Significance
In animals, carotenoid pigments fulfill a number of essential roles in vision, protection from stresses, and signaling. Although carotenoids are synthesized primarily by plants and some bacteria and fungi, carotenoid biosynthetic genes have been acquired by horizontal gene transfer in some insects and mites. In the two-spotted spider mite, Tetranychus urticae, as well as in a related species, we show that a horizontally transferred carotenoid biosynthetic gene is required for normal pigmentation. Spider mites can therefore synthesize their own carotenoids. Further, in a T. urticae strain, the gene is required for the induction of diapause, a key for overwintering of this widespread and important agricultural pest.
Keywords: horizontal gene transfer, xanthophylls, β-carotene, bulked segregant analysis, spider mites
Abstract
Carotenoids underlie many of the vibrant yellow, orange, and red colors in animals, and are involved in processes ranging from vision to protection from stresses. Most animals acquire carotenoids from their diets because de novo synthesis of carotenoids is primarily limited to plants and some bacteria and fungi. Recently, sequencing projects in aphids and adelgids, spider mites, and gall midges identified genes with homology to fungal sequences encoding de novo carotenoid biosynthetic proteins like phytoene desaturase. The finding of horizontal gene transfers of carotenoid biosynthetic genes to three arthropod lineages was unprecedented; however, the relevance of the transfers for the arthropods that acquired them has remained largely speculative, which is especially true for spider mites that feed on plant cell contents, a known source of carotenoids. Pigmentation in spider mites results solely from carotenoids. Using a combination of genetic approaches, we show that mutations in a single horizontally transferred phytoene desaturase result in complete albinism in the two-spotted spider mite, Tetranychus urticae, as well as in the citrus red mite, Panonychus citri. Further, we show that phytoene desaturase activity is essential for photoperiodic induction of diapause in an overwintering strain of T. urticae, consistent with a role for this enzyme in provisioning provitamin A carotenoids required for light perception. Carotenoid biosynthetic genes of fungal origin have therefore enabled some mites to forgo dietary carotenoids, with endogenous synthesis underlying their intense pigmentation and ability to enter diapause, a key to the global distribution of major spider mite pests of agriculture.
Carotenoids are isoprenoid compounds produced by photosynthetic organisms, like plants, as well as by some bacteria, Archaea, and fungi (1). Over 700 structures are found in nature, reflecting modifications to the C40 backbone, such as cyclization or the addition of oxygen-containing groups (1, 2). In animals, carotenoids are widespread and have diverse roles, including signaling to conspecifics and protection from oxidative stress (2–6). Most animals obtain carotenoids from their diet, including carotenes (hydrocarbon carotenoids), like β-carotene, and xanthophylls (carotenoids with oxygen, like lutein or astaxanthin) (7). Many animals are also able to modify carotenes to produce their own brightly colored xanthophylls, and, collectively, carotenoids underlie many of the striking yellows, oranges, and reds observed in the animal kingdom (8), including feather and beak colors in birds (9, 10), as well as body and cocoon colors in insects (8, 11). Further, carotenoids, including β-carotene, are processed in animals to chromophores, including retinal (vitamin A), that, in complexes with opsins, are the chemical transducers in vision (2, 12, 13). Finally, carotenoids, such as lutein and zeaxanthin in human and keto-carotenoids like astaxanthin in other organisms, have been postulated to serve important roles as antioxidants (14, 15).
Despite their ubiquity and essential functions, relatively little is known in animals about carotenoid uptake, transport, and metabolism (8), which contrasts with endogenously synthesized pigments like melanin, for which dozens of genes in the pathway for synthesis and distribution have been characterized (8, 16). Until recently, all animals were thought to lack the ability to synthesize carotenoids de novo. This thinking changed when Moran and Jarvik (11) discovered that the pea aphid (Acyrthosiphon pisum) genome harbors carotenoid biosynthetic genes acquired by horizontal gene transfer. Subsequently, carotenoid biosynthetic genes were identified in other Hemiptera, the adelgids (17), and in two other arthropod lineages, gall midges (Insecta, Diptera) (18) and spider mites (in Tetranychus urticae, a member of Chelicerata, the sister taxon to the mandibulate arthropods) (19, 20). In all cases, the laterally transferred genes consisted of fused cyclase/synthases and phytoene desaturases. The cyclase/synthase fusions suggested a fungal origin of the lateral transfer events, a conclusion confirmed by phylogenetic analyses (11, 18–20). In fungi, cyclase/synthase fusion proteins and phytoene desaturases are required for the synthesis of carotenes, including β-carotene (21), a provitamin A carotenoid and a substrate for the biosynthesis of diverse xanthophylls (7, 22). In aphids, at least one of the carotenoid synthesis genes is active because a spontaneous point mutation in a copy of a phytoene desaturase is associated with loss of red body color (11). Although body color in aphids has been shown to impact interactions with natural enemies (23, 24), the broader significance of the parallel acquisition of fungal carotenoid biosynthesis genes in divergent plant feeding arthropod groups has remained speculative (25).
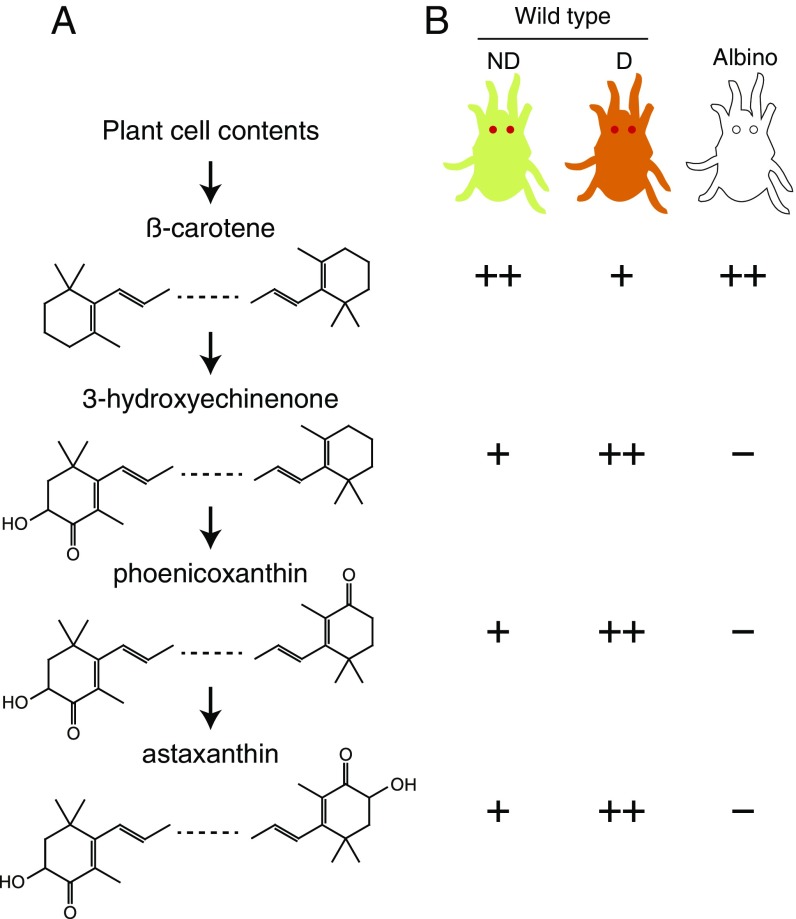
Spider mites are an attractive system to study carotenoid biosynthesis, metabolism, and function. In the two-spotted spider mite, T. urticae, red eyes and yellow body color result solely from carotenoids (Figs. 1 and 2A) (26). In other spider mites, such as the citrus red spider mite, Panonychus citri, the vibrant red body color is also caused by carotenoids (Fig. 2C) (27). Moreover, in response to long nights and lower temperatures in temperate regions, female spider mites of many species, including T. urticae, enter a facultative diapause characterized by cessation of reproduction (egg laying) and a striking change in body color from faint yellow to bright red-orange (26, 28) (Fig. S1A). This body color change results from the accumulation of keto-carotenoids, like astaxanthin, which has been suggested to protect against stresses encountered during overwintering (28, 29). Further, a rich collection of pigmentation mutants of known carotenoid content has been reported in T. urticae and its sister species Tetranychus pacificus (26, 30–33). Albino mutants lack all pigmentation (body color and eyes), the lemon mutant accumulates β-carotene (and therefore is bright yellow), and white-eye mutants are unable to synthesize astaxanthin. As early as 1970, Veerman (34) proposed a pathway for the synthesis of astaxanthin from β-carotene, the latter of which he presumed to be of dietary origin (Fig. 1A). Strikingly, most of the T. urticae pigment mutants diapaused normally with yellow or orange coloration; however, whereas albino mites were observed to diapause as assessed by reproductive cessation, the incidence of induction was reduced and variable compared with wild type (WT) (35).
Fig. 1.
Carotenoids in WT and albino mutants of T. urticae. (A) Carotenoids observed in females of Tetranychus mite species as reported in earlier studies (26, 33, 34). The previously proposed pathway from presumptive dietary β-carotene to the terminal keto-carotenoid astaxanthin is shown (34); intermediates, minor carotenoid species, and carotenoid esters are not indicated. (B) Carotenoid profiles in nondiapausing (ND) and diapausing (D) WT T. urticae females, and in nondiapausing albino mutants as reported by Veerman (26) (compare with A). Plus or minus signs indicate relative levels, with the keto-carotenoids undetectable in albino mites. Schematics of WT and mutant mites are given at the top; for display, the eyes have been enlarged.
Fig. 2.
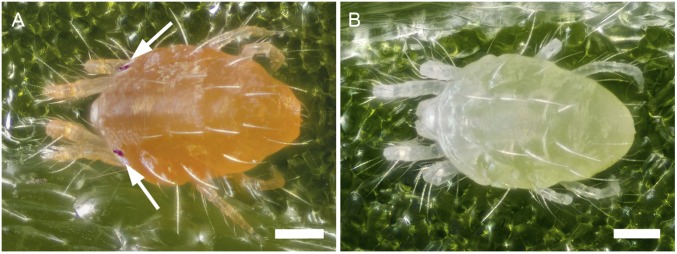
The albino pigment phenotypes of T. urticae and P. citri. Shown are (A) WT T. urticae pigmentation (strains Wasatch, MAR-AB, Foothills, and London; a representative London mite is shown), (B) albino phenotype of T. urticae (strains Alb-NL, Alb-JP, W-Alb-7, -8, -10, -11, and -14; a representative Alb-NL mite is shown), (C) WT P. citri phenotype, and (D) albino phenotype of P. citri. In all cases, adult females are shown. Arrows indicate red eye spots or red-orange in the distal front legs of WT mites that are absent in the albino mutants. The dark regions present in the mite bodies are gut contents that are visible because spider mites are partly translucent. (Scale bars: 0.1 mm.)
Fig. S1.
Diapausing WT and albino T. urticae females. Image of a diapausing (A) WT adult T. urticae female (strain Wasatch) and (B) albino adult T. urticae female (strain Alb-NL). A diapausing WT T. urticae female has a yellow to bright red-orange coloration whereas a diapausing albino T. urticae female has a uniformly white appearance. Note the absence of gut contents (dark regions in the body), reflecting cessation of feeding (compare with feeding mites shown in Fig. 2). Under inducing conditions, females of strain Alb-NL enter diapause at a very low frequency (less than ∼1 in 1,000). Arrows indicate the position of the eyes of the WT T. urticae female. (Scale bars: 0.1 mm.)
T. urticae is an important economic pest of diverse crops and is renowned for its rapid development of pesticide resistance (36, 37). The sequencing of its genome (19) made possible methods for bulked segregant analysis (BSA) genetic mapping that were first applied to clone a monogenic locus for pesticide resistance (38, 39). In this study, we applied these resources and methods to understand the metabolism and roles of carotenoids in spider mites. Albino pigment mutants have been reported to be epistatic to others, indicative of an early and critical step in the carotenoid pathway in spider mites (Fig. 1) (26). We show that disruption of a single horizontally transferred phytoene desaturase is sufficient for complete albinism in T. urticae, as well as in P. citri. Moreover, we show that endogenously synthesized carotenoids are essential for diapause induction in an overwintering strain of T. urticae.
