for future conservation planning, which also would have to account for other factors such as opportunity costs, social/political considerations, and funding for land purchase. Conserving biodiversity beyond species identity (13, 48, 49) is crucial to ensure the provision of ecosystem services and their contribution to human well-being, the evolutionary potential for species to evolve and adapt, and the extraordinary diversity that exists across mammalian lineages.
Methods
Occurrence Data.
We used maps of “extent of suitable habitat” that were generated based on species’ ranges and habitat preferences (50) to calculate mammal species occurrence. We aggregated the occurrence information into distribution maps (presence/absence) of 1° × 1° (∼110 × 110 km at the equator) and identified priority regions for the mammal taxonomic dimension using geographic distribution maps for 4,547 terrestrial mammals.
Traits.
We compiled a species-level traits database for 4,547 terrestrial mammals (17, 51–54). From a total of 23 traits available, we used 14 intrinsic biological traits in the analyses. We chose these traits based on ecological importance, the correlation among them, and the percentage of missing values. Traits were related to resource use (activity cycle, habitat mode, trophic level, diet breadth), speed of life history (body mass, litter size, litters per year, gestation length, weaning age, neonate body mass, maximum life span), and population characteristics (social group size, population density). Because removing species for which some trait data are lacking can cause statistical bias and interpretation error (55), we imputed missing trait values using missForest [Package missForest in R (56)], a nonparametric approach based on random forests. This method performs well on large databases with correlated variables (56, 57). Phylogeny was not used in the imputation analysis. Because some of the traits had high proportions of data missing, we ran the prioritization analysis removing traits with more than 60% of missing data as a robustness check. We found that removing traits that had high proportions of missing data before imputations did not change our main results.
To represent the trait dimension in the prioritization analyses, we followed the framework of Strecker et al. (22), using a grid cells × traits matrix. To create this matrix, all trait variables were converted into binary format. For categorical traits, we assigned the presence/absence of each category. We split quantitative traits into 5% quantiles and then converted them into binary variables. (We choose to use 5% to incorporate more trait variability in our analysis.) Once our dataset was converted to binary format, we created a binary species × traits matrix. By multiplying the species × traits and species × grid cells (obtained from occurrence data) matrices, we obtained a trait × grid cells matrix in which each 1° × 1° grid cell contained the number of species exhibiting a trait value (e.g., the number of nocturnal species in that cell). Then we generated a distribution map for each trait value and used those maps in the prioritization analysis to find the priority regions for mammal trait dimension.
Phylogeny.
We used phylogenetic eigenvectors to represent the phylogenetic dimension in our analyses, thereby avoiding nonindependence issues associated with phylogenetic trees (58). We used an interpolated, smoothed phylogenetic tree of mammals (59) to obtain a phylogenetic distance matrix among all species. Then we synthesized the phylogenetic information in eigenvectors by conducting a principal coordinate analysis (PCoA) based on phylogenetic distances between species (PVR package in R; refs. 58, 60). Eigenvectors from a phylogenetic distance matrix reflect the different phylogenetic relationships among species in independent vectors (60). The first eigenvectors tend to represent larger distances among species, expressing divergences closer to the root of the phylogeny (60), and subsequent eigenvectors tend to capture phylogenetic relationships closer to the terminal nodes. For each species, we generated multiple eigenvector scores that represent relatedness of each species to all other species at different phylogenetic levels. We used only axes that presented eigenvalues larger than 1%, because axes with eigenvalues less than 1% contain only a small fraction of the total variation of the whole phylogeny, and we wanted to avoid including low-representative axes in the analysis. From 4,546 phylogenetic axes generated by the PCoA, we only used 16 eigenvectors in our analyses. These eigenvectors contained 63% of the total variation in the phylogenetic distance matrix. We also tested whether our results were sensitive to the number of eigenvectors by using 16, 100, 200, and 250 eigenvectors. Because the results generated by the different sets were highly correlated, the inclusion of more eigenvectors appeared not to provide any significant new information in the analyses.