Results
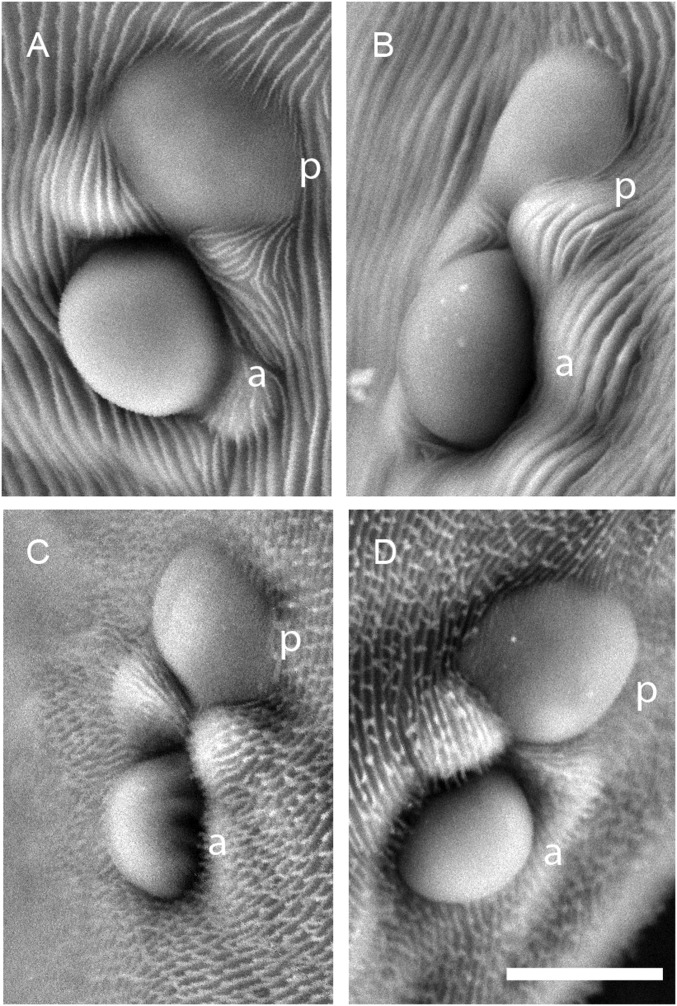
As an entry point for molecular–genetic studies of the albino phenotype in spider mites, we obtained two albino mutant strains of T. urticae, and one of P. citri. One T. urticae strain, hereafter termed Alb-NL, has been maintained at the University of Amsterdam since the late 1960s and is one of the albino 1 or albino 2 mutant lines described earlier (26). Although the exact correspondence is uncertain, albino 1 and albino 2 were reported to have identical phenotypes, including the lack of all mite-produced xanthophyll carotenoids (Fig. 1) (26). The second and previously uncharacterized T. urticae albino strain originated as a spontaneous mutant in a laboratory stock in Japan (hereafter strain Alb-JP). The Alb-NL and Alb-JP strains are phenotypically identical and lack yellow and red colors in the body, anterior and posterior eye spots, and the distal segments of the front legs (Fig. 2 A and B). As opposed to the green forms of T. urticae (which include all of the T. urticae strains used in this study), the body color of P. citri is normally bright red (Fig. 2C). In a spontaneous albino mutant of P. citri that arose in the laboratory, the red body color was absent, as were pigmented eyes (Fig. 2D). At the limit of resolution of scanning electron microscopy, the structure of the eyes in mites of the T. urticae Alb-NL strain and the albino P. citri strain were normal (Fig. S2); therefore, the absence of red eyes results from a lack of pigment as opposed to a defect in eye formation.
Fig. S2.
Scanning electron microscopy (SEM) images of T. urticae and P. citri eyes. SEM images of the anterior (a) and posterior (p) eyes of an (A) WT adult T. urticae female (strain London), (B) adult T. urticae albino female (strain Alb-NL), (C) WT adult P. citri female, and (D) adult P. citri female with an albino phenotype. (Scale bar: 10 μm.)
Inheritance of Albino Phenotypes and Genetic Complementation.
To determine the genetic basis of albino phenotypes, we performed reciprocal crosses between albino and WT strains of T. urticae and P. citri. In all cases, F1s of the resulting crosses had normal body and eye color (Table 1). This result, together with the finding of an ∼1:1 ratio of albino to WT haploid F2 sons produced by virgin F1 females, revealed that albinism in all strains was inherited in a monogenic recessive manner. For Alb-NL, this result confirmed earlier findings and shows that the genetic architecture is unchanged in the more than 40 y the strain has been maintained in the laboratory. As revealed under high magnification and bright light, extremely faint red was apparent in the eyes of some otherwise albino F2 males, a previously reported phenotype attributed to maternal provisioning of carotenoids from phenotypically WT F1 mothers (30, 32). It should be noted that, in one direction, the cross between Alb-NL and a WT strain (MAR-AB) gave a modest but statistically significant deviation from a 1:1 phenotypic ratio in F2 males (Table 1). A similar deviation in F2 ratios was reported in an earlier cross with an albino strain, presumably reflecting a maternal effect (32). Finally, we crossed strain Alb-NL to strain Alb-JP and found that all female F1 progeny were albino. This failure to complement suggests that the pigmentless phenotypes of Alb-NL and Alb-JP result from disruptions in the same gene.
Table 1.
Inheritance of the albino phenotype and genetic complementation
| Crosses and strains | F1 ♀s, % Alb* | F2 haploid ♂s | χ2 | df | P value | |
| Alb | WT | |||||
| Inheritance tests (♀ x ♂) | ||||||
| Tetranychus urticae | ||||||
| Alb-NL × MAR-AB | 0 | 522 | 522 | 0.00 | 1 | 1 |
| MAR-AB × Alb-NL | 0 | 931 | 1,099 | 13.90 | 1 | <0.001 |
| Alb-JP × WT JP | 0 | 455 | 442 | 0.1884 | 1 | 0.6643 |
| WT JP × Alb-JP | 0 | 425 | 413 | 0.1718 | 1 | 0.6785 |
| W-Alb-2 × Wasatch | 0 | 110 | 114 | 0.0714 | 1 | 0.7893 |
| Wasatch × W-Alb-2 | 0 | 218 | 219 | 0.0022 | 1 | 0.9619 |
| W-Alb-14 × Wasatch | 0 | 959 | 830 | 9.16 | 1 | 0.0023 |
| Wasatch × W-Alb-14 | 0 | 493 | 395 | 10.82 | 1 | 0.0010 |
| Panonychus citri | ||||||
| Albino × WT | 0 | 171 | 180 | 0.2308 | 1 | 0.6310 |
| WT × Albino | 0 | 33 | 38 | 0.3521 | 1 | 0.5529 |
| Complementation tests (♀ x ♂) | ||||||
| Tetranychus urticae | ||||||
| Alb-NL × Alb-JP | 100 | |||||
| Alb-JP × Alb-NL | 100 | |||||
| W-Alb-2 × W-Alb-14† | 100 | |||||
| W-Alb-14 × W-Alb-2† | 100 | |||||
| W-Alb-7, -8, -10, -11, -14 × Alb-JP‡ | 100 | |||||
In crosses, all F1 females were either 100% WT (yellowish body coloration and red eyes) or 100% albino (Alb) (white bodies and no eye color). Sample sizes for F1s for all crosses were ≥85.
Although progeny from complementation crosses with W-Alb-2 were albino (white bodies lacking bright red eyes), faint eye color was apparent in F1 adult females, mirroring the phenotype observed in adults of the W-Alb-2 strain.
Denotes that each of W-Alb-7, W-Alb-8, W-Alb-10, W-Alb-11, and W-Alb-14 were crossed to males of Alb-JP (in all cases, progeny were albino in phenotype).
A Locus for Albinism in Strain Alb-NL.
In previous work, we developed bulked segregant analysis (BSA) genetic mapping methods using high-throughput genomic sequencing data to localize a monogenic locus for pesticide resistance in T. urticae (38, 39). In the current study, we adapted this approach to identify the locus for albinism in Alb-NL. Briefly, the Alb-NL strain we obtained was outbred; however, inbred strains are useful in BSA mapping because they facilitate assignment of variants contributed by each parent. Therefore, we inbred, with twofold replication, the Alb-NL strain by sequential rounds of mother–son crosses to yield two strains, PA1 and PA2. Virgin albino females from each of these strains were then crossed to single males of the WT MAR-AB strain, and the resulting populations were allowed to expand for many generations (Materials and Methods) (note that a single male is effectively an inbred line in spider mites because males are haploid). For each of the two resulting populations, and with further twofold replication, pools of unselected and phenotypically albino mites were isolated, and DNA was prepared and sequenced with the Illumina high-throughput method (therefore, in total, four replicates of the selection were preformed). The parental lines (PA1 and PA2, as well as MAR-AB) were also sequenced (Fig. S3).
Fig. S3.
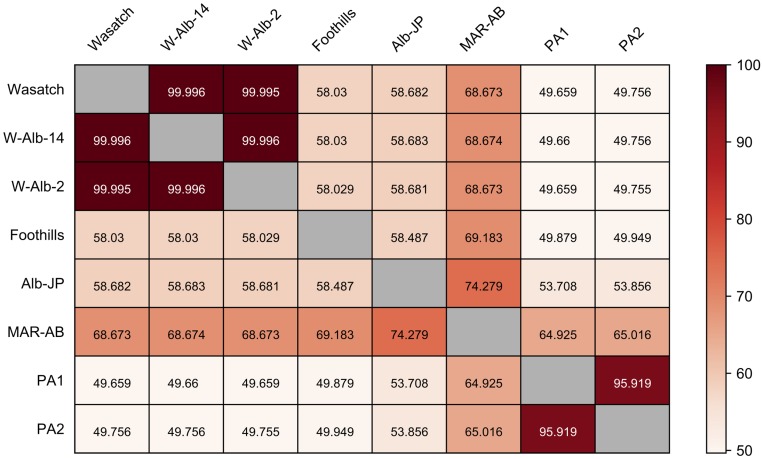
Similarity between strains as assessed with genome-wide SNP data. Values in each rectangle represent the percent similarity of pairs of strains as assessed with 488.6 thousand high quality SNPs (SI Materials and Methods) (percentage similarity for each comparison is color-coded according to the gradient at the far right). Strain names are at the left and across the top. Strains Wasatch, W-Alb-2, and W-Alb-14 are essentially identical, reflecting the inbred nature of Wasatch, and the origin of the two albino mutations on the Wasatch background (W-Alb-2 and W-Alb-14). The high level of similarity between PA1 and PA2 suggests that their progenitor Alb-NL strain was partly, but not completely, inbred at the start of the experiment (the strain had been maintained in the laboratory for more than 40 y and presumably lost genetic variation as a result of bottleneck events). Other strains show much reduced levels of identity to other strains, with Alb-NL and Alb-JP being comparatively dissimilar (reflecting an independent origin of albinism in these two strains). Although most strains are inbred or partly inbred, MAR-AB is highly outbred, potentially reflecting its apparent greater level of similarity to most other strains in the pairwise comparisons (SI Materials and Methods).
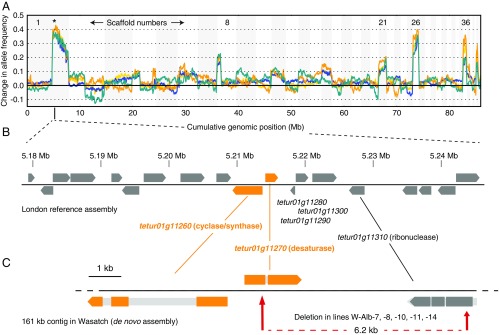
After alignment of genomic reads to the T. urticae reference genome (strain London) and variant detection, we selected informative (segregating) single nucleotide polymorphisms (SNPs) by comparing genotypic data across samples (837.6 and 771.4 thousand high-quality SNPs for the PA1 × MAR-AB and PA2 × MAR-AB crosses, respectively) (Materials and Methods). Subsequently, differences in the frequency of segregating alleles between pools for each of the four replicate selections were assessed across the genome in a sliding window analysis (the largest 44 scaffolds that harbor ∼95% of the T. urticae reference assembly were included in the analysis) (38). Each of the four replicates showed a single, maximal peak of fixation for Alb-NL alleles in the albino pools centered at ∼5.2 Mb on scaffold 1. Several other peak regions exhibited lesser shifts, presumably reflecting linkage to the causal locus (e.g., scaffolds 8, 21, 26, and 36; the scaffolds from the T. urticae genome assembly are unordered (19). Marked discontinuities in the sliding-window analyses were observed upstream of the causal region on scaffold 1, with additional discontinuities apparent on scaffolds 2, 4, and 8. Presumably, these discontinuities reflect misassemblies in several of the larger scaffolds in the T. urticae reference genome assembly (19) but are in regions that did not impact our analyses.
As inferred from the BSA scans, the causal region for recessive albinism in Alb-NL is about 600 kb in length (Fig. 3A). Strikingly, a cluster of laterally transferred carotenoid biosynthesis genes is located at the approximate maxima of the BSA peaks (tetur01g11260 and tetur01g11270). To evaluate these two genes—as well as their close neighbors—as candidates to underlie albinism, we fine-mapped the albino phenotype in a backcross between Alb-NL and strain London with flanking sets of PCR-based markers approximately equidistant to the tetur01g11260/tetur01g11270 region. As inferred from genotyping 478 albino mites, a minimal candidate region of 66.2 kb was identified centered on the carotenoid biosynthesis gene cassette. In addition to tetur01g11260 and tetur01g11270, 17 other coding genes are predicted in the nonrecombining region (Table S1). Other than tetur01g11260 and tetur01g11270, none of these genes encode products with obvious homology to proteins previously implicated in carotenoid synthesis, transport, or sequestration in other animals (e.g., scavenger B receptor or StAR-related transfer domain proteins) (8, 40).
Fig. 3.
A locus on scaffold 1 underlies albinism. (A) The results for bulked segregant analysis (BSA) genetic scans for albinism in Alb-NL are shown (crosses of inbred lines derived from the Alb-NL parent to the WT strain MAR-AB). BSA scans were performed with fourfold replication (colors yellow, orange, green, and blue); changes in allele frequencies between selected (albino) and nonselected populations are shown in a sliding window analysis. Positive changes in allele frequencies denote increased fixation of alleles contributed by the Alb-NL strain in the selected albino offspring. In all cases, the maximal deviation toward Alb-NL alleles was on scaffold 1 (denoted by an asterisk at ∼5.2 Mb). The order of scaffolds is unknown; for display, scaffolds are concatenated from largest to smallest with alternating shading. (B) The minimal candidate region for albinism in strain Alb-NL as established by fine mapping (Table S1). Gene models are indicated in gray or, for the scaffold 1 carotenoid biosynthesis gene cassette, in orange. The genomic structure and annotation are from the London reference strain. (C) Other WT, nonreference strains, including Wasatch, have structural variants 3′ to the phytoene desaturase (tetur01g11270) that remove all or most of three genes (tetur01g11280, tetur01g11290, and tetur01g11300) present in the London reference (see also Fig. S4). The location of a spontaneous 6.2-kb deletion in strain Wasatch associated with the absence of any pigmentation (strains W-Alb-7, -8, -10, -11, and -14) is shown at the bottom (dashed red line, with the beginning and end of the deletion relative to the overlying Wasatch de novo assembly indicated by red arrows). In C, dark or colored block arrows indicate coding regions, with introns and untranslated regions indicated in lighter gray.
Table S1.
Tetranychus urticae genes in the minimal 66.2-kb candidate region for albinism
| Gene name | Beginning* | End* | Functional description† |
| tetur01g11140 | 5179383 | 5180191 | Protein kinase-like domain |
| tetur01g11150 | 5181220 | 5182922 | Similar to 65-kDa Yes-associated protein |
| tetur01g11160 | 5182997 | 5185420 | Hypothetical protein |
| tetur01g11170 | 5185594 | 5189131 | CRAL-TRIO lipid binding domain |
| tetur01g11180 | 5191610 | 5193055 | Leucine-rich repeat domain, L domain-like |
| tetur01g11200 | 5193187 | 5195562 | Ribonuclease Z |
| tetur01g11210 | 5196358 | 5199468 | Translation elongation/initiation factor/ribosomal |
| tetur01g11220 | 5201763 | 5205151 | Amino acid transporter |
| tetur01g11240 | 5207037 | 5209029 | Amino acid transporter |
| tetur01g11260 | 5209423 | 5213678 | Similar to lycopene cyclase/phytoene synthase |
| tetur01g11270 | 5214229 | 5215943 | Phytoene dehydrogenase |
| tetur01g11280 | 5217904 | 5218469 | Endonuclease/exonuclease/phosphatase |
| tetur01g11290 | 5218701 | 5220395 | Hypothetical protein |
| tetur01g11300 | 5221139 | 5224567 | Serine/threonine/dual specificity protein kinase |
| tetur01g11310 | 5226547 | 5228673 | Ribonuclease D |
| tetur01g11320 | 5234400 | 5236409 | Polynucleotidyl transferase; ribonuclease H fold |
| tetur01g11330 | 5236785 | 5238482 | NUC153 |
| tetur01g11340 | 5239599 | 5242040 | Zinc finger, RanBP2-type |
| tetur01g11350 | 5242259 | 5245557 | N-acetyltransferase 10 |
Beginning and end coordinates refer to positions on scaffold 1 of the T. urticae genome assembly that delineate complete gene sequences. Genes in the carotenoid biosynthetic cassette fall roughly in the middle of the region.
Functional annotations are from the Online Resource for Community Annotation of Eukaryotes (bioinformatics.psb.ugent.be/orcae/).
Inactivating Mutations in tetur01g11270 in Albino Strains.
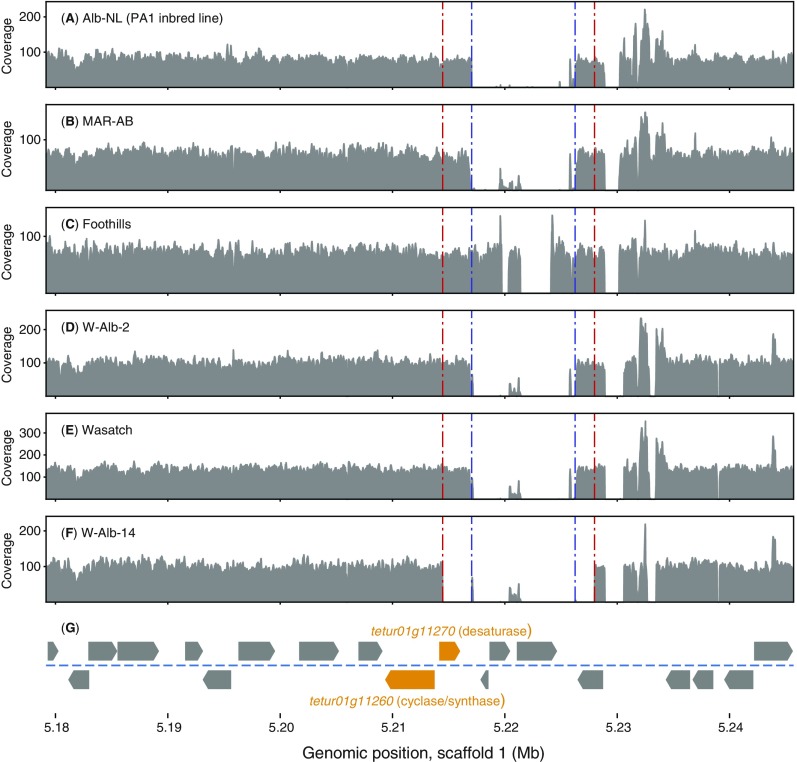
To assess the genic and allelic basis of albinism in the scaffold 1 nonrecombining interval, we examined sequence differences between PA1/PA2 and MAR-AB as assessed from aligned Illumina reads. Further, we extended this analysis with comparable Illumina read data generated for Alb-JP and two other WT strains, Wasatch and Foothills (Figs. S3 and S4). As assessed from read alignments to the London reference sequence, an extended region immediately downstream of (but not including) the carotenoid biosynthesis cluster was devoid of aligned reads in Alb-NL (as assessed from the PA1/PA2 read data), a signature of a large deletion (Fig. S4). Although the region of structural variation included three genes, a nearly identical pattern of read alignments was observed in other WT strains (including MAR-AB, Wasatch, and Foothills), and the deletion of sequences present in the London assembly was subsequently confirmed by a 161-kb contig from a whole-genome de novo assembly of strain Wasatch that spans the entire minimal candidate region (Fig. 3B).
Fig. S4.
Coverage depth of sequence reads aligned to the 66.2-kb minimal candidate region for albinism. (A–F) Coverage variation across the minimal causal region for albinism in strains of T. urticae with the position and direction of transcription of genes in the region shown at bottom (G). Two carotenoid biosynthetic genes in T. urticae of fungal origin are indicated in orange. Coverage (gray) reflects the depth of overlying Illumina genomic DNA reads as aligned to the London reference genome. A region of no read coverage reflects structural variation immediately downstream from the carotenoid biosynthetic genes, including the absence of sequences present in London (this region is defined by dashed blue lines although the genomic differences are not identical across all lines: i.e., compare Wasatch with Foothills). The dashed red lines delineate an additionally deleted region in W-Alb-14, an albino line harboring a spontaneous mutation that arose in the Wasatch strain.
Because the deletion is incidental to the phenotype, we therefore examined other sequence differences among strains. Consistent with other sequencing studies in T. urticae (19, 38, 39), the strains were highly polymorphic (Fig. S3), and many single nucleotide differences were located within the coding regions of the carotenoid biosynthesis genes (no indels were present in WT strains). However, nearly all SNPs were synonymous (42 of 49, or 85.7%, in tetur01g11260, and 46 of 51, or 90.2%, in tetur01g11270), consistent with strong purifying selection at both laterally transferred genes. No unique fixed amino acid changes were observed in Alb-NL in tetur01g11260. However, in the tetur01g11270 gene of Alb-NL, a unique nonsynonymous change affecting codon 220 resulted in a radical threonine (Thr)-to-lysine (Lys) change. The Thr residue was conserved in the sequences of phytoene desaturases in aphids and gall midges, as well as in closely related sequences from fungi, although distantly related fungal and bacterial sequences have several conservative amino acid changes (Fig. S5).
Fig. S5.
Alignment of phytoene desaturases. T. urticae phytoene desaturase tetur01g11270 was aligned with those of other tetranychid species, aphids, and their close relatives adelgids, fungi, and bacteria (11, 18, 20, 74, 78–80). An 80% threshold was used for shading identity (black background) and similarity (gray background). Accession numbers of protein sequences are shown in parentheses. A black triangle indicates the position of the single intron in tetur01g11270. Red asterisks above the alignment indicate the substitutions in tetur01g11270 (Thr220Lys in T. urticae strain Alb-NL and Pro487Leu in T. urticae lines W-Alb-1 and W-Alb-2) that were identified in this study. A red rhombus indicates the position of the insertion into P. citri phytoene desaturase (this study). A blue circle above the alignment indicates the Glu32Lys substitution found in a phytoene desaturase of an A. pisum mutant (11) whereas brown and green circles refer to those substitutions in Phycomyces blakesleeanus (Glu426Lys, Ser444Phe and Leu446Phe, Gly482Ser) and Fusarium fujikuroi (Pro170Leu, Trp449Stop, Gly504Asp) phytoene desaturases that result in lowered desaturase activities (52, 53). The presumed carotenoid binding domain (52) is indicated with an arrow below the alignment.
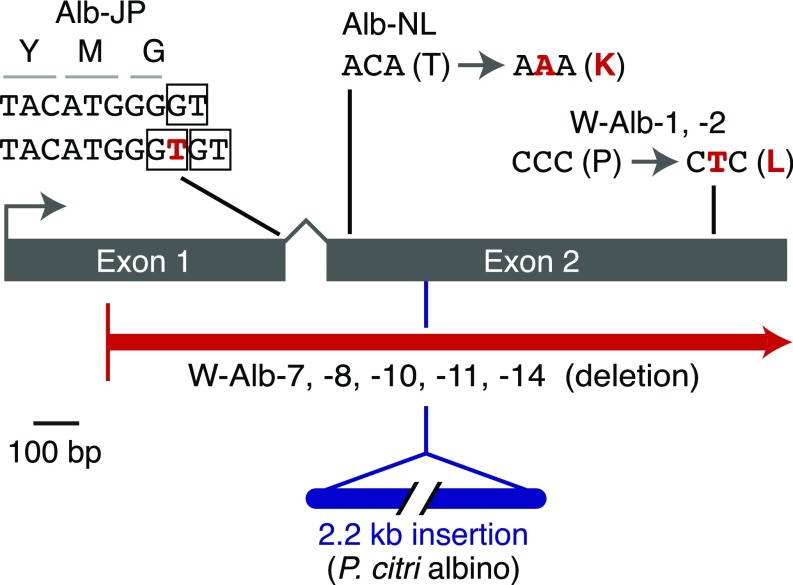
As opposed to the putative nonsynonymous causal change in Alb-NL, dramatic alterations in tetur01g11270 were present in the T. urticae Alb-JP and the P. citri albino strains. In Alb-JP, a thymine was inserted nearby the splice donor of exon 1. This change removed the WT location of the splice donor relative to the end of exon 1, and created two potential “GT” splice donor dinucleotides that are adjacent to each other, but at nonfunctional positions (Fig. 4); use of either of the resulting splice donors was expected to lead to frameshifts in the tetur01g11270 message, a prediction supported by Sanger sequencing of RT-PCR (cDNA) products amplified from Alb-JP with primers in exons 1 and 2 (i.e., no message was observed in which the highly conserved exon 2 was in frame) (Table S2).