We split the first 16 phylogenetic eigenvectors into 5% quantiles, as we did for the continuous traits. For each eigenvector, species were split into 20 same-size phylogenetic groups in which species were grouped based on their phylogenetic affinity in a given phylogenetic level. Then we multiplied the binary matrix of species × phylogenetic groups by the grid cells × species matrix, resulting in a matrix of site × phylogenetic groups in which each 1° × 1° grid cell contained the number of species belonging to a particular phylogenetic group. Next, we generated a distribution map for each phylogenetic group and used those maps in the prioritization analysis to find the priority regions for the mammal phylogenetic dimension.
Prioritization Analyses.
We identified the important areas for mammal conservation across the dimensions of biodiversity using Zonation (61). Zonation produces a hierarchical prioritization of the study region based on the biological value of sites (cells), accounting for complementarity by considering the representation level of all species (or other biodiversity features). Zonation iteratively removes cells whose removal causes the smallest loss in biodiversity representation across the overall remaining region until no cell is left in the region. The hierarchical prioritization of the region is based on the order of cell removal, which is recorded and can be used later to select any given top fraction (e.g., best 10%) of the region. This order of cell removal is called “conservation value” and ranges from 0 to 1, with 0 being the first cell removed (i.e., the least important for retaining biodiversity representation) and 1 being the last cell removed from the region (the most important).
The basic cell-removal rule is the Core-Area Zonation (CAZ) algorithm. The CAZ algorithm calculates the conservation values of each cell based on the marginal loss (i.e., the relative contribution to total diversity) of the species/phylogenetic group/trait value with the higher proportion of its range in that cell. CAZ prioritizes sites by gathering a higher proportion of each dimension distribution, thus favoring rare species/phylogenetic groups/trait values in the final solution, even when they occur in otherwise species-poor regions. We analyzed each dimension separately because we wanted to evaluate the individual solution generated by each dimension and to determine how much they converge. Following the Convention on Biological Diversity (5), which proposed that 17% of the terrestrial areas should be protected by 2020, we focused our analyses on the 17% of the world with the highest conservation value (i.e., cells with conservation value greater than or equal to 0.83). In addition, we provide the full results in Fig. S1.
Protected Areas.
To compare the priority areas across the three dimensions with currently protected areas, we used the International Union for the Conservation of Nature (IUCN) and United Nations Environmental Programme World Conservation Monitoring Center (UNEP-WCMC) data on global protected areas (62). We included only restricted protected areas classified as I–IV by the IUCN in our analyses. We resampled protected areas at the 1° × 1° grid cell level.
Congruence Among Dimensions.
We assessed the congruence among dimensions and between each dimension and species richness and the number of threatened species in two ways: (i) by evaluating how the conservation values generated in our prioritization analysis related between dimensions and (ii) by calculating how much of the global amount of taxonomic/phylogenetic/trait diversity was captured by each spatial priority. First, we calculated the Kendall rank correlation to evaluate how the conservation priority values of the cells for the prioritization based on each dimension (taxonomic/phylogenetic/trait) related to each other and to species richness and to the number of threatened species. Then, to evaluate how much the spatial priorities of each dimension captured the existing taxonomic, functional, and phylogenetic diversity of terrestrial mammals, we calculated the global amount of species richness, the number of threatened mammals, phylogenetic diversity (the PD index) (14), and functional diversity (the FD index) (20). Last, we assessed how much of the global total of each index was captured proportionally by the conservation solution for each dimension and by the overlapping areas among all dimensions. We also performed the same calculation to determine how much of the global total of species richness, number of threatened mammals, phylogenetic diversity, and functional diversity are protected by the currently protected area network. The analyses were performed in R, using the “stats” package (63) for correlation analysis and the “picante” package (64) to calculate phylogenetic and functional diversity indexes.