Fig. 4.
Mutations in the scaffold 1 phytoene desaturase in T. urticae and P. citri albino strains. Coding exons for tetur01g11270 are represented as rectangles. WT sequences are indicated in black, with mutations shown in red (T. urticae) or blue (P. citri); where single nucleotide substitutions are observed, the impact on the coding potential is shown. For the Alb-JP mutation, boxes indicate the exon 1 splice donor in WT (at the top) with two out-of-frame splice donors created by the “T” insertion shown underneath.
Table S2.
Primers used for amplification of sequences in the tetur01g11270 genomic region
| Species | Product name* | Primer names and sequences, 5′ to 3′† | Tm, °C |
| T. urticae | 5′_300_kb‡ | F: ATTGCTTGGTGTCTGTGGTG | 58.7 |
| R: CGAGCAAACTGAGCAAAAGG | 58 | ||
| T. urticae | 5′_50_kb‡ | F: CATACAACATTGGGCCCAGC | 59.5 |
| R: TGATCGCATCCAAAAGCGTTC | 59.9 | ||
| T. urticae | 5′_40_kb‡ | F: AATCCCATTTGTGCCCTCAA | 57.7 |
| R: GGAAACGATCAAGAAAACAGCG | 58.8 | ||
| T. urticae | 5′_30_kb‡ | F: TGTTAAGTTGCTCGATGAATGTC | 57.5 |
| R: CCTGATTGCTTGGATGAGGT | 57.6 | ||
| T. urticae | 5′_20_kb‡ | F: ATATTACATGCCCTGGTCGG | 57.1 |
| R: TGTTAACAAGTGAAATGCGAGAG | 57.7 | ||
| T. urticae | 5′_10_kb‡ | F: GAATTGAAACAACATGGAAAGGC | 57.4 |
| R: AATCATGCATAACACAGTCAAGG | 57.4 | ||
| T. urticae | 3′_300_kb‡ | F: CCTGACAATTTTGCGGCTAAC | 58.4 |
| R: GGGAAATAACGCGAAACATCG | 58.3 | ||
| T. urticae | 3′_50_kb‡ | F: TCGACCCTTTCAACTTCAACTC | 58.5 |
| R: GCTTTGACCTTAGTTGCCCG | 59.5 | ||
| T. urticae | 3′_40_kb‡ | F: TTCTAGGCAAAGTTGATGTTGC | 57.5 |
| R: GAACACGCACAGCAAGATCA | 59.1 | ||
| T. urticae | 3′_30_kb‡ | F: ACGAAATTGGCGATGAAATAGGA | 58.8 |
| R: TCAAGATAGGTAGCGGCTCA | 58 | ||
| T. urticae | 3′_20_kb‡ | F: GTAAGCCCAGGTTCGGTTTC | 58.8 |
| R: ATGCAGGAGAGTTGGCGTAT | 59.1 | ||
| T. urticae | 3′_10_kb‡ | F: CGAGTGATTTATGAAATTGCGCA | 58.7 |
| R: TTCAGTTGACTTGCCAGATGA | 57.5 | ||
| T. urticae | tetur01g11270_part1§ | F: TGATTGTGGGTTCAGTTATCATTACC | 59.4 |
| R: TTTAGTGCCGTACGATGTTGG | 58.7 | ||
| T. urticae | tetur01g11270_part2§ | F: TGGTATAACACTGGAGAGTGGAGAG | 61.1 |
| R: AGGACGAGTGGAATGAAGATACC | 59.6 | ||
| T. urticae | sgRNA_screen | F: ACGAGCAACAAAGTACTTCAAAA | 57.5 |
| R: CAACCTCTCCACTCTCCAGT | 58.4 | ||
| P. citri | Pcitri_01g11270_part1§ | F: TTAACGTTTGAACATCGTAGTAGTT | 57.1 |
| R: CCACCTTGTTGAATCCACCC | 58.7 | ||
| P. citri | Pcitri_01g11270_part2§ | F: CTGACCACATGATCAAAGCGT | 58.9 |
| R: CCACACAAGACAATCGGAACT | 58.5 | ||
| P. citri | Pcitri_shortseq§ | F: AGATTTAAATCAAGGCAAAG | 59.3 |
| R: ACTAGTTTTTGAGAATATAT | 60.2 |
For primer pairs used for genetic mapping, product names refer to the distance either 5′ or 3′ from the ends of the tetur01g11270 gene (i.e., the “5′_300_kb” product is located 300 kb upstream of the 5′-most position in tetur01g11270 on scaffold 1). Locations are rounded off to the nearest 10 kb.
“F” and “R” indicate primer sequences in the forward and reverse orientation, respectively.
Amplicons used for genotyping F2 individuals for genetic mapping.
Amplicons used for variant discovery at the tetur01g11270 locus.
Given the findings for Alb-NL and Alb-JP, we also amplified the tetur01g11270 ortholog in the P. citri albino strain, as well as in its matching parental WT strain. As opposed to amplification products observed in WT, a product corresponding to exon 2 of the P. citri tetur01g11270 ortholog was about 2.2 kb longer in the albino mutant. Sanger sequencing into the region of the inserted sequence revealed homology to the terminal inverted repeat (TIR) of a mutator-like transposon. The insertion site of this sequence was within the tetur01g11270 coding sequence near the beginning of the conserved second exon (Fig. 4). Sequencing of cloned RT-PCR products of the tetur01g11270 ortholog from albino P. citri mites gave several sequences, all of which had part of the inserted sequence (and none of which led to an intact ORF). Using the inserted sequence as a PCR-based marker in a screen of F2 mites from a cross between the WT and albino P. citri strains, the insertion cosegregated invariantly with the albino phenotype, χ2(1, n = 98) = 61.957, P < 10−14, for the null hypothesis of no linkage.
Identification of Additional Mutations in tetur01g11270.
To further establish a requirement for tetur01g11270 in spider mite pigmentation, we attempted to inactivate WT tetur01g11270 by genome editing with the CRISPR-Cas9 system. Multiple single guide RNAs (sgRNAs) directed against a region in exon 2 were preloaded onto Cas9 protein and injected into the abdomens of 5,537 virgin females (for more details, see SI Materials and Methods) of the strain Wasatch (which is highly inbred) (Fig. S3). Although injection of eggs was initially attempted, viable larvae were not recovered. Approximately 20,000 male progeny from injected mothers were then screened for absence of body color and red eyes. Fifteen males were identified that either lacked all pigmentation, or were partially albino (perhaps reflecting mosaicism). From these males, we recovered seven stable albino lines (the lines were recovered in backcrosses to Wasatch, thus maintaining the otherwise isogenic background) (Fig. S3). For five of these lines (W-Alb-7, -8, -10, -11, and -14), the phenotype was complete lack of pigmentation whereas, for two lines (W-Alb-1 and -2), some red color was apparent in the eyes of older adults (although developing stages lacked pigment). In all cases, albinism was recessive, and, in crosses of all lines to Alb-JP, failure to complement was observed (Table 1), consistent with new mutations in tetur01g11270.
Unexpectedly, attempts to amplify by PCR across the region in tetur01g11270 with sequence identity to the sgRNAs gave no products in strains W-Alb-7, -8, -10, -11, and -14, or a product that was unaltered as assessed by Sanger sequencing for strains W-Alb-1 and -2. We therefore sequenced these strains with the Illumina method. De novo assembly of each strain gave single contigs that spanned the carotenoid biosynthesis gene cluster and flanking regions. Compared with Wasatch, the W-Alb-7, -8, -10, -11, and -14 strains harbored an identical 6.2-kb deletion (with an associated small insertion of 13 bp) that removed the majority of tetur01g11270, as well as much of the adjacent downstream gene (tetur01g11310, a gene of unknown function that is putatively annotated as a ribonuclease) (Figs. 3C and 4). Moreover, tetur01g11270 differed in strains W-Alb-1 and -2 from the parental Wasatch sequence by the presence of a single nucleotide change in exon 2. This mutation caused a radical amino acid substitution (proline to leucine) at position 487. The proline at 487 was invariant among the phytoene desaturases we analyzed and was located in a sequence suggested to be involved in carotenoid binding in a fungal phytoene desaturase (Fig. S5). Nevertheless, the red in the eyes of older adults in the W-Alb-1 and -2 strains suggests that the Pro487Leu change is hypomorphic (at least at postdeveloping stages).
The mutations we recovered are not typical of CRISPR-Cas9 editing events, which are usually present as small indels at sgRNA target sites (41). For example, the location of the substitution in W-Alb-1/2 was more than 700 nucleotides away from the sgRNA target sites. Moreover, identical lesions were recovered multiple times, in some cases from females injected on different days, and whose progeny were kept on different plates, which suggests that the screen recovered two spontaneous mutations that had been segregating at very low frequency in the large inbred source population used for the injections (the inbred Wasatch line had been maintained previously in the laboratory for several years).
Tetur01g11270 Is Required for Diapause Induction in an Overwintering Strain.
The genetic control of diapause in spider mites is complex, with many genetic and environmental factors affecting induction (42–47). Previously, albino mites were shown to be capable of entering diapause, as assessed by cessation of feeding and reproductive arrest, but the frequency of induction varied dramatically among mutants (35). Consistent with the previous reports, we were able to induce diapause in Alb-NL mites (Fig. S1) (these mites are completely white with no gut contents; compare with Fig. 2B), but at a very low frequency. However, many genetically diverse but normally pigmented T. urticae populations either do not enter diapause or are polymorphic for diapause entry, especially at low latitudes (48). Further, even after 40 y in the laboratory, we found that Alb-NL was not fully inbred (Fig. S3), confounding inferences about the requirement of carotenoids for diapause induction.
To test the role of the laterally transferred carotenoid biosynthetic pathway in diapause induction in spider mites, we turned to albino mutants on the Wasatch background. We chose Wasatch because this strain was field-collected from a region of harsh winters where diapause inducibility is expected to be essential for year-to-year persistence and because it was inbred soon after collection (Materials and Methods) (of the albino mutants in T. urticae and P. citri used in our study, those on the Wasatch background uniquely satisfied these conditions). To test induction, we subjected developing Wasatch, W-Alb-14, and W-Alb-2 females on detached kidney bean leaves to diapause-inducing conditions (8 h light; 16 h of dark at 17 °C) and transferred the resulting adult 1- to 2-d-old females to individual bean leaves in the same inducing environment (W-Alb-14 and W-Alb-2 were chosen because they represent each of the two mutations in tetur01g11270 identified on the Wasatch background). After 11 d, we scored mite color and eggs laid.
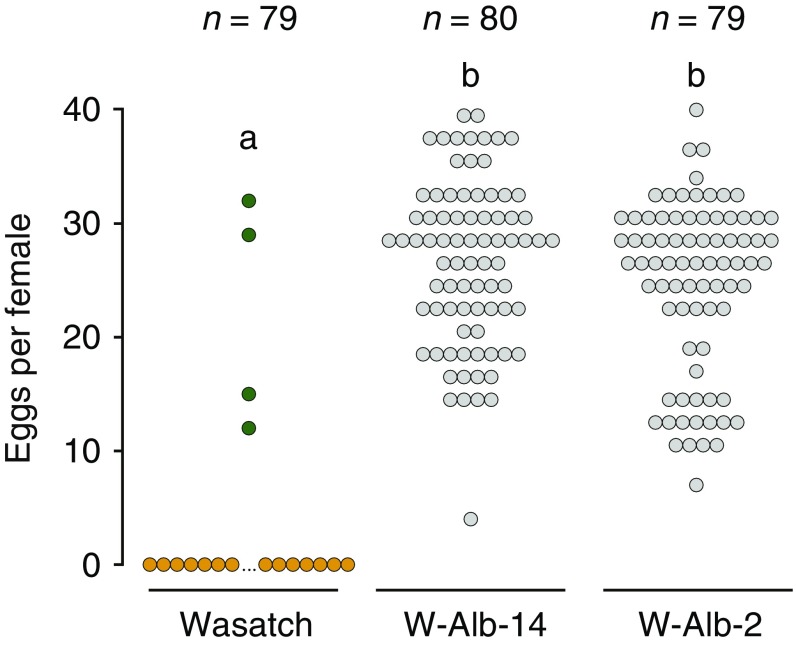
As expected for strain Wasatch, most females turned orange (94.9%), the stereotypical visual indicator of diapause induction, and orange Wasatch mites invariably produced no eggs (Fig. 5). In contrast, W-Alb-14 and W-Alb-2 fed actively, and 100% laid eggs (color change is irrelevant as an indicator of diapause in albino lines because they cannot produce red-orange keto-carotenoids) (Fig. 1 and Discussion). As assessed with the Kruskal–Wallis test, differences in the number of eggs laid per female among genotypes was highly significant (P < 10−15). In follow-up comparisons, eggs laid per female for both the W-Alb-14 and W-Alb-2 strains was found to be significantly higher compared with Wasatch (P < 10−15 in each case; Wilcoxon rank-sum test). In contrast, the analogous test between the W-Alb-14 and W-Alb-2 strains revealed no statistically significant difference (P = 0.51).
Fig. 5.
Impact of inactivating mutations in tetur01g11270 on diapause incidence. The incidence of diapause under inducing conditions, as revealed by coloration (orange or green circles for WT, diapausing and nondiapausing colors, respectively, and gray for albino mites) and egg laying (y axis), in WT Wasatch and the albino W-Alb-14 and W-Alb-2 mutant strains. For egg laying, the total number per female after 11 d is shown (that is, each circle represents a single female’s total oviposition). Sample sizes are indicated at the top, and differing letter designations denote significant differences (P < 10−15 in pairwise comparisons, Wilcoxon rank-sum tests).
SI Materials and Methods
CRISPR-Cas9 Targeting of tetur01g11270.
The CRISPR-Cas9 system was attempted to introduce nonhomologous end-joining mutations in the T. urticae tetur01g11270 gene. sgRNAs were designed with the sgRNAcas9 software package (www.biootools.com) (77). This package designs sgRNAs and evaluates the potential off-target sites. The three highest ranked sgRNAs for tetur01g11270 were selected (sgRNA-S33, sgRNA-A32, and sgRNA-A30) (Table S3), and the first two were amplified according to the Guide-it sgRNA In Vitro Transcription and Screening Systems kit (Clontech Laboratories, Inc.) and subsequently purified with the RNeasy Mini Kit (Qiagen) following the standard protocol for RNA cleanup. Quality and quantity of the sgRNA were checked on a DS-11 FX spectrophotometer (DeNovix) and by running an aliquot on a 2% agarose gel. Measured concentrations of sgRNA-S33 and sgRNA-A32 were 711 ng/µL and 1,200 ng/µL, respectively. Lyophilized Cas9 protein (50 µg, “CP01, Cas9 protein with nuclear localization signal”) and the third selected sgRNA (sgRNA-A30, 50 µg) were ordered from PNA Bio, Inc. We used two injection mixtures: The first was a combination of sgRNA-A30 with Cas9 protein, and the second mixture contained two sgRNAs (S33 and A32) with Cas9 protein. The injection mixture of the sgRNAs and Cas9 was prepared as follows: the Cas9 protein was dissolved in 5µL of dH2O for the mixture of sgRNAs and 10 µL of dH2O for the sgRNA-A30 and diluted with one volume of protein storage buffer (1× 20 mM Hepes-KOH, 150 mM KCl, and 1 mM DTT, pH 7.5, final concentration of Cas9 protein 5µg/µL and 2.5 µg/µL, respectively); next, 2 µL of both sgRNAs (sgRNA-S33 and sgRNA-A32) and 1 µL of sgRNA-A30 (dissolved to a concentration of 3 µg/µL in protein storage buffer) were added to a 2-µL and 4-µL aliquot of the dissolved Cas9 protein, respectively, and incubated at 37 °C for 10 min on the day of injection. To attain a final concentration (Cas9, 833 ng/µL; sgRNA-S33, 120 ng/µL; and sgRNA-A32, 200 ng/µL) for the mixtures, the volume was two times diluted with 10× Tris-EDTA (TE) buffer. The other injection mix (with the single sgRNA-A30) obtained a final concentration (Cas9, 770 ng/µL; and sgRNA-A30, 230 ng/µL) and was diluted 2.6 times with the same buffer. Subsequently, both injection mixtures were centrifuged for 10 min at 4 °C and kept on ice until injection. Female mites of the strain Wasatch were allowed to lay eggs on the upper part of a detached bean leaf on wet cotton in a Petri dish. After 8 d, all males were removed from the plates (so that only unfertilized female offspring were left), and resulting 3- to 5-d-old females were used for injections. In total 5,537 females were injected in the proximity of the ovaria with an IM 300 Microinjector (Narishige); the survival percentage was about 70%. These females were allowed to lay eggs for 24 h, with an average of 4.9 eggs per female. The male haploid progeny of these injected females were visually screened for the albino phenotype beginning 3 d after egg deposition, and 15 albino males were identified from the total injected population. These males were transferred to separate detached bean leaves and were allowed to cross with 9 to 12 unfertilized Wasatch females to start new albino lines. In total, seven stable albino lines were recovered as homozygous stocks: W-Alb-1, W-Alb-2, W-Alb-7, W-Alb-8, W-Alb-10, W-Alb-11, and W-Alb-14. After the initial crosses, DNA was extracted from the original albino males with 20 µL of a direct PCR buffer (10 mM Tris⋅HCl, 100 mM NaCl, 1 mM EDTA, pH 8) with 2 µL of proteinase K (10 mg/mL), followed by an incubation of 30 min at 37 °C and 5 min at 95 °C. PCR reactions with primers flanking the regions targeted by the sgRNAs (named sgRNA_screen) (Table S2) were then performed, followed by Sanger sequencing as previously described.
Table S3.
Single guide RNAs used for the CRISPR-Cas9 experiment with injections of adult females
| Name sgRNA | Sequences, 5′ to 3′ |
| sgRNA-A30 | TTAGTGCCGTACGATGTTGG |
| sgRNA-S33 | GGTATTTGGTATCCCAAAGG |
| sgRNA-A32 | AAGGTGGATTTCACAGAGTT |
De Novo Assembly of T. urticae Genomes.
Using the Illumina sequence data generated for strain Wasatch, as well as the albino lines recovered in the CRISPR-Cas9 screen, we performed de novo genome assembly using CLC Genomics Workbench 8.5 (https://www.qiagenbioinformatics.com/). Reads were imported into CLC and trimmed using the program’s default settings (quality score limit of 0.05 and a maximum of two ambiguous nucleotides). De novo assembly of each strain was carried out with the following parameters: automatic word and bubble size, a minimum contig length of 200, auto-detect paired distances, perform scaffolding, and map reads back to contigs (mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.5; similarity fraction, 0.8; and “Update contigs” checked).
Alignment of Phytoene Desaturase Sequences.
Phytoene desaturase protein sequences were aligned using MUSCLE (75). T. urticae phytoene desaturase tetur01g11270 was aligned with those of other tetranychid species [P. citri, Panonychus ulmi (74), and Tetranychus evansi (78)], other animals (11, 18, 79, 80), representative species of a group of fungi that clustered with tetur01g11270 in a previously published phylogenetic analysis [Phycomyces blakeeslanus, Blakeslea trispora, Mucor circinelloides, Xanthophyllomyces dendrorhous, Ustilago maydis, Fusarium fujikuroi, and Neurospora crassa (20)], and two bacterial species (Rhodobacter sphaeroides and Pantoeae ananatis), of which phytoene desaturases have been extensively characterized (81, 82).
Interstrain Genetic Comparisons Using Genome-Wide SNP Data.
To select high-confidence variants for interstrain genetic comparisons, SNPs predicted by GATK version 3.60 were required to show homozygosity in every strain included in the analysis (Fig. S3 and Materials and Methods) and to have a minimum Phred-scaled quality score of 100, a minimum genotype score of 65 across all samples, a mean read rms (root-mean-square) mapping score of at least 52, and read depth coverage within 0.5× to 2× of the strain’s genome-wide coverage mean.
Discussion
The acquisition of carotenoid biosynthetic genes in divergent arthropod lineages—aphids and their close relatives the adelgids (11, 17), spider mites (19, 20), and gall midges (18)—suggests that de novo carotenoid biosynthesis in animals can be highly advantageous. As opposed to aphids, which feed on phloem that is expected to be largely devoid of lipophilic carotenoids (11), spider mites feed on the contents of mesophyll cells, including carotenoid-rich chloroplasts (49). High levels of plant carotenoids, including β-carotene, have been reported in feeding albino mites, much of which likely reflects gut contents (26, 33). Based on this observation, and given that no animals were known to synthesize carotenoids at the time, Veerman proposed that albino strains of T. urticae were compromised in the transport or metabolism of dietary β-carotene that accumulates in the lemon mutant that disrupts the next step in the putative pathway to astaxanthin (Fig. 1) (26, 33, 34).
The unexpected discovery of carotenoid biosynthesis genes in the T. urticae genome (19, 20) raised the alternative possibility that spider mites rely instead on endogenously synthesized β-carotene. Consistent with this possibility, we show that four albino mutants in T. urticae and one mutant in P. citri have inactivating mutations in a single laterally transferred phytoene desaturase. This gene, tetur01g11270, is adjacent to a carotenoid cyclase/synthase on scaffold 1 and comprises one of two carotenoid biosynthetic gene clusters in the T. urticae genome (19). The second cluster is located on scaffold 11 and harbors two putatively intact phytoene desaturases and one cyclase/synthase (along with one phytoene desaturase pseudogene). The expression of the T. urticae carotenoid biosynthetic genes in nondiapausing and diapausing females, or in red forms of T. urticae, is robust for genes in the scaffold 1 cluster (20, 50). However, low or no expression was observed for genes in the scaffold 11 cluster (50). Further, in a phylogenetic analysis, one protein from the scaffold 11 cluster fell outside a clade with other arthropod and fungal carotenoid biosynthetic protein sequences, and another had a long branch length (18). These prior observations, coupled with our current finding that disruption of tetur01g11270 is sufficient for albinism, raise the possibility that only the scaffold 1 phytoene desaturase is active in carotenoid synthesis (18, 50). Whether all genes in the scaffold 11 cluster are inactive, have more specific roles in carotenoid biosynthesis, or have evolved to perform other functions remains uncertain.
In fungi, inactivating mutations in phytoene desaturases (51–54), as well as the fused cyclase/synthases, abolish carotenoid synthesis (54–57). All five albino mutations characterized in our study resided in tetur01g11270 or its P. citri ortholog. Previously, two T. urticae albino mutants studied by Ballantyne (32) complemented each other but were extraordinarily tightly linked (∼2 to 3 recombination events in 10,000 F2 males). It is tempting to speculate that these previously studied mutations disrupted each of the genes in the scaffold 1 carotenoid biosynthetic cluster, and, as additional albino mutants are identified, they should be assessed for mutations in the scaffold 1 cyclase/synthase (tetur01g11260). Alternatively, our identification of mutations solely in tetur01g11270 in the T. urticae and P. citri strains we examined might reflect functional redundancy between multiple copies of the cyclase/synthase fusion genes (e.g., tetur01g11260 and tetur11g04840 in the T. urticae scaffold 1 and 11 clusters, respectively).
The most striking visible cue of diapausing mites is their intense red-orange color that occurs after suspension of reproduction (egg laying) and eventual cessation of feeding (58). Recently, Kawaguchi et al. (29) showed that imaginal feeding (feeding after the last molt) was required for WT incidence of diapause, as well as for accumulation of red-orange xanthophyll carotenoids like astaxanthin (28). Based on these findings, the authors suggested that dietary β-carotene, which is rich in plant cells, may serve as the precursor for the bulk keto-carotenoid synthesis that occurs at the onset of diapause. They did not exclude the possibility, however, that imaginal feeding might be required to obtain energy or a nutritional state required for diapause and endogenous carotenoid synthesis. Our findings that mutations in tetur01g11270 in T. urticae underlie albinism, coupled with the observation that diapausing albino mites are uniformly white (Fig. S1B and refs. 26 and 59), are consistent with the latter explanation (i.e., endogenously synthesized β-carotene is needed for the red-orange color development of diapausing mites, not dietary β-carotene as previously proposed) (Fig. 1A). Moreover, WT mites at the larval stage fed on an artificial diet with no added carotenes diapaused with bright red-orange color, a finding interpreted at the time as a maternal effect or contamination in the artificial diet (59). In light of our findings, endogenously synthesized carotenoids likely explain this result. This conclusion is supported as well by our findings with P. citri, where inactivation of the tetur01g11270 ortholog associates with the complete absence of the vibrant red pigmentation of WT mites.