Acknowledgments
We thank Antonin Machac, Ben Weinstein, and Brunno Oliveira for helpful discussions and the two anonymous reviewers for their valuable suggestions to improve this paper. Our research was funded by the National Science Foundation Dimensions of Biodiversity Program Grant DEB-1136586. We thank Bruce Young and Tom Brooks, co-PIs in the NSF DEB-1136586 grant that helped to support this research. F.T.B. was supported by a Coordination for the Improvement of Higher Education Personnel (CAPES) doctorate scholarship, received CAPES Doctorate Scholarship BEX 7910/13-4 to visit C.H.G.’s laboratory at Stony Brook University, and currently holds a postdoctoral scholarship from O Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Grant 152172/2016-5. R.L. was supported by CNPq Grants 308532/2014-7, 479959/2013-7, 407094/2013-0, and 563621/2010-9 and by O Boticário Group Foundation for Nature Protection Grant PROG_0008_2013. This paper is a contribution of the Instituto Nacional de Ciência e Tecnologia in Ecology, Evolution, and Biodiversity Conservation founded by Ministério da Ciência, Tecnologia, Inovações e Comunicações/CNPq and Fundação de Amparo a Pesquisa do Estado de Goiás (Grant 465610/2014-5). V.C.R. received funding from the Terra and Aqua Program of the National Aeronautics and Space Administration Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.C.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706461114/-/DCSupplemental.
References
- 1.Ellis EC, Ramankutty N. Putting people in the map: Anthropogenic biomes of the world. Front Ecol Environ. 2008;6:439–447. [Google Scholar]
- 2.Barnosky AD, et al. Has the Earth’s sixth mass extinction already arrived? Nature. 2011;471:51–57. doi: 10.1038/nature09678. [DOI] [PubMed] [Google Scholar]
- 3.Pimm SL, et al. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344:1246752. doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- 4.Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. Human domination of Earth’s ecosystems. Science. 1997;277:494–499. [Google Scholar]
- 5. Convention on Biological Diversity (2010) COP 10 Decision X/2: Strategic Plan for Biodiversity 2011–2020. Available at https://www.cbd.int/decision/cop/?id=12268. Accessed April 15, 2014.
- 6.Kullberg P, Moilanen A. How do recent spatial biodiversity analyses support the convention on biological diversity in the expansion of the global conservation area network? Nat Conserv. 2014;12:3–10. [Google Scholar]
- 7.Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- 8.Rondinini C, Rodrigues ASL, Boitani L. The key elements of a comprehensive global mammal conservation strategy. Philos Trans R Soc Lond B Biol Sci. 2011;366:2591–2597. doi: 10.1098/rstb.2011.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks TM, et al. Global biodiversity conservation priorities. Science. 2006;313:58–61. doi: 10.1126/science.1127609. [DOI] [PubMed] [Google Scholar]
- 10.Jenkins CN, Pimm SL, Joppa LN. Global patterns of terrestrial vertebrate diversity and conservation. Proc Natl Acad Sci USA. 2013;110:E2602–E2610. doi: 10.1073/pnas.1302251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CJ, et al. Effective conservation requires clear objectives and prioritizing actions, not places or species. Proc Natl Acad Sci USA. 2015;112:E4342. doi: 10.1073/pnas.1509189112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kukkala AS, Moilanen A. Core concepts of spatial prioritisation in systematic conservation planning. Biol Rev Camb Philos Soc. 2013;88:443–464. doi: 10.1111/brv.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz S, et al. Incorporating plant functional diversity effects in ecosystem service assessments. Proc Natl Acad Sci USA. 2007;104:20684–20689. doi: 10.1073/pnas.0704716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 15.Mazel F, et al. Multifaceted diversity-area relationships reveal global hotspots of mammalian species, trait and lineage diversity. Glob Ecol Biogeogr. 2014;23:836–847. doi: 10.1111/geb.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaac NJB, Redding DW, Meredith HM, Safi K. Phylogenetically-informed priorities for amphibian conservation. PLoS One. 2012;7:e43912. doi: 10.1371/journal.pone.0043912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson AD, Hamilton MJ, Boyer AG, Brown JH, Ceballos G. Multiple ecological pathways to extinction in mammals. Proc Natl Acad Sci USA. 2009;106:10702–10705. doi: 10.1073/pnas.0901956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv Biol. 2010;24:1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 19.Purvis A, Agapow PM, Gittleman JL, Mace GM. Nonrandom extinction and the loss of evolutionary history. Science. 2000;288:328–330. doi: 10.1126/science.288.5464.328. [DOI] [PubMed] [Google Scholar]