Apart from the putative involvement of keto-carotenoids in protecting against stresses experienced during overwintering (28, 60), the role of carotenoids in diapause induction itself has attracted great interest. WT T. urticae strains or populations range from 0 to 100% in their diapause induction, presumably reflecting variation at diverse steps in the pathway for diapause entry (48). Our recovery of two mutations in tetur01g11270 in strain Wasatch allowed us to test the role of endogenous carotenoid synthesis in diapause induction in an isogenic, diapausing background. Under inducing conditions, diapause induction was completely abolished in the resulting albino strains. In W-Alb-14, a large deletion removed most of tetur01g11270 and an adjacent gene of unknown function. However, in W-Alb-2, the lesion for albinism was a nonsynonymous point mutation in tetur01g11270, specifically linking tetur01g11270 activity to diapause induction.
Entry into diapause does not occur in constant darkness (61), and the requirement of tetur01g11270 activity for diapause in Wasatch presumably reflects vitamin A deficiency (resulting in an inability to perceive inductive photoperiods, although our findings do not formally exclude other or additional roles for carotenoids). β-carotene and its derivative 3-hydroxyechinenone (Fig. 1) are processed to vitamin A (retinoids) in animals, and vitamin A is the apparent chromophore for light perception in T. urticae (28). In an earlier study, Bosse and Veerman (59) examined diapause incidence in albino mites grown under inductive photoperiods on an artificial diet devoid of β-carotene and vitamin A, and artificial diets supplemented with either compound. While on an artificial diet lacking β-carotene and vitamin A, no albino mites entered diapause. In contrast, 99% of albino mites raised on the artificial diet supplemented with vitamin A entered diapause. However, when fed a high concentration of β-carotene in the artificial diet, only 47% of albino mites entered diapause, mirroring the observation that albino mites fed on leaves (a source of β-carotene) have intermediate frequencies of diapause induction compared with WT strains on which the mutants arose (35). These earlier findings suggested that T. urticae can use dietary β-carotene for diapause induction, albeit inefficiently. They also contrast with our observation that albino mutants in strain Wasatch completely lose the ability to enter diapause. It is possible that the ability to take up dietary β-carotene or to respond to secondary signals that also influence the diapause induction threshold (e.g., temperature and leaf quality) (62, 63) varies between strains. Regardless, even if dietary uptake occurs in some strains in the field, it seems unlikely that dietary sources of carotenoids are physiologically relevant to T. urticae, given their ability to synthesize their own carotenoids in abundance.
To our knowledge, de novo carotenoid biosynthetic genes are absent from the genomes of all mites outside Tetranychidae (spider mites) (37). In insects and other animals, it is now well-documented that carotenoid delivery to target tissues is not passive, but instead receptor-mediated (8, 13), and active uptake from the gut is presumably the ancestral state in chelicerates. For instance, when the predatory mite Amblyseius potentillae (family Phytoseiidae) was fed on pollen low in β-carotene, or on developing eggs laid by albino T. urticae females, no diapause occurred; in contrast, when fed on pollen supplemented with β-carotene, or when fed on T. urticae eggs from WT females, full diapause was restored (64). Therefore, in mites distantly related to T. urticae and P. citri, dietary β-carotene seems to be essential. As more becomes known about carotenoid metabolism in related chelicerates, it should be possible to understand the fate of ancestral genes and pathways involved in dietary carotenoid uptake. In particular, an outstanding question is whether such pathways were lost in spider mites or perhaps repurposed for other roles.
Conclusion and Future Directions
A finding of our study is that lateral acquisition of carotenoid biosynthetic genes from fungi has effectively eliminated the requirement for dietary carotenoids in spider mites, including for diapause that is essential for survival in temperate regions (and therefore underlies the global distribution of many agronomically important spider mite pests). In addition to albino mutants, other T. urticae pigmentation mutants have been reported that affect the synthesis, intraorganismal transport, and esterification of keto-carotenoids (26). To our knowledge, these mutant lines no longer exist. However, we recovered two spontaneous mutations in ∼2 × 104 haploid males, a frequency consistent with the ∼1 in 104 incidence of recessive mutations observed at some pigmentation loci in a closely related sister species, T. pacificus (65). Reisolation of mutants that act downstream of the carotenoid biosynthetic genes, and that could be characterized with the methods used in the current study, should afford diverse insights into molecular aspects of carotenoid metabolism and transport that have been difficult to study in most animals.
Materials and Methods
Mite Strains, Inbreeding, and Husbandry.
Albinism in a strain collected from The Netherlands (strain NL) arose spontaneously several times (26, 32). We obtained one of the resulting albino strains, hereafter Albino-NL (Alb-NL), that had been continually maintained in The Netherlands (University of Amsterdam). Although Alb-NL is one of the albino 1 (“a”) or the albino 2 (“p”) mutant strains characterized by Veerman in 1974 (26), the exact correspondence is no longer known. Albino strain Alb-JP was isolated from a WT T. urticae strain found in Tsukuba, Japan in 2011, and the albino strain of P. citri originated from a WT population from Minamishimabara, Nagasaki Prefecture, Japan in 2006. As WT references for genetic and genomic studies, we used several nonalbino strains. T. urticae strains Wasatch and Foothills were collected in the Salt Lake Valley of Utah state in 2012, and the MAR-AB strain was collected in Greece in 2009. The reference strain, London, which originates from London, Ontario, Canada, was also used (19).
As reported in several studies that analyzed high-throughput sequencing data, T. urticae strains are often highly heterozygous (19, 38), a confounding factor for classical genetic studies. Therefore, we inbred a subset of the T. urticae strains by sequential rounds of mother–son matings; strain Alb-NL was inbred for nine rounds, and strains Foothills and Wasatch were each inbred for six rounds (strain Alb-JP was inbred for one generation). For strain London, an inbred line was used that had been inbred for seven generations (66). For Alb-NL, two independent inbred lines (PA1 and PA2) were generated in parallel from the single starting Alb-NL population. Unless otherwise noted, the resulting inbred lines for the respective strains were used in the current study. For inbreeding, teliochrysalis (virgin) females were placed on detached bean leaves and allowed to lay eggs (unmated females lay haploid eggs that develop as males). When the males reached adulthood, they were allowed to fertilize their female parent, at which time remaining eggs harbored diploid females, and the process was repeated.
Both T. urticae and P. citri strains were maintained on whole plants or detached leaves (on wet cotton in Petri dishes) of the kidney bean (Phaseolus vulgaris L.) under 16:8 light:dark photoperiods after methods published previously (67). For genetic crosses and BSA analyses, mites were maintained in chambers at 26 °C and 60% relative humidity.
Microscopy.
Scanning electron microscope (SEM) images of spider mite eyes were taken with a Hitachi TM-1000 Tabletop SEM whereas the photographs of female adult mites on bean leaves were acquired by a Leica Photar 25/2 lens on bellows attached to a Nikon D7000 camera with a Nikon SB-800 flash. Enhanced depth of field (image stack) was created using the Helicon Focus and Adobe Photoshop software.
Inheritance and Complementation.
To assess the mode of inheritance of the albino phenotype in spider mites, 20 teliochrysalis virgin females of an albino strain and 30 adult males of a WT strain (Table 1 for the different crosses) were placed on the upper part of a detached bean leaf. Twenty of the resulting teliochrysalis unfertilized F1 females were then placed on each of four fresh detached bean leaves, and the F1 phenotypes (WT or albino) were noted. Upon hatching, the females were allowed to lay eggs for one day before being transferred to another bean leaf (the transfers were repeated eight times). Ten days after hatching began, the resulting F2 males were phenotyped. The mode of inheritance observed in F2 individuals was assessed with χ2 goodness-of-fit tests (specifically, the hypothesis of monogenic recessive inheritance, for which the expected F2 ratio is 1:1 WT to albino, was tested). Additionally, complementation tests were performed for a subset of albino T. urticae strains. Briefly, 16 teliochrysalis females from a given albino strain were crossed with 30 males from a second albino strain on detached bean leaves, and at least 85 resulting F1 females were assessed for albinism.
Collection and Sequencing of Segregating Populations.
To genetically map the Alb-NL phenotype using bulked segregant analysis (BSA) methods (38, 39), we crossed the derived inbred lines PA1 and PA2 with males from the MAR-AB strain. For each of the crosses, single haploid males selected from the MAR-AB population were crossed to multiple females of the PA1 and PA2 strains on detached leaves (different single males were used in each of the PA1 and PA2 crosses). From each cross, 120 F1 females were allowed to grow in bulk on whole plants. After approximately the fourth generation (as assessed by the typical generation time under our standard growth conditions), two phenotypes were selected from the segregating population: a complete albino phenotype with no body color or eye color (complete albino, or “CA”) and an albino phenotype with no body color but very faint red eyes (incomplete albino, or “IA”). For each of the PA1 and PA2 cross, ∼100 CA and IA teliochrysalis females were selected and placed on detached bean leaves to create large populations. These four derived populations were then expanded for an additional approximately four to five generations to confirm the stability of the albino phenotypes. The faint eye color observed in the IA selections disappeared during the expansions (that is, complete albinism was observed in the following generations); we therefore concluded that the IA phenotype resulted from maternal pigment contribution (faint eye color in the otherwise albino progeny of heterozygous mothers has been reported previously) (30). Afterward, at generations approximately 9 to 11, mites from the four populations (PA1-CA, PA1-IA, PA2-CA, and PA2-IA) were collected en masse for genomic DNA (gDNA) extraction. Additionally, mites for the parental inbred albino lines PA1 and PA2, the WT parental MAR-AB population, and mixed offspring from the segregating populations derived from the respective PA1/PA2 × MAR-AB crosses were collected (PA1-Mix and PA2-Mix; these collections were performed at the 13th to 15th generation). In each case, ∼3,000 adult females were collected per sample.
DNA Preparation, Genomic Sequencing, and Variant Detection.
Genomic DNA for BSA samples and parental controls, as well as for other strains used in this study, was extracted according to Van Leeuwen et al. (38), purified using an EZNA Cycle Pure Kit (Omega Bio-tek) according to the manufacturer’s protocol, and quantified using an ND-1000 Nanodrop spectrophotometer (Nanodrop Technologies). An exception was for strains Wasatch and Foothills, for which DNA was prepared using the 5-Prime ArchivePure DNA Cell/Tissues Kit (5-Prime) using the manufacturer’s Protocol 13 with the following modifications: 7.5 μL of proteinase K, 6 μL of RNase A, and 65 μL of DNA Hydration Solution were added. Illumina genomic DNA libraries were then constructed, and sequencing was performed to generate paired-end reads of either 101 or 125 bp. Library construction and sequencing was performed at the Centro Nacional de Análisis Genómico (CNAG, Barcelona) or the High-throughput Genomics Core at the Huntsman Cancer Institute of the University of Utah (Salt Lake City). Illumina reads from each sample were aligned to the reference T. urticae genome from strain London (19) using the default settings of the Burrows–Wheeler Aligner (BWA) versions 0.7.10 or 0.7.15 (68), and variant detection was performed with the Genome Analysis Toolkit (GATK) versions 3.30 or 3.60 (69). In accordance with GATK best practices recommendations (70, 71), duplicate reads were marked using Picard versions 1.126 or 2.6.0 (broadinstitute.github.io/picard/), and indel realignment was undertaken (for variant predictions for BSA scans, base quality score recalibration was additionally performed). Genotypes at positions of single nucleotide polymorphisms (SNPs), as well as associated quality information and read support for each allele, were recovered across all samples using GATK’s Unified Genotyper tool. Potential impacts of variants on coding sequences were assessed with SNPeff version 4.1 (72) using the T. urticae genome annotation release of July 29, 2015 as downloaded from the Online Resource for Community Annotation of Eukaryotes (73); this annotation was used for all analyses in the current study. Illumina sequencing datasets generated as part of this project have been deposited to the sequence read archive (SRA) under accession numbers SAMN07138853–SAMN07138871.
Bulked Segregant Analysis Mapping.
The recessive albino locus from the Alb-NL strain was localized in the T. urticae genome using BSA methods adapted from our earlier studies (38, 39). Briefly, at sites segregating for single nucleotide differences (SNPs) in the BSA populations, we assessed the frequency of alleles from the PA1 and PA2 parents compared with those contributed by the respective MAR-AB male parents on a per sample basis (counts of each base at a polymorphic site, and associated quality information, were from the GATK output). In a sliding window analysis across the T. urticae genome, we then compared allele frequencies of the selected albino versus unselected samples to identify a genomic region of fixation of PA1 and PA2 alleles (such a region is expected to identify the monogenic, recessive albino locus). Briefly, samples PA1-CA and PA1-IA were each compared with PA1-Mix, and samples PA2-CA and PA2-IA were each compared with PA2-Mix (note that the PA1 and PA2 inbred lines were derived independently from the starting noninbred Alb-NL population and thus can differ in genotypes across the genome). To avoid using incorrect variant calls, only SNPs with a minimum Phred-scaled quality score of 100 across all samples were included in the analysis. Moreover, only SNP positions supported by at least 20 aligned Illumina reads in each sample in respective comparisons were included. Further, we considered only sites that were fixed in the PA1 and PA2 parents and different from the respective MAR-AB male parents (sites with support for multiple bases in the PA1 and PA2 parents might reflect residual heterozygosity or, alternatively, copy variation). The noninbred MAR-AB strain was used as the source of single males for the BSA crosses, and the individual genotypes of the single haploid MAR-AB males could not be determined directly (single males are tiny and don’t provide enough material for library construction); therefore, the respective male genotypes were inferred by comparing the genotypic data for PA1 and PA2 with that of the respective segregating populations. For example, if a site was homozygous for T in the PA1 parent, but segregated as both T and A in PA1-CA/PA1-IA or PA1-Mix with at least 3% minor allele frequency, the genotype of the haploid MAR-AB male parent used in the PA1 crosses was inferred to be an A (this analysis was preconditioned on the A being present in the MAR-AB population as predicted by GATK; fixed differences between MAR-AB and PA1 or PA2 as assessed by GATK were included in the respective analyses without applying the minor allele frequency filter). In the sliding window analyses, the allele frequency differences between selected and unselected samples were averaged over all included positions within sliding windows of 150 kb (sequential window offsets of 15 kb were used).
Fine Mapping of BSA Region.
Fine mapping of the common albino locus identified in BSA scans with PA1 and PA2 was performed by crossing 35 virgin females of the albino line PA2 to 80 males of the London inbred line on a detached bean leaf. After 3 d, the London males were removed, and the fertilized females were allowed to deposit eggs for three more days on the original bean leaf, and then were allowed to lay more eggs on a second bean leaf. PA2 albino males were then allowed to fertilize the phenotypically WT heterozygous F1 females, and, subsequently, 280 of the F1 females were transferred to new bean leaves. The F2 generation resulted in a mixture of albino and WT females, of which 478 F2 albino females were selected. From each of these F2 females, DNA was immediately extracted by homogenizing the single mite directly in 20 µL of PCR buffer (10 mM Tris⋅HCl, 100 mM NaCl, 1 mM EDTA, pH 8) to which 2 µL of proteinase K (10 mg/mL) was added; the homogenate was then incubated at 37 °C for 30 min, followed by 95 °C for 5 min to inactivate the proteinase K. For each of the F2 individuals, short fragments were amplified at distances of ∼300, 50, 40, 30, 20, and 10 kb 5′ and 3′ of the ends of tetur01g11270, which is approximately at the center of the observed BSA peak for albinism (PCR primer sequences and locations are provided in Table S2). The fragments were selected, based on Illumina sequencing data, to harbor variants that distinguish PA2 from London. Genotyping of resulting amplicons was performed by direct Sanger sequencing across variant sites or, in the case of the 3′ 300-kb marker, by digestion with the restriction enzyme DpnII (New England Biolabs, Inc.) followed by gel electrophoresis (the segregating variant in this amplicon changed a DpnII site). PCR amplifications were all performed with Jumpstart Taq DNA polymerase (Sigma-Aldrich), and Sanger sequencing was performed by LGC Genomics (Germany). Amplification and genotyping at the internal markers were performed only when a recombinant was detected between the 5′ and 3′ flanking 300-kb markers. As controls, DNA samples from the PA2 and London inbred parents were included as homozygous references.
Characterization of tetur01g11270 by Sanger Sequencing.
To confirm putative mutations identified by Illumina sequencing in tetur01g11270 in T. urticae albino strains, PCR and Sanger sequencing were performed as described for fine mapping (primers are listed in Table S2). Additionally, amplification was also performed from cDNA for the Alb-JP strain to assess the impact of the putative causal variant on splicing; RNA was extracted from 100 adult females with the RNeasy mini kit (Qiagen) and reverse transcribed using the Maxima First Strand cDNA synthesis kit for RT-qPCR (reverse transcription quantitative polymerase chain reaction) (Thermo Fisher Scientific).
The sequence of the tetur01g11270 ortholog in P. citri was recovered from the transcriptome assembly of P. citri (74) with tBLASTn using tetur01g11270 as the query. DNA and cDNA from both the P. citri albino and parental WT strains were produced as described above, and an amplicon of the first part of the orthologous gene was generated by PCR, purified, and Sanger-sequenced as previously described. The second part of the gene was amplified by long distance PCR (Expand Long Range dNTPack; Roche Diagnostics) with the following program: 92 °C for 2 min; 10 cycles of 92 °C for 10 s, 55 °C for 15 s, and 58 °C for 5 min; 27 cycles of 92 °C for 10 s, 55 °C for 15 s, 58 °C for 5 min plus 10 additional s per cycle; and, finally, 58 °C for 5 min. Primers for long-PCR were designed with Primer3Plus 4.0.0 (Table S2). When this PCR generated multiple amplicons, a gel extraction was carried out (E.Z.N.A. Gel Extraction Kit; Omega Bio-tek) followed by cloning (CloneJet PCR Cloning Kit; Life Technologies, Thermo Fisher Scientific) and plasmid extraction (E.Z.N.A. Plasmid mini Kit; Omega Bio-tek), and the inserts were Sanger-sequenced (LGC Genomics).
Linkage Characterization of P. citri Albinism.
To test whether albinism in P. citri cosegregated with variation in the tetur01g11270 ortholog, four albino P. citri teliochrysalis females were crossed to 10 WT males, and both WT and albino F2 males were then obtained from the resulting segregating population as previously described for T. urticae. DNA of the males was extracted on a single mite level. Subsequently, the long distance PCR protocol already described to amplify the second half of the tetur01g11270 ortholog—and that distinguishes the albino P. citri sequence variant in the tetur01g11270 ortholog from WT—was used for genotyping. In the case that a PCR failed (DNA from single mites is low quality and quantity), a different primer (P. citri_shortseq) (Table S2) in the insertion that gave a shorter product was used to test for the presence of the inserted sequence in the second exon. PCR products were purified and Sanger-sequenced as previously described.
CRISPR-Cas9 Targeting of tetur01g11270.
The CRISPR-Cas9 system was attempted to introduce nonhomologous end-joining mutations in the T. urticae tetur01g11270 gene. We used two injection mixtures: The first was a combination of one sgRNA in combination with the Cas9 protein, and the second mixture contained two different sgRNAs and the Cas9 protein (sgRNA sequences are provided in Table S3). See SI Materials and Methods for more detailed information.
De Novo Assembly of T. urticae Genomes.
Using the Illumina sequence data generated for strain Wasatch, as well as the albino lines recovered in the CRISPR-Cas9 screen, we performed de novo genome assembly using CLC Genomics Workbench 8.5 (https://www.qiagenbioinformatics.com/). See SI Materials and Methods for more detailed information.
Alignment of Phytoene Desaturase Sequences.
T. urticae phytoene desaturase tetur01g11270 was aligned with phytoene desaturase protein sequences of other tetranychid species, aphids, gall midges, fungi, and bacteria using MUSCLE (75). See SI Materials and Methods for more detailed information.
Assessment of Diapause Incidence in Albino Mutants.
To evaluate diapause incidence of albino lines derived on the Wasatch isogenic background, for W-Alb-2, W-Alb-14, and Wasatch (WT control), 100 adult females were allowed to lay eggs for 24 h on the upper part of a detached kidney bean leaf (Phaseolus vulgaris L.) on wet cotton in a Petri dish with fivefold replication (500 females in total). When larvae hatched, Petri dishes were placed in diapause-inducing conditions (17 °C, L:D 8 h:16 h) until adult females were observed. Eighty 1- to 2-d-old females were separately placed on 9-cm2 square kidney bean leaf discs placed on wet cotton (the age of 1 to 2 d was selected because it was before initiation of egg laying or color changes under these conditions). Thereafter, the individual fecundity and the body color of the females were monitored daily until 11 to 12 d, an age at which body color has been reported to reliably indicate reproductive arrest in WT mites (76).
Acknowledgments
We thank Jan Van Arkel for technical assistance in photography, Ioannis Livadaras for injecting spider mites, and René Feyereisen for helpful comments on the manuscript. This work was supported by Research Foundation Flanders (FWO) Grant G009312N (to L.T. and T.V.L.) and Grant G053815N (to L.T., T.V.L., and W.D.); incentive funds from the University of Utah (to R.M.C.); National Science Foundation Grant DEB-1457346 (to R.M.C.); Ontario Research Fund–Research 753 Excellence Round 8 Grant RE08-067 (to M.G.); and the Government of Canada through Genome Canada and the Ontario Genomics Institute (Grant OGI-046) (to M.G.). R.G. was funded by National Institutes of Health Genetics Training Grant T32GM007464, and W.D. is a postdoctoral fellow of the Research Foundation Flanders (FWO). M.O. was supported by Japan Society for the Promotion of Science Kakenhi Grant 26292029. The statements herein represent the views of the authors and not necessarily those of the funding agencies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The following sequences reported in this paper have been deposited in the GenBank database (accession nos. MF167354–MF167357, MF190365, MF190366, and MF190367). The following sequences reported in this paper have been deposited in the Sequence Read Archive (accession nos. SAMN07138853–SAMN07138871).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706865114/-/DCSupplemental.
References
- 1.Britton G. Structure and properties of carotenoids in relation to function. FASEB J. 1995;9:1551–1558. [PubMed] [Google Scholar]
- 2.Heath JJ, Cipollini DF, Stireman JO. The role of carotenoids and their derivatives in mediating interactions between insects and their environment. Arthropod-Plant Interact. 2013;7:1–20. [Google Scholar]
- 3.Bendich A, Olson JA. Biological actions of carotenoids. FASEB J. 1989;3:1927–1932. [PubMed] [Google Scholar]
- 4.Sies H, Stahl W. Vitamins E and C, beta-carotene, and other carotenoids as antioxidants. Am J Clin Nutr. 1995;62(6) Suppl:1315S–1321S. doi: 10.1093/ajcn/62.6.1315S. [DOI] [PubMed] [Google Scholar]
- 5.Stahl W, Ale-Agha N, Polidori MC. Non-antioxidant properties of carotenoids. Biol Chem. 2002;383:553–558. doi: 10.1515/BC.2002.056. [DOI] [PubMed] [Google Scholar]
- 6.Stahl W, Sies H. Antioxidant activity of carotenoids. Mol Aspects Med. 2003;24:345–351. doi: 10.1016/s0098-2997(03)00030-x. [DOI] [PubMed] [Google Scholar]
- 7.Avalos J, Carmen Limón M. Biological roles of fungal carotenoids. Curr Genet. 2015;61:309–324. doi: 10.1007/s00294-014-0454-x. [DOI] [PubMed] [Google Scholar]
- 8.Toews DPL, Hofmeister NR, Taylor SA. The evolution and genetics of carotenoid processing in animals. Trends Genet. 2017;33:171–182. doi: 10.1016/j.tig.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Lopes RJ, et al. Genetic basis for red coloration in birds. Curr Biol. 2016;26:1427–1434. doi: 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mundy NI, et al. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr Biol. 2016;26:1435–1440. doi: 10.1016/j.cub.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 11.Moran NA, Jarvik T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science. 2010;328:624–627. doi: 10.1126/science.1187113. [DOI] [PubMed] [Google Scholar]
- 12. Britton G, Liaaen-Jensen S, Pfander H, eds. (1995) Natural Functions, Carotenoids (Birkhäuser, Basel), Vol 4.
- 13.von Lintig J. Metabolism of carotenoids and retinoids related to vision. J Biol Chem. 2012;287:1627–1634. doi: 10.1074/jbc.R111.303990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miki W. Biological functions and activities of animal carotenoids. Pure Appl Chem. 1991;63:141–146. [Google Scholar]
- 15.Mortensen A, Skibsted LH, Sampson J, Rice-Evans C, Everett SA. Comparative mechanisms and rates of free radical scavenging by carotenoid antioxidants. FEBS Lett. 1997;418:91–97. doi: 10.1016/s0014-5793(97)01355-0. [DOI] [PubMed] [Google Scholar]
- 16.Hoekstra HE. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity (Edinb) 2006;97:222–234. doi: 10.1038/sj.hdy.6800861. [DOI] [PubMed] [Google Scholar]
- 17.Nováková E, Moran NA. Diversification of genes for carotenoid biosynthesis in aphids following an ancient transfer from a fungus. Mol Biol Evol. 2012;29:313–323. doi: 10.1093/molbev/msr206. [DOI] [PubMed] [Google Scholar]
- 18.Cobbs C, Heath J, Stireman JO, 3rd, Abbot P. Carotenoids in unexpected places: Gall midges, lateral gene transfer, and carotenoid biosynthesis in animals. Mol Phylogenet Evol. 2013;68:221–228. doi: 10.1016/j.ympev.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Grbić M, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altincicek B, Kovacs JL, Gerardo NM. Horizontally transferred fungal carotenoid genes in the two-spotted spider mite Tetranychus urticae. Biol Lett. 2012;8:253–257. doi: 10.1098/rsbl.2011.0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz C, Shahriari M, Eslava AP. 2012. The genetics and molecular biology of carotenoid biosynthesis in Mucorales. Biotechnology of Fungal Genes, eds Gupta VK, Ayyachamy M (Science Publishers, St. Helier, Jersey, British Channel Islands), pp 21–52.