- 20.Petchey OL, Gaston KJ. Extinction and the loss of functional diversity. Proc Biol Sci. 2002;269:1721–1727. doi: 10.1098/rspb.2002.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobral FL, et al. Spatial conservation priorities for top predators reveal mismatches among taxonomic, phylogenetic and functional diversity. Nat Conserv. 2014;12:150–155. [Google Scholar]
- 22.Strecker AL, Olden JD, Whittier JB, Paukert CP. Defining conservation priorities for freshwater fishes according to taxonomic, functional, and phylogenetic diversity. Ecol Appl. 2011;21:3002–3013. [Google Scholar]
- 23.Huang S, Stephens PR, Gittleman JL. Traits, trees and taxa: Global dimensions of biodiversity in mammals. Proc Biol Sci. 2012;279:4997–5003. doi: 10.1098/rspb.2012.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safi K, et al. Understanding global patterns of mammalian functional and phylogenetic diversity. Philos Trans R Soc Lond B Biol Sci. 2011;366:2536–2544. doi: 10.1098/rstb.2011.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreiss LM, et al. Taxonomic, functional, and phylogenetic dimensions of rodent biodiversity along an extensive tropical elevational gradient. Ecography. 2015;38:876–888. [Google Scholar]
- 26.Devictor V, et al. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: The need for integrative conservation strategies in a changing world. Ecol Lett. 2010;13:1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- 27.Guilhaumon F, et al. Representing taxonomic, phylogenetic and functional diversity: New challenges for Mediterranean marine-protected areas. Divers Distrib. 2015;21:175–187. [Google Scholar]
- 28.Rodrigues ASL, et al. Global gap analysis: Priority regions for expanding the global protected-area network. Bioscience. 2004;54:1092–1100. [Google Scholar]
- 29.Schipper J, et al. The status of the world’s land and marine mammals: Diversity, threat, and knowledge. Science. 2008;322:225–230. doi: 10.1126/science.1165115. [DOI] [PubMed] [Google Scholar]
- 30.Di Marco M, et al. A novel approach for global mammal extinction risk reduction. Conserv Lett. 2012;5:134–141. [Google Scholar]
- 31.Dobrovolski R, Loyola RD, Guilhaumon F, Gouveia SF, Diniz-Filho JAF. Global agricultural expansion and carnivore conservation biogeography. Biol Conserv. 2013;165:162–170. [Google Scholar]
- 32.Dobrovolski R, Loyola R, da Fonseca GAB, Diniz-Filho JAF, Araujo MB. Globalizing conservation efforts to save species and enhance food production. Bioscience. 2014;64:539–545. [Google Scholar]
- 33.Di Minin E, et al. Global priorities for national carnivore conservation under land use change. Sci Rep. 2016;6:23814. doi: 10.1038/srep23814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergamin RS, et al. Linking beta diversity patterns to protected areas: Lessons from the Brazilian Atlantic Rainforest. Biodivers Conserv. 2017;26:1557–1568. [Google Scholar]
- 35.Carwardine J, et al. Cost-effective priorities for global mammal conservation. Proc Natl Acad Sci USA. 2008;105:11446–11450. doi: 10.1073/pnas.0707157105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albuquerque F, Beier P. Global patterns and environmental correlates of high-priority conservation areas for vertebrates. J Biogeogr. 2015;42:1397–1405. [Google Scholar]