- 22.Álvarez V, et al. The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of β-carotene into astaxanthin and other xanthophylls. Fungal Genet Biol. 2006;43:261–272. doi: 10.1016/j.fgb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Losey JE, Harmon J, Ballantyne F, Brown C. A polymorphism maintained by opposite patterns of parasitism and predation. Nature. 1997;388:269–272. [Google Scholar]
- 24.Harmon JP, Losey JE, Ives AR. The role of vision and color in the close proximity foraging behavior of four coccinellid species. Oecologia. 1998;115:287–292. doi: 10.1007/s004420050518. [DOI] [PubMed] [Google Scholar]
- 25.Wybouw N, Pauchet Y, Heckel DG, Van Leeuwen T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol Evol. 2016;8:1785–1801. doi: 10.1093/gbe/evw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veerman A. Carotenoid metabolism in Tetranychus urticae koch (Acari: Tetranychidae) Comp Biochem Physiol B. 1974;47:101–116. doi: 10.1016/0305-0491(74)90095-9. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf RL, Newell IM. Investigation of the biochromes of mites. Ann Entomol Soc Am. 1962;55:350–353. [Google Scholar]
- 28.Goto SG. Physiological and molecular mechanisms underlying photoperiodism in the spider mite: Comparisons with insects. J Comp Physiol B. 2016;186:969–984. doi: 10.1007/s00360-016-1018-9. [DOI] [PubMed] [Google Scholar]
- 29.Kawaguchi S, Manabe Y, Sugawara T, Osakabe M. Imaginal feeding for progression of diapause phenotype in the two-spotted spider mite (Acari: Tetranychidae) Environ Entomol. 2016;45:1568–1573. doi: 10.1093/ee/nvw127. [DOI] [PubMed] [Google Scholar]
- 30.Zon AQ, Helle W. Albinism as a marker in Tetranychus pacificus. Entomol Exp Appl. 1966;9:205–208. [Google Scholar]
- 31.Zon AQ, Helle W. Pigment mutations in Tetranychus pacificus. Entomol Exp Appl. 1966;9:402–403. [Google Scholar]
- 32.Ballantyne GH. Genetic fine structure and complementation at the albino locus in spider mites (Tetranychus species: Acarina) Genetica. 1969;40:289–323. [Google Scholar]
- 33.Veerman A. Carotenoids of wild-type and mutant strains of Tetranychus pacificus McGregor (Acari: Tetranychidae) Comp Biochem Physiol Part B Comp Biochem. 1972;42:329–340. [Google Scholar]
- 34.Veerman A. The pigments of Tetranychus cinnabarinus boisd (Acari: Tetranychidae) Comp Biochem Physiol. 1970;36:749–763. [Google Scholar]
- 35.Veerman A. Functional involvement of carotenoids in photoperiodic induction of diapause in the spider mite, Tetranychus urticae. Physiol Entomol. 1980;5:291–300. [Google Scholar]
- 36.Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem Mol Biol. 2010;40:563–572. doi: 10.1016/j.ibmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Van Leeuwen T, Dermauw W. The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu Rev Entomol. 2016;61:475–498. doi: 10.1146/annurev-ento-010715-023907. [DOI] [PubMed] [Google Scholar]
- 38.Van Leeuwen T, et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc Natl Acad Sci USA. 2012;109:4407–4412. doi: 10.1073/pnas.1200068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demaeght P, et al. High resolution genetic mapping uncovers chitin synthase-1 as the target-site of the structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae. Insect Biochem Mol Biol. 2014;51:52–61. doi: 10.1016/j.ibmb.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horvath MP, et al. Structure of the lutein-binding domain of human StARD3 at 1.74 Å resolution and model of a complex with lutein. Acta Crystallogr F Struct Biol Commun. 2016;72:609–618. doi: 10.1107/S2053230X16010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golic KG. RNA-guided nucleases: A new era for engineering the genomes of model and nonmodel organisms. Genetics. 2013;195:303–308. doi: 10.1534/genetics.113.155093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ignatowicz S, Helle W. Genetics of diapause supression in the two-spotted spider mite, Tetranychus urticae Koch. Exp Appl Acarol. 1986;2:161–172. [Google Scholar]
- 43.Goka K, Takafuji A. Genetical studies on the diapause of the two-spotted spider mite, Tetranychus urticae Koch (1) Appl Entomol Zool (Jpn) 1990;25:119–125. [Google Scholar]
- 44.Goka K, Takafuji A. Genetical studies on the diapause of the two-spotted spider mite, Tetranychus urticae Koch (2) Appl Entomol Zool (Jpn) 1991;26:77–84. [Google Scholar]
- 45.Koveos DS, Kroon A, Veerman A. Geographic variation of diapause intensity in the spider mite Tetranychus urticae. Physiol Entomol. 1993;18:50–56. [Google Scholar]
- 46.Takafuji A, Goka K. Mode of diapause inheritance in the Kanzawa spider mite, Tetranychus kanzawai (Acari: Tetranychidae) Appl Entomol Zool (Jpn) 1999;34:299–302. [Google Scholar]
- 47.Kawakami Y, Numata H, Ito K, Goto SG. Dominant and recessive inheritance patterns of diapause in the two-spotted spider mite Tetranychus urticae. J Hered. 2010;101:20–25. doi: 10.1093/jhered/esp085. [DOI] [PubMed] [Google Scholar]
- 48.Takafuji A, So P-M, Tsuno N. Inter- and intra-population variations in diapause attribute of the two-spotted spider mite, Tetranychus urticae Koch, in Japan. Res Popul Ecol (Kyoto) 1991;33:331–344. [Google Scholar]
- 49.Bensoussan N, et al. Plant-herbivore interaction: Dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Front Plant Sci. 2016;7:1105. doi: 10.3389/fpls.2016.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryon A, Wybouw N, Dermauw W, Tirry L, Van Leeuwen T. Genome wide gene-expression analysis of facultative reproductive diapause in the two-spotted spider mite Tetranychus urticae. BMC Genomics. 2013;14:815–834. doi: 10.1186/1471-2164-14-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernández-Martín R, Cerdá-Olmedo E, Avalos J. Homologous recombination and allele replacement in transformants of Fusarium fujikuroi. Mol Gen Genet. 2000;263:838–845. doi: 10.1007/s004380000249. [DOI] [PubMed] [Google Scholar]
- 52.Sanz C, et al. Interallelic complementation provides genetic evidence for the multimeric organization of the Phycomyces blakesleeanus phytoene dehydrogenase. Eur J Biochem. 2002;269:902–908. doi: 10.1046/j.0014-2956.2001.02724.x. [DOI] [PubMed] [Google Scholar]
- 53.Prado-Cabrero A, et al. Deviation of the neurosporaxanthin pathway towards β-carotene biosynthesis in Fusarium fujikuroi by a point mutation in the phytoene desaturase gene. FEBS J. 2009;276:4582–4597. doi: 10.1111/j.1742-4658.2009.07164.x. [DOI] [PubMed] [Google Scholar]
- 54.Alcalde E, Fraser PD. Metabolite profiling of Phycomyces blakesleeanus carotene mutants reveals global changes across intermediary metabolism. Microbiology. 2016;162:1963–1971. doi: 10.1099/mic.0.000376. [DOI] [PubMed] [Google Scholar]
- 55.Arrach N, Fernández-Martín R, Cerdá-Olmedo E, Avalos J. A single gene for lycopene cyclase, phytoene synthase, and regulation of carotene biosynthesis in Phycomyces. Proc Natl Acad Sci USA. 2001;98:1687–1692. doi: 10.1073/pnas.021555298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linnemannstöns P, Prado MM, Fernández-Martín R, Tudzynski B, Avalos J. A carotenoid biosynthesis gene cluster in Fusarium fujikuroi: The genes carB and carRA. Mol Genet Genomics. 2002;267:593–602. doi: 10.1007/s00438-002-0690-5. [DOI] [PubMed] [Google Scholar]
- 57.Díaz-Sánchez V, et al. Analysis of al-2 mutations in Neurospora. PLoS One. 2011;6:e21948. doi: 10.1371/journal.pone.0021948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veerman A. Diapause. In: Sabelis M, Helle W, editors. Spider Mites: Their Biology, Natural Enemies and Control. Elsevier; Amsterdam: 1985. pp. 279–317. [Google Scholar]
- 59.Bosse TC, Veerman A. Involvement of vitamin A in the photoperiodic induction of diapause in the spider mite Tetranychus urticae is demonstrated by rearing an albino mutant on a semi-synthetic diet with and without β-carotene or vitamin A. Physiol Entomol. 1996;21:188–192. [Google Scholar]
- 60.Suzuki T, Watanabe M, Takeda M. UV tolerance in the two-spotted spider mite, Tetranychus urticae. J Insect Physiol. 2009;55:649–654. doi: 10.1016/j.jinsphys.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Veerman A. Aspects of the induction of diapause in a laboratory strain of the mite Tetranychus urticae. J Insect Physiol. 1977;23:703–711. [Google Scholar]
- 62.Helle W. 1962 Genetics of Resistance to Organophosphorus Compounds and Its Relation to Diapause in Tetranychus urticae Koch (Acari) (H. Veenman, Wageningen, The Netherlands). Available at https://catalog.hathitrust.org/Record/009171119. Accessed April 11, 2017.
- 63.Ito K. Effect of host plants on diapause induction in immature and adult Tetranychus kanzawai (Acari: Tetranychidae) Exp Appl Acarol. 2010;52:11–17. doi: 10.1007/s10493-010-9342-3. [DOI] [PubMed] [Google Scholar]
- 64.Veerman A. Physiological aspects of diapause in plant-inhabiting mites. In: Schuster R, Murphy PW, editors. The Acari: Reproduction, Development and Life-History Strategies. Springer; Dordrecht, The Netherlands: 1991. pp. 245–265. [Google Scholar]
- 65.Helle W, Van Zon AQ. Rates of spontaneous mutation in certain genes of an arrhenotokous, Tetranychus pacificus. Entomol Exp Appl. 1967;10:189–193. [Google Scholar]
- 66.Díaz-Riquelme J, et al. Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae. BMC Genomics. 2016;17:74. doi: 10.1186/s12864-016-2401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Leeuwen T, Stillatus V, Tirry L. Genetic analysis and cross-resistance spectrum of a laboratory-selected chlorfenapyr resistant strain of two-spotted spider mite (Acari: Tetranychidae) Exp Appl Acarol. 2004;32:249–261. doi: 10.1023/b:appa.0000023240.01937.6d. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sterck L, Billiau K, Abeel T, Rouzé P, Van de Peer Y. ORCAE: Online resource for community annotation of eukaryotes. Nat Methods. 2012;9:1041. doi: 10.1038/nmeth.2242. [DOI] [PubMed] [Google Scholar]
- 74.Bajda S, et al. Transcriptome profiling of a spirodiclofen susceptible and resistant strain of the European red mite Panonychus ulmi using strand-specific RNA-seq. BMC Genomics. 2015;16:974. doi: 10.1186/s12864-015-2157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ito K, Fukuda T, Hayakawa H, Arakawa R, Saito Y. Relationship between body colour, feeding, and reproductive arrest under short-day development in Tetranychus pueraricola (Acari: Tetranychidae) Exp Appl Acarol. 2013;60:471–477. doi: 10.1007/s10493-013-9660-3. [DOI] [PubMed] [Google Scholar]
- 77.Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: A software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Villarroel CA, et al. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016;86:119–131. doi: 10.1111/tpj.13152. [DOI] [PubMed] [Google Scholar]
- 79.Zhao C, et al. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr Biol. 2015;25:613–620. doi: 10.1016/j.cub.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 80.Mathers TC, et al. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 2017;18:27. doi: 10.1186/s13059-016-1145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schaub P, et al. On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase. PLoS One. 2012;7:e39550. doi: 10.1371/journal.pone.0039550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chi SC, et al. Assembly of functional photosystem complexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim Biophys Acta. 2015;1847:189–201. doi: 10.1016/j.bbabio.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]