- 37.Forget P, Vander Wall SB. Scatter-hoarding rodents and marsupials: Convergent evolution on diverging continents. Trends Ecol Evol. 2001;16:65–67. doi: 10.1016/s0169-5347(00)02072-3. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins CN, Van Houtan KS, Pimm SL, Sexton JO. US protected lands mismatch biodiversity priorities. Proc Natl Acad Sci USA. 2015;112:5081–5086. doi: 10.1073/pnas.1418034112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montesino Pouzols F, et al. Global protected area expansion is compromised by projected land-use and parochialism. Nature. 2014;516:383–386. doi: 10.1038/nature14032. [DOI] [PubMed] [Google Scholar]
- 40.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 41.Mittermeier RA, et al. Wilderness and biodiversity conservation. Proc Natl Acad Sci USA. 2003;100:10309–10313. doi: 10.1073/pnas.1732458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Saout S, et al. Protected areas and effective biodiversity conservation. Science. 2013;342:803–805. doi: 10.1126/science.1239268. [DOI] [PubMed] [Google Scholar]
- 43.Nori J, et al. Amphibian conservation, land-use changes and protected areas: A global overview. Biol Conserv. 2015;191:367–374. [Google Scholar]
- 44.Venter O, et al. Targeting global protected area expansion for imperiled biodiversity. PLoS Biol. 2014;12:e1001891. doi: 10.1371/journal.pbio.1001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devillers R, et al. Reinventing residual reserves in the sea: Are we favouring ease of establishment over need for protection? Aquat Conserv. 2015;25:480–504. [Google Scholar]
- 46.United Nations Environment Programme World Conservation Monitoring Center; International Union for the Conservation of Nature . Protected Planet Report 2016. UNEP-WCMC; Cambridge, UK: 2016. [Google Scholar]
- 47.Pressey RL, Cabeza M, Watts ME, Cowling RM, Wilson KA. Conservation planning in a changing world. Trends Ecol Evol. 2007;22:583–592. doi: 10.1016/j.tree.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Forest F, et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- 49.Hidasi-Neto J, Loyola RD, Cianciaruso MV. Global and local evolutionary ecological distinctiveness of terrestrial mammals: Identifying priorities across scales. Divers Distrib. 2015;21:548–559. [Google Scholar]
- 50.Rondinini C, et al. Global habitat suitability models of terrestrial mammals. Philos Trans R Soc Lond B Biol Sci. 2011;366:2633–2641. doi: 10.1098/rstb.2011.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones KE, et al. PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009;90:2648–2648. [Google Scholar]
- 52.Tacutu R, et al. Human ageing genomic resources: Integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pacifici M, et al. Generation length for mammals. Nat Conserv. 2013;5:87–94. [Google Scholar]
- 54.Verde Arregoitia LD, Blomberg SP, Fisher DO. Phylogenetic correlates of extinction risk in mammals: Species in older lineages are not at greater risk. Proc R Soc B Biol Sci. 2013;280:20131092. doi: 10.1098/rspb.2013.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakagawa S, Freckleton RP. Missing inaction: The dangers of ignoring missing data. Trends Ecol Evol. 2008;23:592–596. doi: 10.1016/j.tree.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Stekhoven DJ, Bühlmann P. MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 57.Penone C, et al. Imputation of missing data in life-history trait datasets: Which approach performs the best? Methods Ecol Evol. 2014;5:961–970. [Google Scholar]
- 58.Diniz-Filho JAF, de Sant’Ana CER, Bini LM. An eigenvector method for estimating phylogenetic inertia. Evolution. 1998;52:1247–1262. doi: 10.1111/j.1558-5646.1998.tb02006.x. [DOI] [PubMed] [Google Scholar]
- 59.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diniz-Filho JAF, et al. On the selection of phylogenetic eigenvectors for ecological analyses. Ecography. 2012;35:239–249. [Google Scholar]
- 61.Moilanen A, et al. 2012. Zonation Spatial Conservation Planning Framework and Software, User Manual (Edita, Helsinki), Version 3.1.
- 62. International Union for the Cconservation of Nature; United Nations Environment Programme (2016) World Database on Protected Areas. Available at https://www.protectedplanet.net/. Accessed June 15, 2016.
- 63. R Development Core Team (2016) R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna)
- 64.Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]